Insight into the High-Efficiency Benzo(a)pyrene Degradation Ability of Pseudomonas benzopyrenica BaP3 and Its Application in the Complete Bioremediation of Benzo(a)pyrene
Abstract
1. Introduction
2. Results
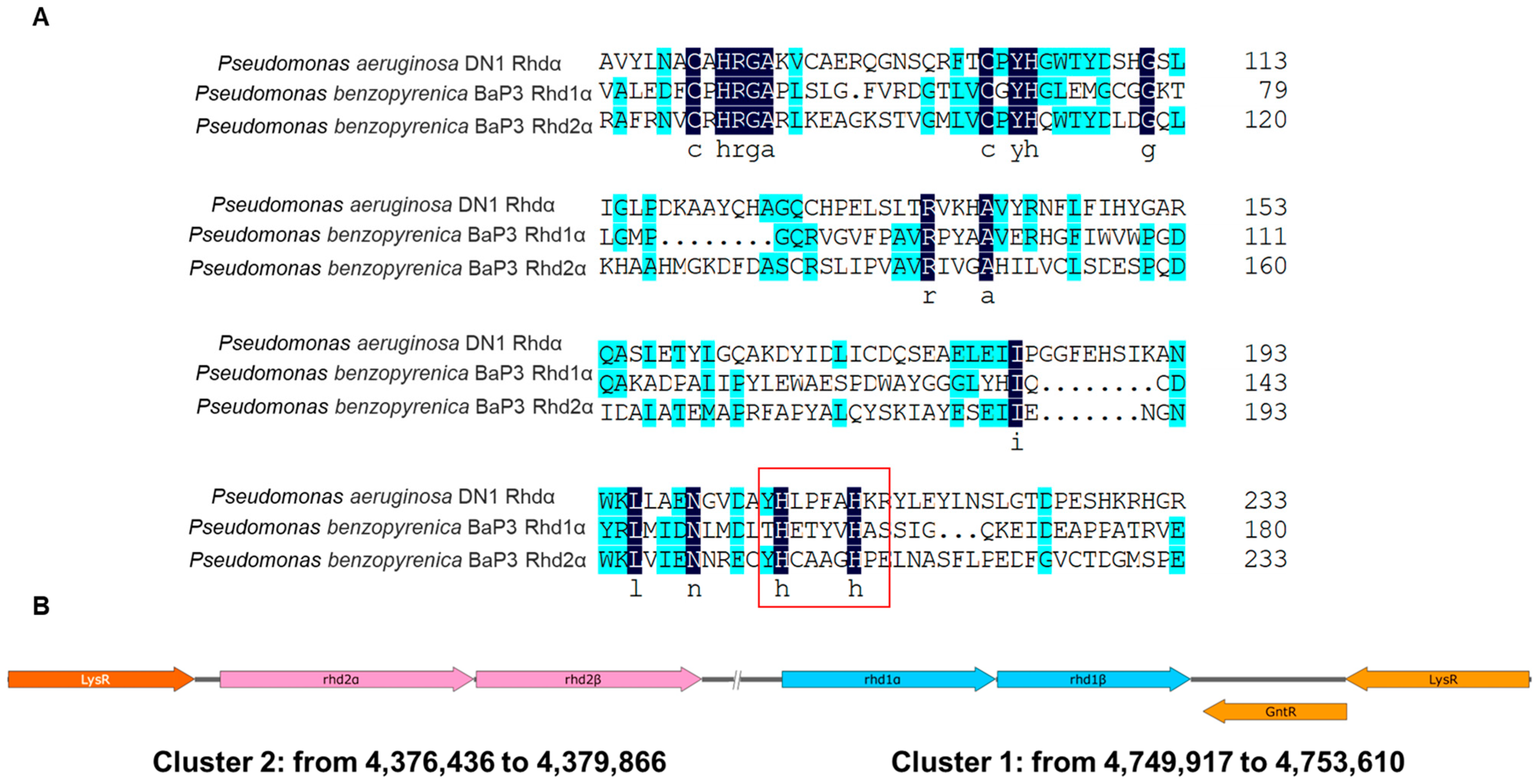
2.1. Annotation of Two RHDs above the Genome Sequence of Strain BaP3
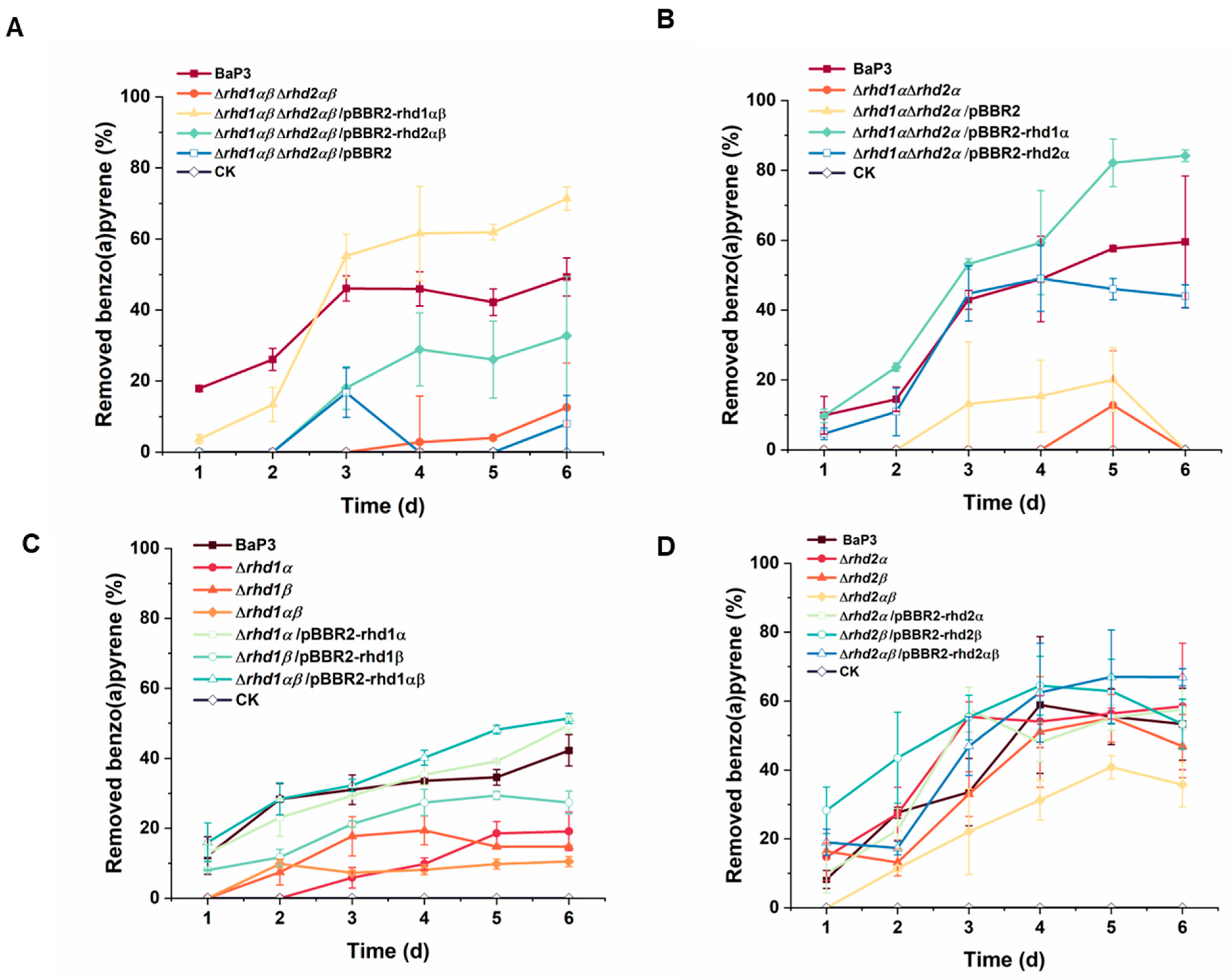
2.2. RHDs Are the Main Genes Involved in the Degradation of Benzo(a)pyrene in Strain BaP3
2.3. Rhd1α Is Suitable for the Degradation of Benzo(a)pyrene
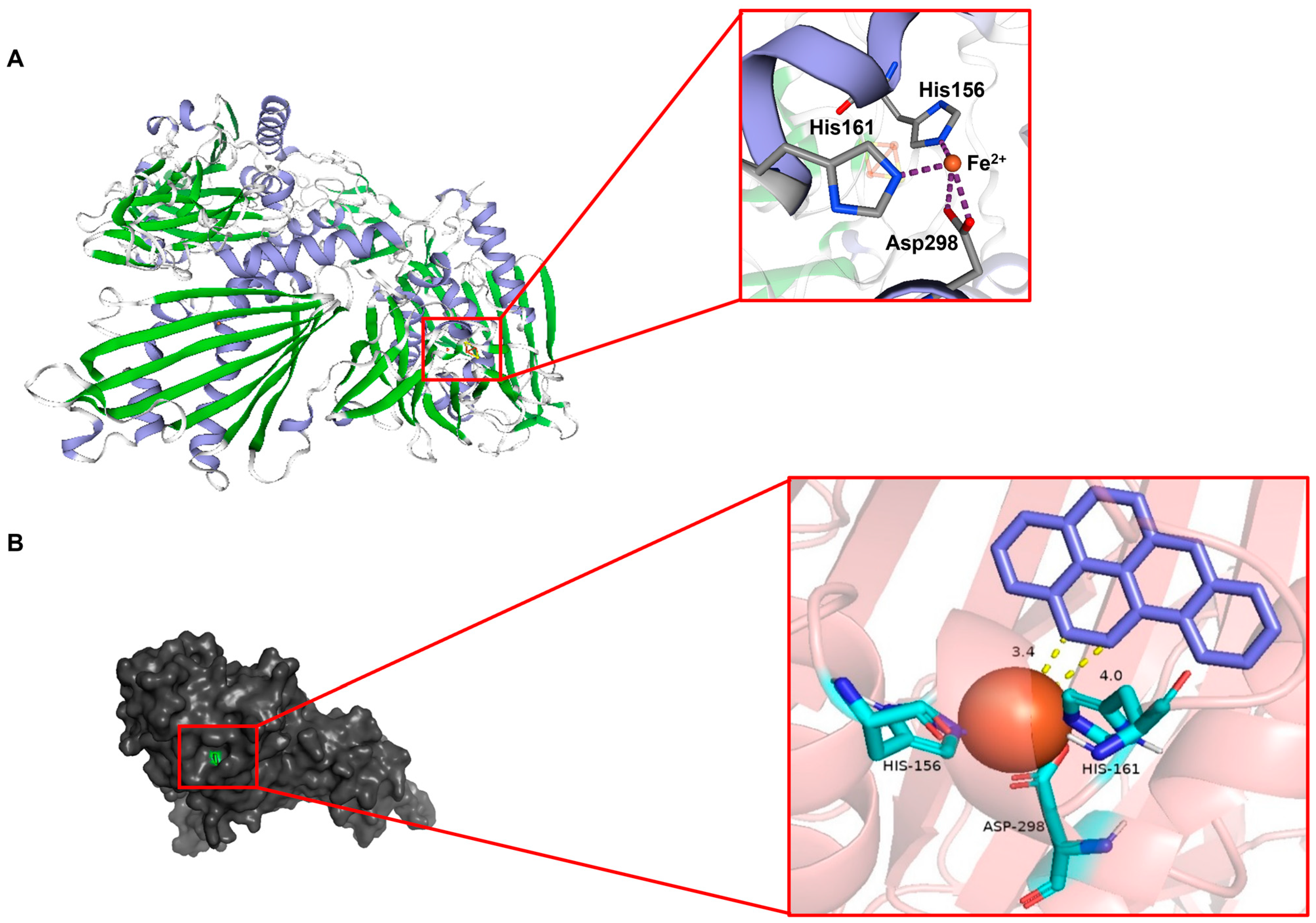
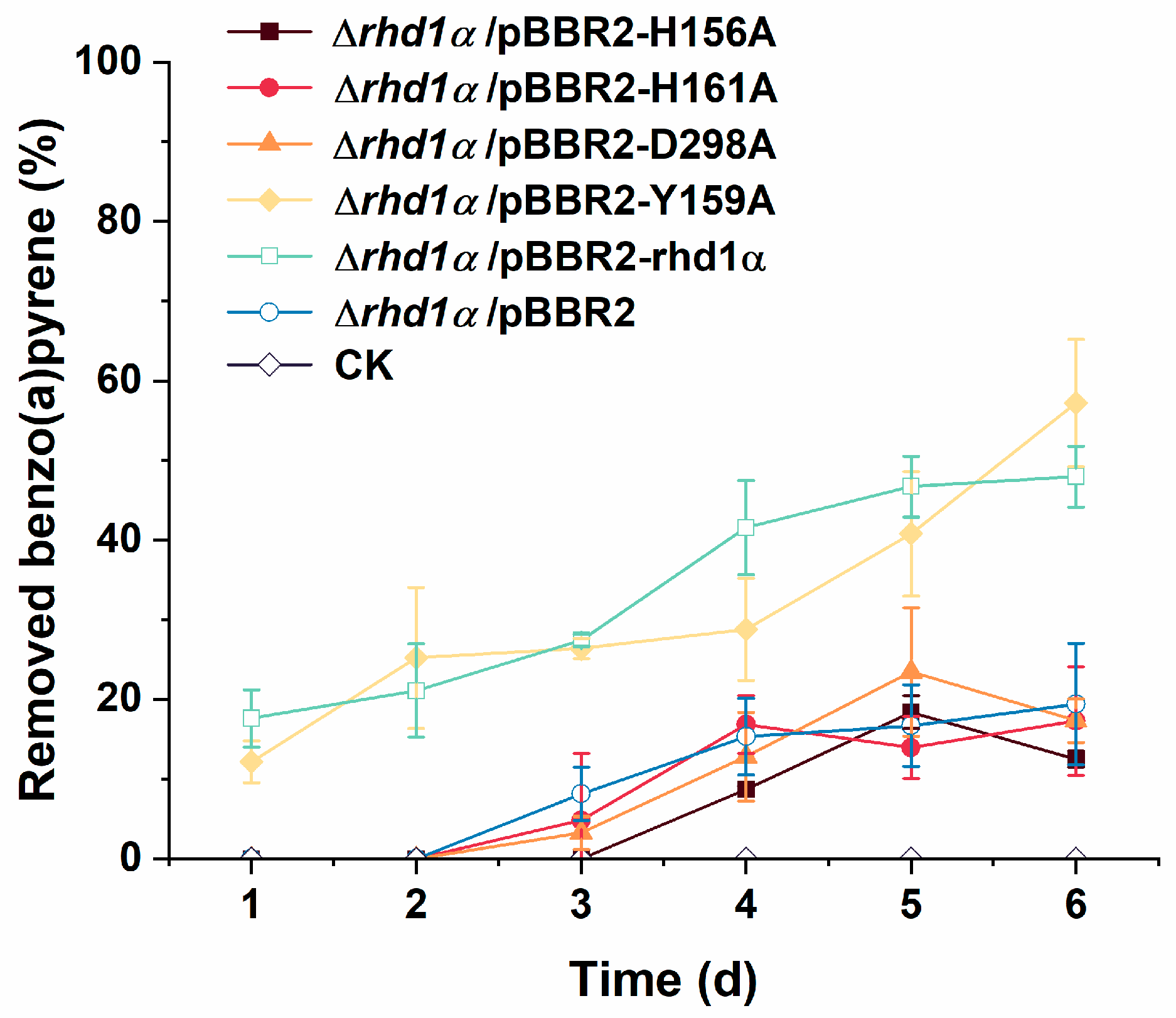
2.4. Residues H156, H161, and D298 Are Essential for Rhd1α
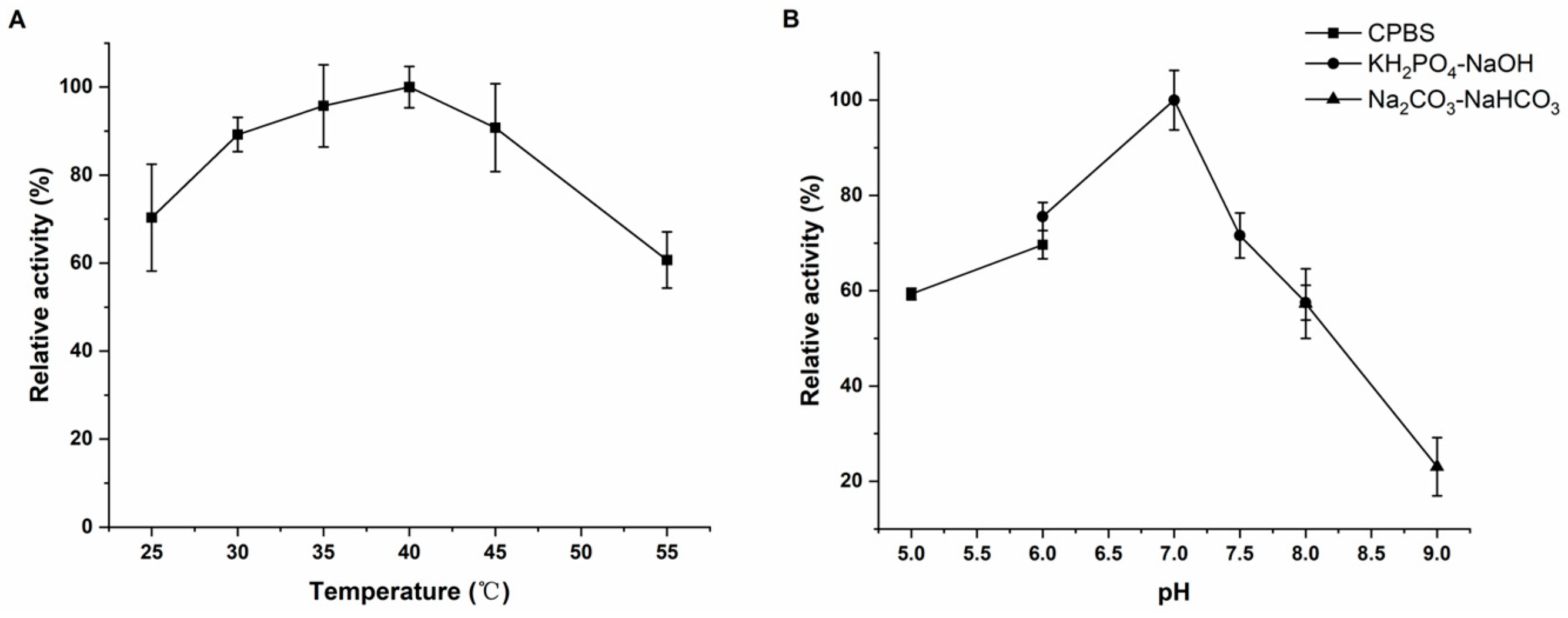
2.5. The In Vitro Active Detection of Rhd1
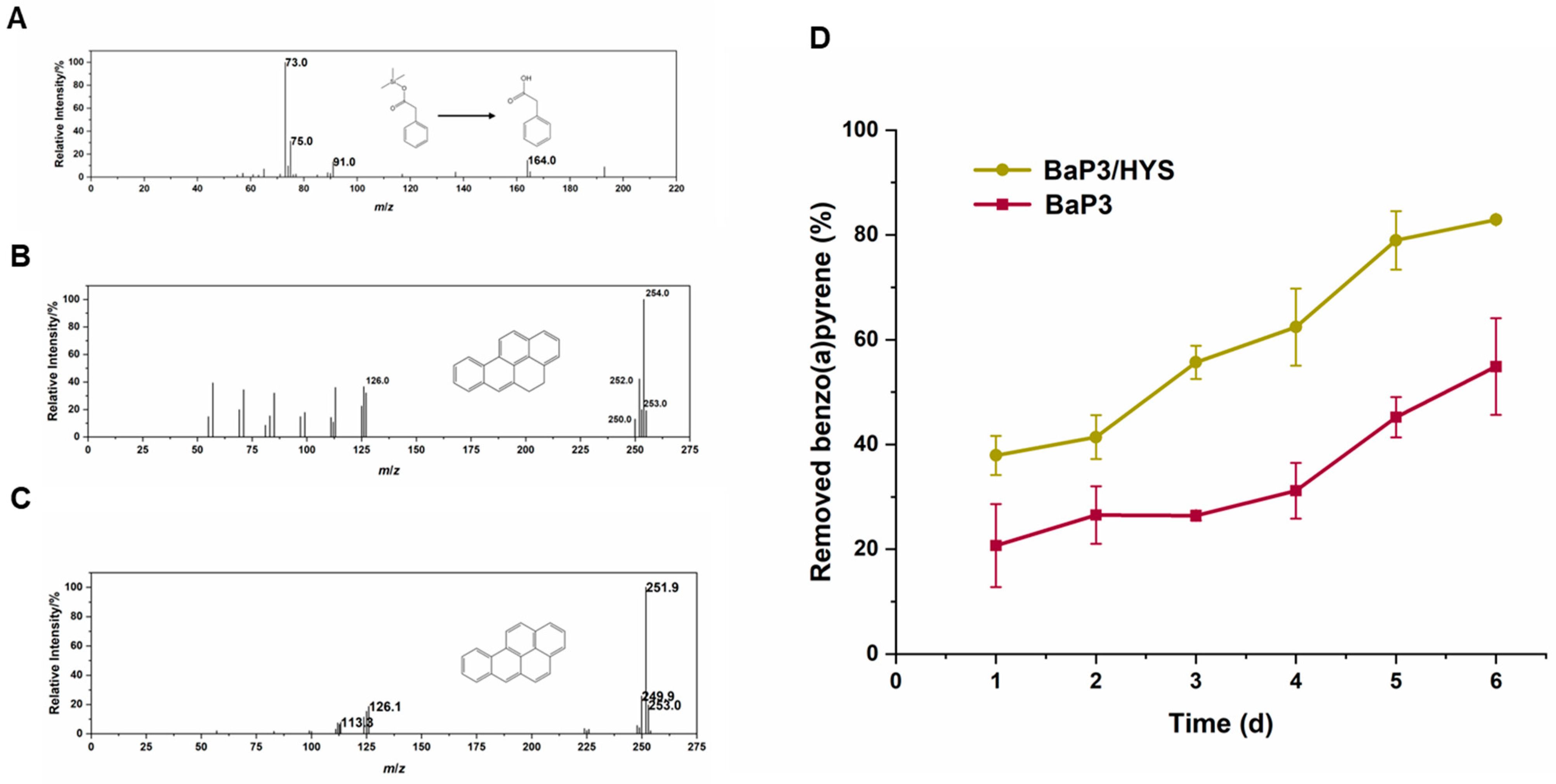
2.6. The Accumulation of Phenylacetic Acid May Be the Reason Why Benzo(a)pyrene Could Not Be Completely Degraded by Strain BaP3
2.7. Synthetic Biology Is a Beneficial Tool to Increase the Degradation Efficiency of PAHs
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, and Culture Media
4.2. Whole-Genome Analysis
4.3. Gene Deletion and Complement
4.4. Determination of Degradability and Growth Curve
4.5. Homology Modeling and Molecular Docking of Rhd1α
4.6. Site-Directed Mutation
4.7. Purification of Rhd1α and Rhd1β Protein
4.8. Characteristic Absorption Peaks and Enzyme Activity
4.9. GC-MS Analysis
4.10. Co-Metabolism and Heterologous Expression
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marris, C.R.; Kompella, S.N.; Miller, M.R.; Incardona, J.P.; Brette, F.; Hancox, J.C.; Sørhus, E.; Shiels, H.A. Polyaromatic hydrocarbons in pollution: A heart-breaking matter. J. Physiol. 2020, 598, 227–247. [Google Scholar] [CrossRef] [PubMed]
