Comparative Metabolomic and Transcriptomic Analyses Reveal Distinct Ascorbic Acid (AsA) Accumulation Patterns between PCA and PCNA Persimmon Developing Fruit
Abstract
1. Introduction
2. Results
2.1. Content of AsA in Persimmon Fruit
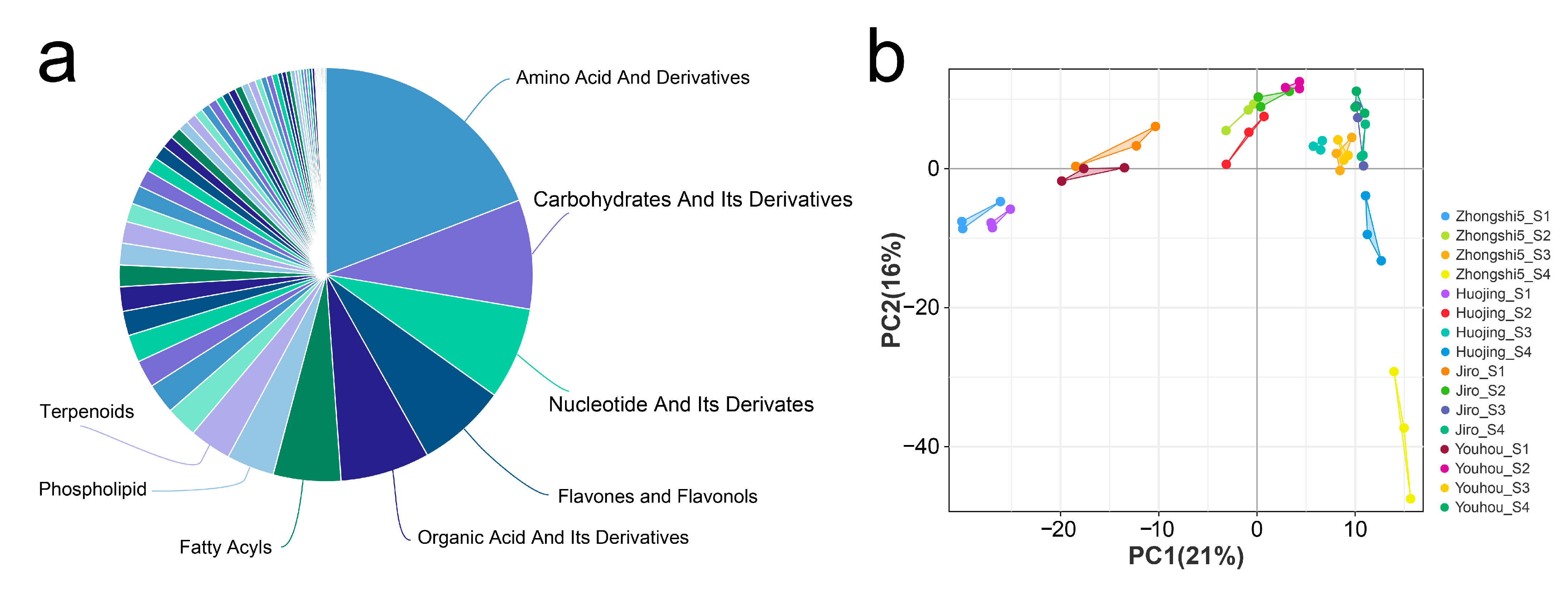
2.2. Metabolic Features of PCA and PCNA Persimmon Developing Fruit
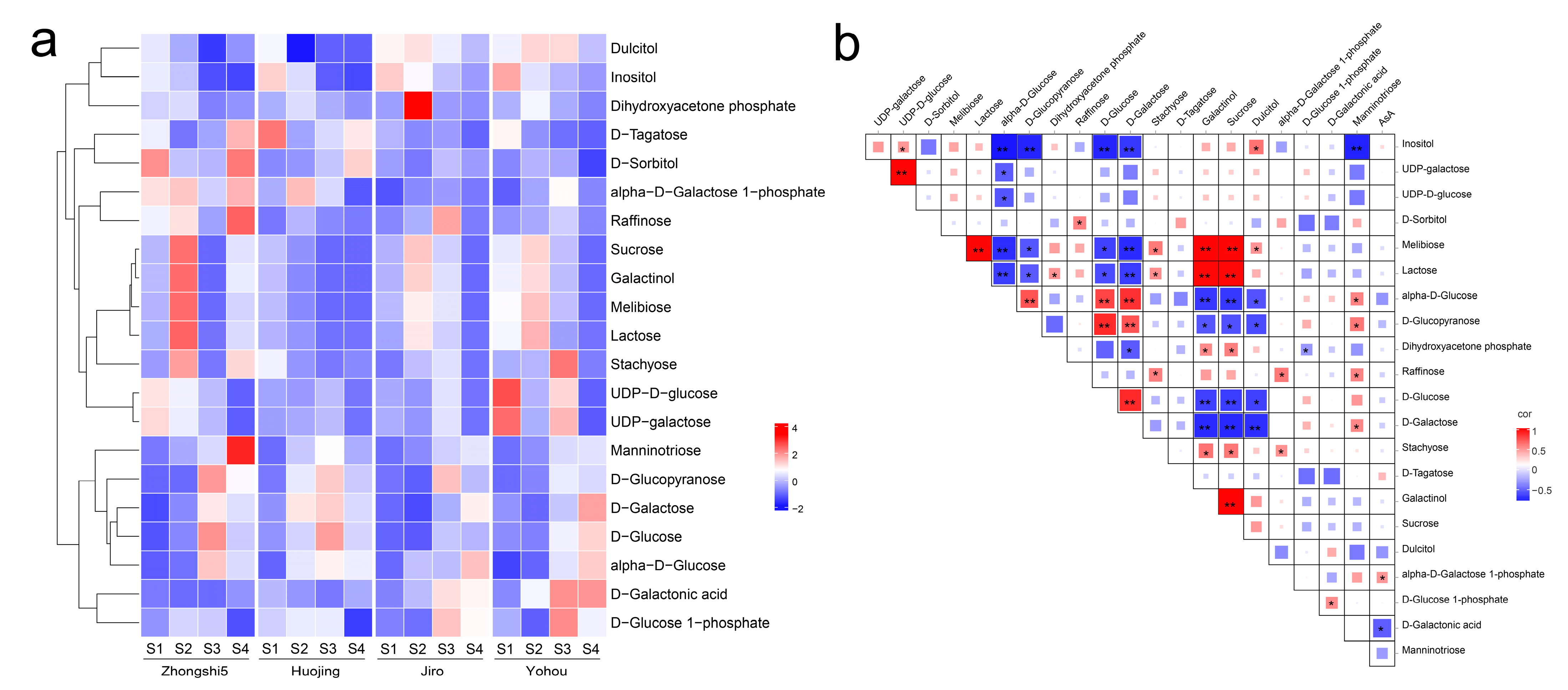
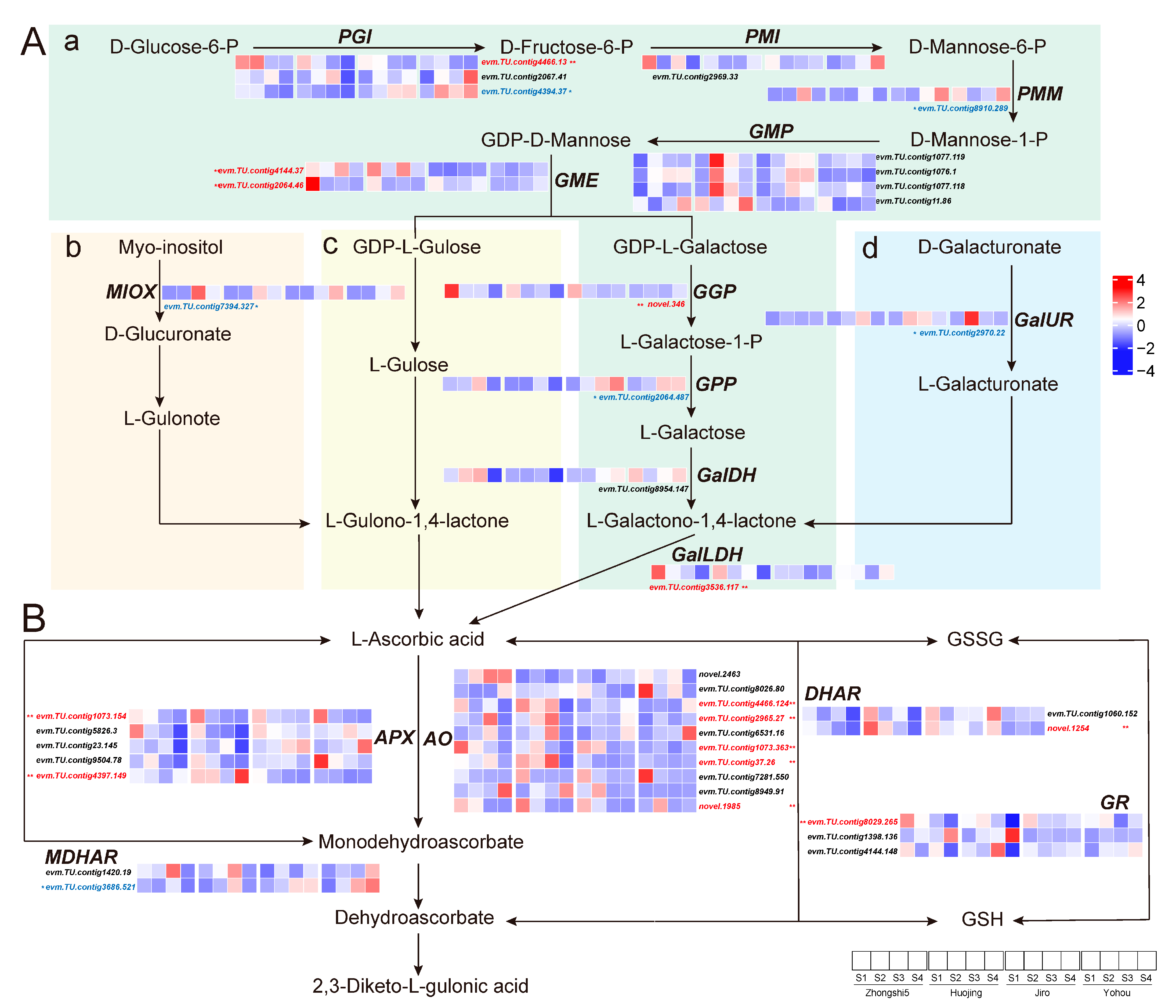
2.3. KEGG Enrichment Analysis of DAMs and Galactose Metabolism Pathway Analysis
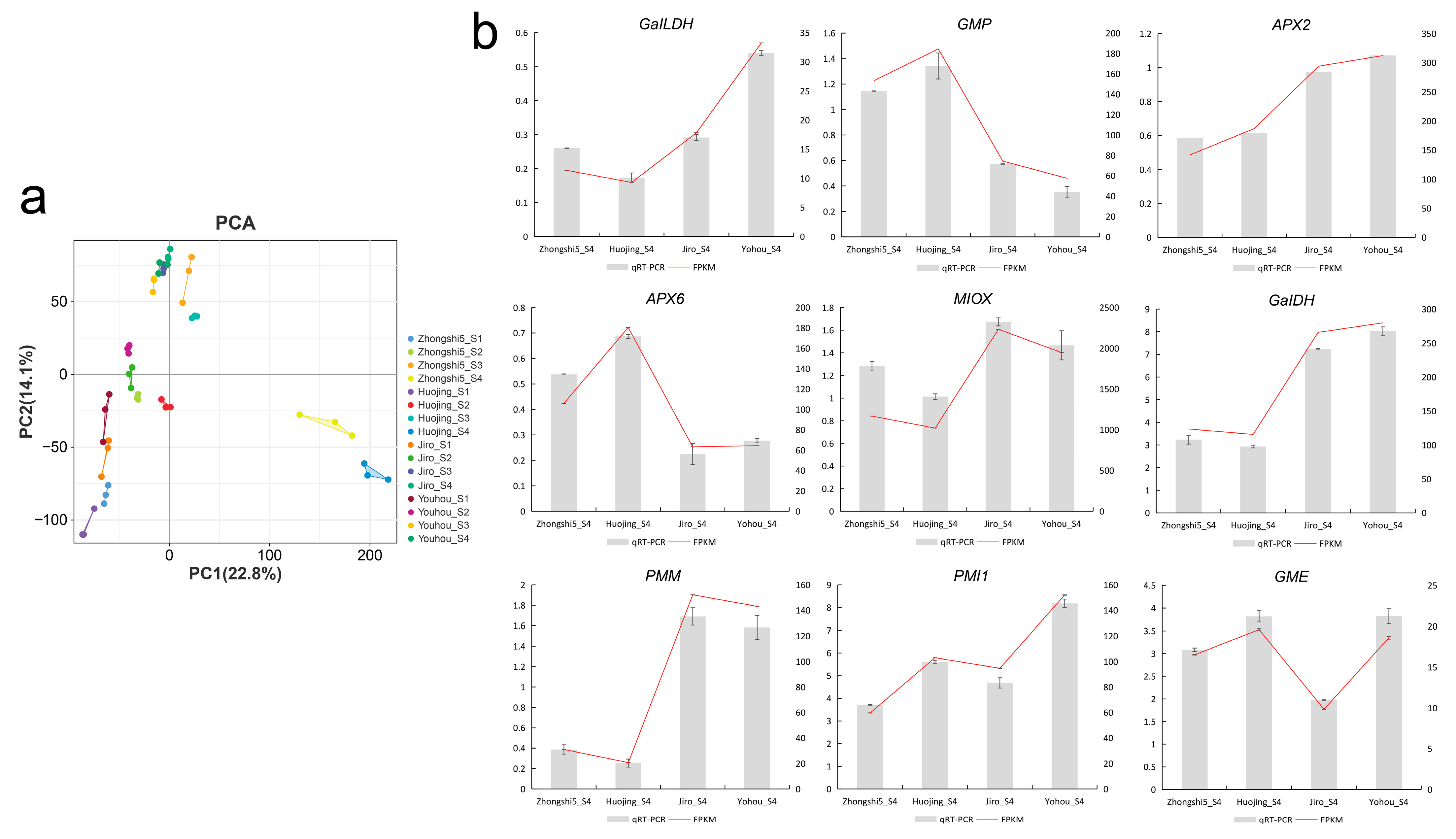
2.4. Transcriptomic Analysis of PCA and PCNA Persimmon Developing Fruit
2.5. Analysis of the AsA Biosynthesis and Recycling Pathway Genes
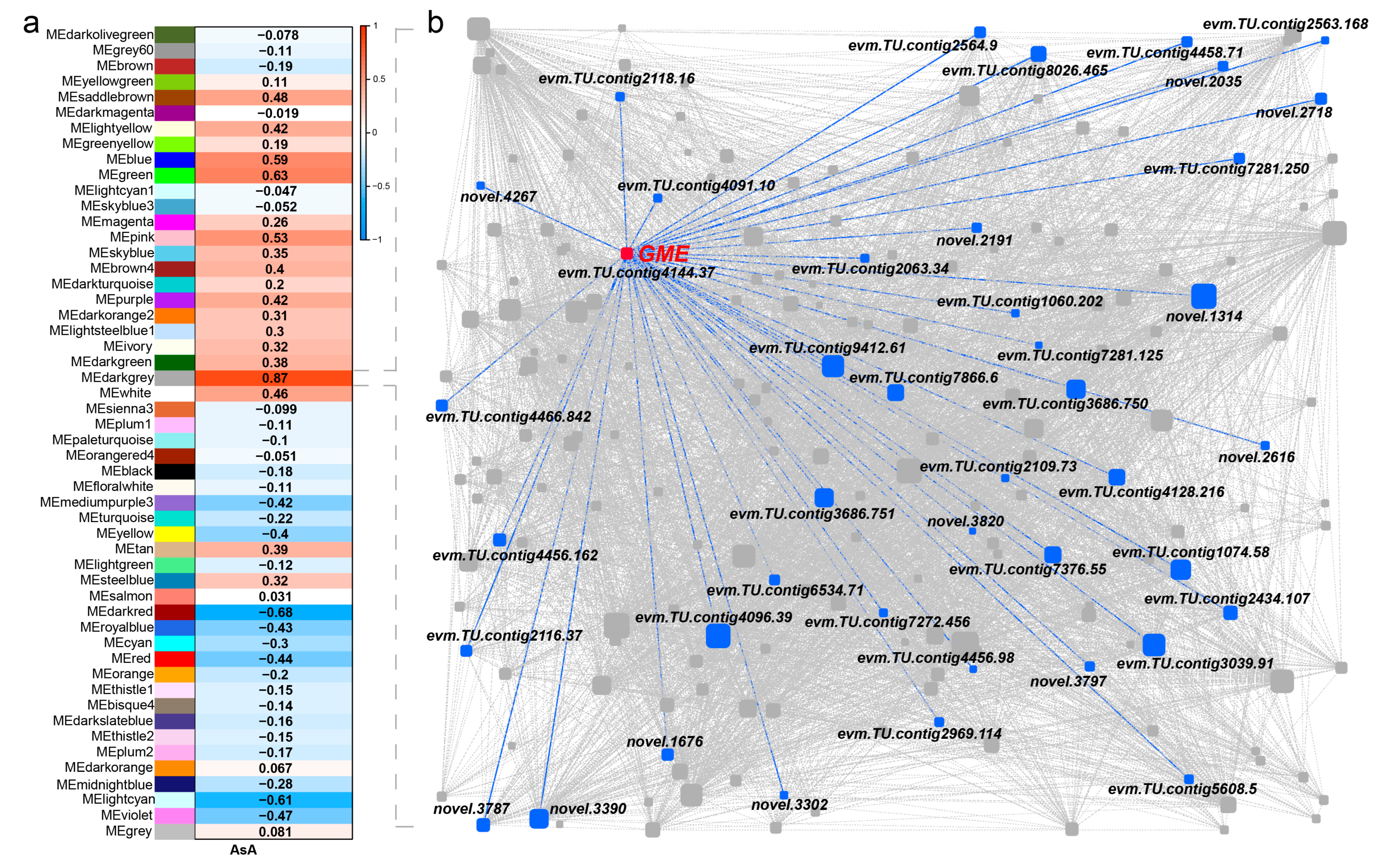
2.6. Weighted Gene Co-Expression Network (WGCNA)
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Extraction and Determination of AsA
4.3. Transcriptome Data Analysis
4.4. Metabolome Data Analysis
4.5. Gene Expression Analysis by qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, M.; Ma, F.; Zhang, M.; Pu, F. Distribution and metabolism of ascorbic acid in apple fruits (Malus domestica Borkh cv. Gala). Plant Sci. 2008, 174, 606–612. [Google Scholar] [CrossRef]
- Smirnoff, N. Ascorbic acid metabolism and functions: A comparison of plants and mammals. Free Radic. Biol. Med. 2018, 122, 116–129. [Google Scholar] [CrossRef]
- Shenoy, N.; Creagan, E.; Witzig, T.; Levine, M. Ascorbic Acid in Cancer Treatment: Let the Phoenix Fly. Cancer Cell 2018, 34, 700–706. [Google Scholar] [CrossRef]
- Santos, K.; Bragança, V.; Pacheco, L.; Ota, S.; Aguiar, C.; Borges, R. Essential features for antioxidant capacity of ascorbic acid (vitamin C). J. Mol. Model. 2021, 28, 1. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R. L-Ascorbic Acid: A Multifunctional Molecule Supporting Plant Growth and Development. Scientifica 2013, 2013, 795964. [Google Scholar] [CrossRef]