- Haritash, A.; Kaushik, C. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): A review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jira, W.; Ziegenhals, K.; Speer, K. Gas chromatography-mass spectrometry (GC-MS) method for the determination of 16 European priority polycyclic aromatic hydrocarbons in smoked meat products and edible oils. Food Addit. Contam. Part A 2008, 25, 704–713. [Google Scholar] [CrossRef] [PubMed]
- García, M.M.; de Llasera, M.P.G. A review on the enzymes and metabolites identified by mass spectrometry from bacteria and microalgae involved in the degradation of high molecular weight PAHs. Sci. Total Environ. 2021, 797, 149035. [Google Scholar] [CrossRef] [PubMed]
- Moorthy, B.; Chu, C.; Carlin, D.J. Polycyclic aromatic hydrocarbons: From metabolism to lung cancer. Toxicol. Sci. 2015, 145, 5–15. [Google Scholar] [CrossRef]
- Patel, A.B.; Shaikh, S.; Jain, K.R.; Desai, C.; Madamwar, D. Polycyclic aromatic hydrocarbons: Sources, toxicity, and remediation approaches. Front. Microbiol. 2020, 11, 562813. [Google Scholar] [CrossRef]
- Pysh, E.S.; Yang, N.C. Polarographic oxidation potentials of aromatic compounds. J. Am. Chem. Soc. 1963, 85, 2124–2130. [Google Scholar] [CrossRef]
- Mazarji, M.; Minkina, T.; Sushkova, S.; Mandzhieva, S.; Fedorenko, A.; Bauer, T.; Soldatov, A.; Barakhov, A.; Dudnikova, T. Biochar-assisted Fenton-like oxidation of benzo[a]pyrene-contaminated soil. Environ. Geochem. Health 2021, 44, 195–206. [Google Scholar] [CrossRef]
- Zhang, T.; Lowry, G.V.; Capiro, N.L.; Chen, J.; Chen, W.; Chen, Y.; Dionysiou, D.D.; Elliott, D.W.; Ghoshal, S.; Hofmann, T.; et al. In situ remediation of subsurface contamination: Opportunities and challenges for nanotechnology and advanced materials. Environ. Sci. Nano 2019, 6, 1283–1302. [Google Scholar] [CrossRef]
- Usman, M.; Hanna, K.; Haderlein, S. Fenton oxidation to remediate PAHs in contaminated soils: A critical review of major limitations and counter-strategies. Sci. Total. Environ. 2016, 569–570, 179–190. [Google Scholar] [CrossRef]
- Wang, Z.; Song, J.; Yuan, W.; Li, Y.; Wu, N.; Li, X.; Rong, L.; Wang, H.; Zhang, J.; Wei, W.; et al. Quantitative monitoring and potential mechanism of the secondary corrosion risk of PAH-contaminated soil remediated by persulfate oxidation. J. Environ. Manag. 2023, 325, 116407. [Google Scholar] [CrossRef] [PubMed]
- Gou, Y.; Ma, J.; Yang, S.; Song, Y. Insights into the effects of Fenton oxidation on PAH removal and indigenous bacteria in aged subsurface soil. Environ. Pollut. 2022, 298, 118872. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Huang, Y.; Wang, H. pahE, a functional marker gene for polycyclic aromatic hydrocarbon-degrading bacteria. Appl. Environ. Microbiol. 2019, 85, e02399-18. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Luo, C.; Jiang, L.; Zhang, D.; Wang, Y.; Zhang, G. Identification of benzo[a]pyrene-metabolizing bacteria in forest soils by using DNA-based stable-isotope probing. Appl. Environ. Microbiol. 2015, 81, 7368–7376. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Zhu, Q.; Li, Y.; Dai, Y.; Wu, Y.; Sun, Y.; Miu, L.; Chen, H.; Lin, X. Isolation of diverse pyrene-degrading bacteria via introducing readily utilized phenanthrene. Chemosphere 2019, 222, 534–540. [Google Scholar] [CrossRef]
- Molina, M.C.; González, N.; Bautista, L.F.; Sanz, R.; Simarro, R.; Sánchez, I.; Sanz, J.L. Isolation and genetic identification of PAH degrading bacteria from a microbial consortium. Biodegradation 2009, 20, 789–800. [Google Scholar] [CrossRef]
- Oyehan, T.A.; Al-Thukair, A.A. Isolation and characterization of PAH-degrading bacteria from the Eastern Province, Saudi Arabia. Mar. Pollut. Bull. 2017, 115, 39–46. [Google Scholar] [CrossRef]
- Yuan, H.; Yao, J.; Masakorala, K.; Wang, F.; Cai, M.; Yu, C. Isolation and characterization of a newly isolated pyrene-degrading Acinetobacter strain USTB-X. Environ. Sci. Pollut. Res. 2014, 21, 2724–2732. [Google Scholar] [CrossRef]