- Pastori, G.; Kiddle, G.; Antoniw, J.; Bernard, S.; Veljovic-Jovanovic, S.; Verrier, P.; Noctor, G.; Foyer, C. Leaf Vitamin C Contents Modulate Plant Defense Transcripts and Regulate Genes That Control Development through Hormone Signaling. Plant Cell 2003, 15, 939–951. [Google Scholar] [CrossRef]
- Maruta, T.; Ichikawa, Y.; Mieda, T.; Takeda, T.; Tamoi, M.; Yabuta, Y.; Ishikawa, T.; Shigeoka, S. The Contribution of Arabidopsis Homologs of l-Gulono-1,4-lactone Oxidase to the Biosynthesis of Ascorbic Acid. Biosci. Biotechnol. Biochem. 2010, 74, 1494–1497. [Google Scholar] [CrossRef]
- Visser, D.; Jansen, N.; Ijland, M.; de Koning, T.; van Hasselt, P. Intracranial bleeding due to vitamin K deficiency: Advantages of using a pediatric intensive care registry. Intens. Care Med. 2011, 37, 1014–1020. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chaturvedi, S.; Khan, S.; Bhunia, R.; Kaur, K.; Tiwari, S. Metabolic engineering in food crops to enhance ascorbic acid production: Crop biofortification perspectives for human health. Physiol. Mol. Biol. Pla. 2022, 28, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.; Montagu, M.; Inzé, D.; Sanmartin, M.; Kanellis, A.; Smirnoff, N.; Benzie, I.; Strain, J.; Favell, D.; Fletcher, J. Plant L-ascorbic acid: Chemistry, function, metabolism, bioavailability and effects of processing. J. Sci. Food Agric. 2000, 80, 825–860. [Google Scholar] [CrossRef]
- Li, H.; Huang, W.; Wang, G.; Wang, W.; Cui, X.; Zhuang, J. Transcriptomic analysis of the biosynthesis, recycling, and distribution of ascorbic acid during leaf development in tea plant (Camellia sinensis (L.) O. Kuntze). Sci. Rep. 2017, 7, 46212. [Google Scholar] [CrossRef] [PubMed]
- Mellidou, I.; Koukounaras, A.; Chatzopoulou, F.; Kostas, S.; Kanellis, A.K. Plant Vitamin C: One Single Molecule with a Plethora of Roles. In Fruit and Vegetable Phytochemicals; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 463–498. [Google Scholar]