- Kumari, S.; Regar, R.K.; Manickam, N. Improved polycyclic aromatic hydrocarbon degradation in a crude oil by individual and a consortium of bacteria. Bioresour. Technol. 2018, 254, 174–179. [Google Scholar] [CrossRef]
- Hennessee, C.T.; Li, Q.X. Effects of polycyclic aromatic hydrocarbon mixtures on degradation, gene expression, and metabolite production in four Mycobacterium species. Appl. Environ. Microbiol. 2016, 82, 3357–3369. [Google Scholar] [CrossRef]
- Kweon, O.; Kim, S.-J.; Holland, R.D.; Chen, H.; Kim, D.-W.; Gao, Y.; Yu, L.-R.; Baek, S.; Baek, D.-H.; Ahn, H.; et al. Polycyclic aromatic hydrocarbon metabolic network in Mycobacterium vanbaalenii PYR-1. J. Bacteriol. 2011, 193, 4326–4337. [Google Scholar] [CrossRef] [PubMed]
- Punetha, A.; Saraswat, S.; Rai, J.P.N. An insight on microbial degradation of benzo[a]pyrene: Current status and advances in research. World J. Microbiol. Biotechnol. 2022, 38, 61. [Google Scholar] [CrossRef] [PubMed]
- Nzila, A.; Musa, M.M. Current status of and future perspectives in bacterial degradation of benzo[a]pyrene. Int. J. Environ. Res. Public Health 2020, 18, 262. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Huang, Y.; Wang, Y.; Ye, Q.; Zhang, Z.; Wang, H. Distribution of bacterial polycyclic aromatic hydrocarbon (PAH) ring-hydroxylating dioxygenases genes in oilfield soils and mangrove sediments explored by gene-targeted metagenomics. Appl. Microbiol. Biotechnol. 2019, 103, 2427–2440. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Malagnoux, L.; Violet, F.; Jakoncic, J.; Jouanneau, Y. Diversity and catalytic potential of PAH-specific ring-hydroxylating dioxygenases from a hydrocarbon-contaminated soil. Appl. Microbiol. Biotechnol. 2013, 97, 5125–5135. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peng, T.; Luo, A.; Kan, J.; Liang, L.; Huang, T.; Hu, Z. Identification of A ring-hydroxylating dioxygenases capable of anthracene and benz[a]anthracene oxidization from Rhodococcus sp. P14. Microb. Physiol. 2018, 28, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Zhou, N.-Y. Purification and characterization of salicylate 5-hydroxylase, a three-component monooxygenase from Ralstonia sp. strain U2. Appl. Microbiol. Biotechnol. 2014, 98, 671–679. [Google Scholar] [CrossRef]
- Xue, S.-W.; Tian, Y.-X.; Pan, J.-C.; Liu, Y.-N.; Ma, Y.-L. Binding interaction of a ring-hydroxylating dioxygenase with fluoranthene in Pseudomonas aeruginosa DN1. Sci. Rep. 2021, 11, 21317. [Google Scholar] [CrossRef]
- Dong, X.; Rao, Z.; Wu, S.; Peng, F.; Xie, Z.; Long, Y. Pseudomonas benzopyrenica sp. nov., isolated from soil, exhibiting high-efficiency degradation of benzo(a)pyrene. Int. J. Syst. Evol. Microbiol. 2023, 73, 006034. [Google Scholar] [CrossRef]
- Qian, Z.; Peng, T.; Huang, T.; Hu, Z. Oxidization of benzo[a]pyrene by CYP102 in a novel PAHs-degrader Pontibacillus sp. HN14 with potential application in high salinity environment. J. Environ. Manag. 2023, 321, 115922. [Google Scholar] [CrossRef]
- Gao, J.; Xie, G.; Peng, F.; Xie, Z. Pseudomonas donghuensis sp. nov., exhibiting high-yields of siderophore. Antonie Van Leeuwenhoek 2015, 107, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, L.; Shao, Z. Polycyclic aromatic hydrocarbon (PAH) degradation pathways of the obligate marine PAH degrader Cycloclasticus sp. Strain P1. Appl. Environ. Microbiol. 2018, 84, e01261-18. [Google Scholar] [CrossRef] [PubMed]
- Martí-Renom, M.A.; Stuart, A.C.; Fiser, A.; Sánchez, R.; Melo, F.; Šali, A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 291–325. [Google Scholar] [CrossRef] [PubMed]
- Jakoncic, J.; Jouanneau, Y.; Meyer, C.; Stojanoff, V. The crystal structure of the ring-hydroxylating dioxygenase from Sphingomonas CHY-1. FEBS J. 2007, 274, 2470–2481. [Google Scholar] [CrossRef]