- Wheeler, G.; Jones, M.; Smirnoff, N. The biosynthetic pathway of vitamin C in higher plants. Nature 1998, 393, 365–369. [Google Scholar] [CrossRef]
- Ishikawa, T.; Dowdle, J.; Smirnoff, N. Progress in manipulating ascorbic acid biosynthesis and accumulation in plants. Physiol. Plant. 2006, 126, 343–355. [Google Scholar] [CrossRef]
- Wang, Z.; Xiao, Y.; Chen, W.; Tang, K.; Zhang, L. Increased Vitamin C Content Accompanied by an Enhanced Recycling Pathway Confers Oxidative Stress Tolerance in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Agius, F.; González-Lamothe, R.; Caballero, J.L.; Muñoz-Blanco, J.; Botella, M.A.; Valpuesta, V. Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat. Biotechnol. 2003, 21, 177–181. [Google Scholar] [CrossRef]
- Jimenez, A.; Creissen, G.; Kular, B.; Firmin, J.; Robinson, S.; Verhoeyen, M.; Mullineaux, P. Changes in oxidative processes and components of the antioxidant system during tomato fruit ripening. Planta 2002, 214, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Pateraki, I.; Sanmartin, M.; Kalamaki, M.; Gerasopoulos, D.; Kanellis, A. Molecular characterization and expression studies during melon fruit development and ripening of l-galactono-1,4-lactone dehydrogenase. J. Exp. Bot. 2004, 55, 1623–1633. [Google Scholar] [CrossRef]
- Wang, Y.; Suo, Y.; Han, W.; Li, H.; Wang, Z.; Diao, S.; Sun, P.; Fu, J. Comparative transcriptomic and metabolomic analyses reveal differences in flavonoid biosynthesis between PCNA and PCA persimmon fruit. Front. Plant Sci. 2023, 14, 1130047. [Google Scholar] [CrossRef]
- Saleem, M.; Ejaz, S.; Anjum, M.; Nawaz, A.; Naz, S.; Hussain, S.; Ali, S.; Canan, İ. Postharvest application of gum arabic edible coating delays ripening and maintains quality of persimmon fruits during storage. J. Food Process Preserv. 2020, 44, e14583. [Google Scholar] [CrossRef]
- Suo, Y.; Sun, P.; Cheng, H.; Han, W.; Diao, S.; Li, H.; Mai, Y.; Zhao, X.; Li, F.; Fu, J. A high-quality chromosomal genome assembly of Diospyros oleifera Cheng. GigaScience 2020, 9, giz164. [Google Scholar] [CrossRef]
- Han, W.; Wang, Y.; Li, H.; Diao, S.; Suo, Y.; Li, T.; Sun, P.; Li, F.; Fu, J. Transcriptome and Metabolome Reveal Distinct Sugar Accumulation Pattern between PCNA and PCA Mature Persimmon Fruit. Int. J. Mol. Sci. 2023, 24, 8599. [Google Scholar] [CrossRef]
- Akagi, T.; Ikegami, A.; Suzuki, Y.; Yoshida, J.; Yamada, M.; Sato, A.; Yonemori, K. Expression balances of structural genes in shikimate and flavonoid biosynthesis cause a difference in proanthocyanidin accumulation in persimmon (Diospyros kaki Thunb.) fruit. Planta 2009, 230, 899–915. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Kui, L.; Tang, M.; Li, D.; Wei, K.; Chen, W.; Miao, J.; Dong, Y. High-Throughput Transcriptome Profiling in Drug and Biomarker Discovery. Front. Genet. 2020, 11, 19. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Funct. Genom. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Yang, C.; Fang, X.; Wu, X.; Mao, Y.; Wang, L.; Chen, X. Transcriptional Regulation of Plant Secondary Metabolism. F. J. Integr. Plant Biol. 2012, 54, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Cui, R.; Wang, D.; Huang, H.; Rui, C.; Malik, W.A.; Wang, J.; Zhang, H.; Xu, N.; Liu, X. Combined transcriptomic and metabolomic analyses elucidate key salt-responsive biomarkers to regulate salt tolerance in cotton. BMC Plant Biol. 2023, 23, 245. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, W.; Elango, D.; Liu, H.; Jin, D.; Wang, X.; Wu, Y. Metabolome and Transcriptome Analysis Reveals Molecular Mechanisms of Watermelon under Salt Stress. Environ. Exp. Bot. 2023, 206, 105200. [Google Scholar] [CrossRef]
- Smirnoff, N.; Wheeler, G. Ascorbic Acid in Plants: Biosynthesis and Function. Crit. Rev. Biochem. Mol. 2000, 35, 291–314. [Google Scholar] [CrossRef]