- Weber, D.J.; Mullen, G.P.; Mildvan, A.S. Conformation of an enzyme-bound substrate of staphylococcal nuclease as determined by NMR. Biochemistry 1991, 30, 7425–7437. [Google Scholar] [CrossRef]
- Ledesma, A.E.; Taranto, M.P.; Bustos, A.Y. Characterization of substrate specificity and inhibitory mechanism of bile salt hydrolase from Lactobacillus reuteri CRL 1098 using molecular docking analysis. Biotechnol. Lett. 2021, 43, 1063–10730. [Google Scholar] [CrossRef]
- Kadri, T.; Rouissi, T.; Brar, S.K.; Cledon, M.; Sarma, S.; Verma, M. Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by fungal enzymes: A review. J. Environ. Sci. 2017, 51, 52–74. [Google Scholar] [CrossRef]
- Yabukarski, F.; Biel, J.T.; Pinney, M.M.; Doukov, T.; Powers, A.S.; Fraser, J.S.; Herschlag, D. Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray–derived conformational ensembles. Proc. Natl. Acad. Sci. USA 2020, 117, 33204–33215. [Google Scholar] [CrossRef]
- Zeng, X.-H.; Du, H.; Zhao, H.-M.; Xiang, L.; Feng, N.-X.; Li, H.; Li, Y.-W.; Cai, Q.-Y.; Mo, C.-H.; Wong, M.-H.; et al. Insights into the binding interaction of substrate with catechol 2,3-dioxygenase from biophysics point of view. J. Hazard. Mater. 2020, 391, 122211. [Google Scholar] [CrossRef]
- Leng, Q.; Mu, J.; Yang, G. Efficient anaerobic bioremediation of high-concentration benzo[a]pyrene in marine environments. Environ. Pollut. 2021, 284, 117210. [Google Scholar] [CrossRef]
- O’Leary, N.D.; Duetz, W.A.; Dobson, A.D.; O’Connor, K.E. Induction and repression of the sty operon in Pseudomonas putida CA-3 during growth on phenylacetic acid under organic and inorganic nutrient-limiting continuous culture conditions. FEMS Microbiol. Lett. 2002, 208, 263–268. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Teufel, R.; Gantert, C.; Voss, M.; Eisenreich, W.; Haehnel, W.; Fuchs, G. Studies on the mechanism of ring hydrolysis in phenylacetate degradation. J. Biol. Chem. 2011, 286, 11021–11034. [Google Scholar] [CrossRef] [PubMed]
- Cameron, D.E.; Bashor, C.J.; Collins, J.J. A brief history of synthetic biology. Nat. Rev. Genet. 2014, 12, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Hwang, S.; Lee, Y.; Cho, S.; Palsson, B.; Cho, B.-K. Synthetic biology tools for novel secondary metabolite discovery in Streptomyces. J. Microbiol. Biotechnol. 2019, 29, 667–686. [Google Scholar] [CrossRef]
- Chen, R.P.; Gaynor, A.S.; Chen, W. Synthetic biology approaches for targeted protein degradation. Biotechnol. Adv. 2019, 37, 107446. [Google Scholar] [CrossRef] [PubMed]
- Hanson, A.D.; Jez, J.M. Synthetic biology meets plant metabolism. Plant Sci. 2018, 273, 1–2. [Google Scholar] [CrossRef]
- Jiang, Z.; Chen, M.; Yu, X.; Xie, Z. 7-Hydroxytropolone produced and utilized as an iron-scavenger by Pseudomonas donghuensis. BioMetals 2016, 29, 817–826. [Google Scholar] [CrossRef]
- Kim, S.-J.; Kweon, O.; Freeman, J.P.; Jones, R.C.; Adjei, M.D.; Jhoo, J.-W.; Edmondson, R.D.; Cerniglia, C.E. Molecular cloning and expression of genes encoding a novel dioxygenase involved in low- and high-molecular-weight polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Appl. Environ. Microbiol. 2006, 72, 1045–1054. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A Broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Bio/Technology 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Palma, T.L.; Magno, G.; Costa, M.C. Biodegradation of paracetamol by some gram-positive bacterial isolates. Curr. Microbiol. 2021, 78, 2774–2786. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.T.; Karkhoff-Schweizer, R.R.; Kutchma, A.J.; Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998, 212, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wang, P.; Zhu, X.; Xie, Z. Pseudomonas donghuensis HYS gtrA/B/II gene cluster contributes to its pathogenicity toward Caenorhabditis elegans. Int. J. Mol. Sci. 2021, 22, 10741. [Google Scholar] [CrossRef] [PubMed]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef] [PubMed]