- Dun, W.; Wei, X.; Wang, L.; Liu, J.; Zhao, J.; Sun, P.; Fang, C.; Xie, X. Over-expression of FaGalLDH Increases Ascorbic Acid Concentrations and Enhances Salt Stress Tolerance in Arabidopsis thaliana. J. Plant Biol. 2023, 66, 35–46. [Google Scholar] [CrossRef]
- Di Matteo, A.; Sacco, A.; Anacleria, M.; Pezzotti, M.; Delledonne, M.; Ferrarini, A.; Frusciante, L.; Barone, A. The ascorbic acid content of tomato fruits is associated with the expression of genes involved in pectin degradation. BMC Plant Biol. 2010, 10, 163. [Google Scholar] [CrossRef]
- Xu, Y.; Huang, B. Exogenous Ascorbic Acid Mediated Abiotic Stress Tolerance in Plants. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Hossain, M., Munné-Bosch, S., Burritt, D., Diaz-Vivancos, P., Fujita, M., Lorence, A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 233–253. [Google Scholar]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Liu, X.; Zhang, Q.; Wu, H.; Yan, D.; Liu, Y.; Zhu, X.; Ren, X.; Yang, Y. Carotenoid accumulation and gene expression in fruit skins of three differently colored persimmon cultivars during fruit growth and ripening. Sci. Hortic. 2019, 248, 282–290. [Google Scholar] [CrossRef]
- Vincente, A.R.; Manganaris, G.A.; Ortiz, C.M.; Sozzi, G.O.; Crisosto, C.H. Chapter 5-Nutritional Quality of Fruits and Vegetables. In Postharvest Handling, 3rd ed.; Florkowski, W., Shewfelt, R., Brueckner, B., Prussia, S., Eds.; Academic Press: San Diego, CA, USA, 2014; pp. 69–122. [Google Scholar]
- Lee, S.K.; Kader, A.A. Preharvest and postharvest factors influencing vitamin C content of horticultural crops. Postharvest Biol. Tec. 2000, 20, 207–220. [Google Scholar] [CrossRef]
- Harrison, F.E. A critical review of vitamin C for the prevention of age-related cognitive decline and Alzheimer’s disease. J. Alzheimer’s Dis 2012, 29, 711–726. [Google Scholar] [CrossRef] [PubMed]
- George, A.; Mowat, A.; Collins, R.; Morley-Bunker, M. The pattern and control of reproductive development in non-astringent persimmon (Diospyros kaki L.): A review. Sci. Hortic. 1997, 70, 93–122. [Google Scholar] [CrossRef]