- Porter, L.L.; Rose, G.D. Redrawing the Ramachandran plot after inclusion of hydrogen-bonding constraints. Proc. Natl. Acad. Sci. USA 2010, 108, 109–113. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Li, Y.; Qu, J.; Lin, Y.; Lu, G.; You, Y.; Jiang, G.; Wu, Y. Visible post-data analysis protocol for natural mycotoxin production. J. Agric. Food Chem. 2020, 68, 9603–9611. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.; Wu, S.; Rao, Z.; Xiao, Y.; Long, Y.; Xie, Z. Insight into the High-Efficiency Benzo(a)pyrene Degradation Ability of Pseudomonas benzopyrenica BaP3 and Its Application in the Complete Bioremediation of Benzo(a)pyrene. Int. J. Mol. Sci. 2023, 24, 15323. https://doi.org/10.3390/ijms242015323
Dong X, Wu S, Rao Z, Xiao Y, Long Y, Xie Z. Insight into the High-Efficiency Benzo(a)pyrene Degradation Ability of Pseudomonas benzopyrenica BaP3 and Its Application in the Complete Bioremediation of Benzo(a)pyrene. International Journal of Molecular Sciences. 2023; 24(20):15323. https://doi.org/10.3390/ijms242015323
Chicago/Turabian StyleDong, Xingchen, Siyi Wu, Zihuan Rao, Yaqian Xiao, Yan Long, and Zhixiong Xie. 2023. "Insight into the High-Efficiency Benzo(a)pyrene Degradation Ability of Pseudomonas benzopyrenica BaP3 and Its Application in the Complete Bioremediation of Benzo(a)pyrene" International Journal of Molecular Sciences 24, no. 20: 15323. https://doi.org/10.3390/ijms242015323
APA StyleDong, X., Wu, S., Rao, Z., Xiao, Y., Long, Y., & Xie, Z. (2023). Insight into the High-Efficiency Benzo(a)pyrene Degradation Ability of Pseudomonas benzopyrenica BaP3 and Its Application in the Complete Bioremediation of Benzo(a)pyrene. International Journal of Molecular Sciences, 24(20), 15323. https://doi.org/10.3390/ijms242015323