- Shaya, F.; David, I.; Yitzhak, Y.; Izhaki, A. Hormonal interactions during early physiological partenocarpic fruitlet abscission in persimmon (Diospyros Kaki Thunb.) ‘Triumph’ and ‘Shinshu’ cultivars. Sci. Hortic. 2019, 243, 575–582. [Google Scholar] [CrossRef]
- Linster, C.; Clarke, S. L-Ascorbate biosynthesis in higher plants: The role of VTC2. Trends Plant Sci. 2008, 13, 567–573. [Google Scholar] [CrossRef]
- Ren, J.; Chen, Z.; Duan, W.; Song, X.; Liu, T.; Wang, J.; Hou, X.; Li, Y. Comparison of ascorbic acid biosynthesis in different tissues of three non-heading Chinese cabbage cultivars. Plant Physiol. Biochem. 2013, 73, 229–236. [Google Scholar] [CrossRef]
- Imai, T.; Ban, Y.; Terakami, S.; Yamamoto, T.; Moriguchi, T. L-Ascorbate biosynthesis in peach: Cloning of six l-galactose pathway-related genes and their expression during peach fruit development. Physiol. Plant. 2009, 136, 139–149. [Google Scholar] [CrossRef]
- Li, X.; Ye, J.; Munir, S.; Yang, T.; Chen, W.; Liu, G.; Zheng, W.; Zhang, Y. Biosynthetic Gene Pyramiding Leads to Ascorbate Accumulation with Enhanced Oxidative Stress Tolerance in Tomato. Int. J. Mol. Sci. 2019, 20, 1558. [Google Scholar] [CrossRef]
- Kunert, K.J.; Foyer, C.H. Chapter Three-The ascorbate/glutathione cycle. In Advances in Botanical Research; Mittler, R.O.N., Breusegem, F.V., Eds.; Academic Press: Cambridge, MA, USA, 2023; Volume 105, pp. 77–112. [Google Scholar]
- Blauer, J.; Kumar, G.; Knowles, L.; Dhingra, A.; Knowles, N. Changes in ascorbate and associated gene expression during development and storage of potato tubers (Solanum tuberosum L.). Postharvest Biol. Tec. 2013, 78, 76–91. [Google Scholar] [CrossRef]
- Mounet-Gilbert, L.; Dumont, M.; Ferrand, C.; Bournonville, C.; Monier, A.; Jorly, J.; Lemaire-Chamley, M.; Mori, K.; Atienza, I.; Hernould, M.; et al. Two tomato GDP-D-mannose epimerase isoforms involved in ascorbate biosynthesis play specific roles in cell wall biosynthesis and development. J. Exp. Bot. 2016, 67, 4767–4777. [Google Scholar] [CrossRef]
- Ma, L.; Wang, Y.; Liu, W.; Liu, Z. Overexpression of an alfalfa GDP-mannose 3, 5-epimerase gene enhances acid, drought and salt tolerance in transgenic Arabidopsis by increasing ascorbate accumulation. Biotechnol. Lett. 2014, 36, 2331–2341. [Google Scholar] [CrossRef]
- Major, L.; Wolucka, B.; Naismith, J. Structure and Function of GDP-Mannose-3′,5′-Epimerase: An Enzyme which Performs Three Chemical Reactions at the Same Active Site. J. Am. Chem. Soc. 2005, 127, 18309–18320. [Google Scholar] [CrossRef] [PubMed]
- Beerens, K.; Gevaert, O.; Desmet, T. GDP-Mannose 3,5-Epimerase: A View on Structure, Mechanism, and Industrial Potential. Front. Mol. Biosci. 2022, 8, 784142. [Google Scholar] [CrossRef]
- Gilbert, L.; Alhagdow, M.; Nunes-Nesi, A.; Quemener, B.; Guillon, F.; Bouchet, B.; Faurobert, M.; Gouble, B.; Page, D.; Garcia, V.; et al. GDP-d-mannose 3,5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J. 2009, 60, 499–508. [Google Scholar] [CrossRef]
- Li, H.; Sun, P.; Wang, Y.; Zhang, Z.; Yang, J.; Suo, Y.; Han, W.; Diao, S.; Li, F.; Fu, J. Allele-aware chromosome-level genome assembly of the autohexaploid Diospyros kaki Thunb. Sci. Data 2023, 10, 270. [Google Scholar] [CrossRef]
- Zhang, Y.; Park, C.; Bennett, C.; Thornton, M.; Kim, D. Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 2021, 31, 1290–1295. [Google Scholar] [CrossRef]
- Shumate, A.; Wong, B.; Pertea, G.; Pertea, M. Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLOS Comput. Bio. 2022, 18, e1009730. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Li, J.; Miao, B.; Wang, S.; Dong, W.; Xu, H.; Si, C.; Wang, W.; Duan, S.; Lou, J.; Bao, Z.; et al. Hiplot: A comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization. Brief. Bioinform. 2022, 23, bbac261. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Du, Q.; Du, H.; Wang, Q.; Wang, L.; Du, L.; Liu, P. Analysis of chemotypes and their markers in leaves of core collections of Eucommia ulmoides using metabolomics. Front. Plant Sci. 2023, 13, 1029907. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Wang, L.; Li, H.; Sun, P.; Fu, J.; Suo, Y.; Han, W.; Diao, S.; Mai, Y.; Li, F. Selection and validation of reference genes for quantitative gene expression analyses in persimmon (Diospyros kaki thunb.) using real-time quantitative PCR. Biol. Fu. 2019, 70, 261–267. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Diao, S.; Li, H.; Ye, L.; Suo, Y.; Zheng, Y.; Sun, P.; Han, W.; Fu, J. Comparative Metabolomic and Transcriptomic Analyses Reveal Distinct Ascorbic Acid (AsA) Accumulation Patterns between PCA and PCNA Persimmon Developing Fruit. Int. J. Mol. Sci. 2023, 24, 15362. https://doi.org/10.3390/ijms242015362
Wang Y, Diao S, Li H, Ye L, Suo Y, Zheng Y, Sun P, Han W, Fu J. Comparative Metabolomic and Transcriptomic Analyses Reveal Distinct Ascorbic Acid (AsA) Accumulation Patterns between PCA and PCNA Persimmon Developing Fruit. International Journal of Molecular Sciences. 2023; 24(20):15362. https://doi.org/10.3390/ijms242015362
Chicago/Turabian StyleWang, Yiru, Songfeng Diao, Huawei Li, Lingshuai Ye, Yujing Suo, Yanhao Zheng, Peng Sun, Weijuan Han, and Jianmin Fu. 2023. "Comparative Metabolomic and Transcriptomic Analyses Reveal Distinct Ascorbic Acid (AsA) Accumulation Patterns between PCA and PCNA Persimmon Developing Fruit" International Journal of Molecular Sciences 24, no. 20: 15362. https://doi.org/10.3390/ijms242015362
APA StyleWang, Y., Diao, S., Li, H., Ye, L., Suo, Y., Zheng, Y., Sun, P., Han, W., & Fu, J. (2023). Comparative Metabolomic and Transcriptomic Analyses Reveal Distinct Ascorbic Acid (AsA) Accumulation Patterns between PCA and PCNA Persimmon Developing Fruit. International Journal of Molecular Sciences, 24(20), 15362. https://doi.org/10.3390/ijms242015362




