Recent Advances in In Vivo Somatic Cell Gene Modification in Newborn Pups
Abstract
1. Introduction
2. Concept for In Vivo Organ/Tissue Genome Editing
3. Various Methods and Routes for In Vivo Gene Delivery in Newborn Pups
3.1. Correction of Genetic Disorders through i.v. Introduction of Therapeutic Viral Vectors
3.2. Tissue Tropism among rAAV Serotypes as Revealed by i.v. Introduction of rAAVs to Neonates
3.3. Gene Delivery to Peripheral Tissues through i.v. Introduction of rAAVs in Neonates
3.4. Neonatal i.m. Injection
3.5. Neonatal i.p. Injection
3.6. Gene Delivery via the Retro-Orbital Sinus or toward Retinal Cells
3.6.1. Retinal Gene Delivery into the Subretinal Space
3.6.2. Gene Delivery via the Retro-Orbital Sinus
3.7. Intracerebral Injection
3.8. Gene Delivery to the Skin Cells of Newborn Mice
4. Genome Editing at Neonatal Stages
4.1. Neonatal Gene Correction in Genetic Liver Disorders
4.2. Neonatal Gene Correction in Genetic Blood Clotting Disorders
4.3. Neonatal Gene Correction in Muscular Dysfunction
4.4. Neonatal Gene KO in the CNS
4.5. Clinical Trials Using CRISPR/Cas9-Based Genome Editing
5. Advantages and Limitations of Using Newborns
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAV | Adeno-associated virus |
| ABEs | Adenine base editors |
| AD | Adenoviral vector |
| APOE3 | Apolipoprotein E3 |
| BBB | Blood–brain barrier |
| BEs | Base editors |
| Cas9 | CRISPR-associated protein 9 |
| CBEs | Cytosine base editors |
| CNS | Central nervous system |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| dCas9 | Dead Cas9 |
| ddPCR | Droplet digital PCR |
| DG | Dentate gyrus |
| DMD | Duchenne muscular dystrophy |
| DRG | Dorsal root ganglia |
| DSB | Double-stranded DNA break |
| EGFP | Enhanced green fluorescent protein |
| ENS | Enteric nervous system |
| EP | Electroporation |
| ER | Estrogen receptor |
| ERT2 | Mutated ligand-binding domain of ER |
| ET | Egg transfer |
| F8 (9) | Factor 8 (9) |
| GAG | Glycosaminoglycan |
| GALC | β-Galactocerebrosidase |
| GCs | Genome copies |
| GFP | Green fluorescent protein |
| GOI | Gene of interest |
| GONAD/i-GONAD | Genome Editing via Oviductal Nucleic Acids Delivery/improved GONAD |
| gRNA | Guide RNA |
| GUSB | β-Glucuronidase |
| HDR | Homology-directed repair |
| HA | Hemophilia A |
| HB | Hemophilia B |
| HGD | Hydrodynamics-based gene delivery |
| ICV | Intracerebroventricular |
| IDUA | α-L-iduronidase |
| i.m. | Intramuscular |
| i.p. | Intraperitoneal |
| IT | Intrathecal |
| ITRs | Inverted terminal repeats |
| i.v. | Intravenous |
| KI | Knock-in |
| KO | Knockout |
| LMNs | Lower motor neurons |
| LPS | Lipopolysaccharide |
| LV | Lentivirus |
| MMEJ | Microhomology-mediated end-joining |
| MPS I | Mucopolysaccharidosis type I |
| MPS VII | Mucopolysaccharidosis type VII |
| NCAM (hNCAM-140) | Neural cell adhesion molecule |
| nCas9 | Cas9 nickase |
| NF-κB | Nuclear factor-κB |
| NHEJ | Non-homologous end-joining |
| NSCs | Neural stem cells |
| OB | Olfactory bulb |
| 4OHT | 4-Hydroxytamoxifen |
| OTC | Ornithine transcarbamylase |
| PAM | Protospacer adjacent motif |
| PBS | Phosphate-buffered saline |
| PEs | Prime editors |
| pegRNA | Prime editing guide RNA |
| PFV | Foamy virus |
| PG | Periglomerular |
| RGCs | Radial glial cells |
| RMS | Rostral migratory stream |
| RNAi | RNA interference |
| RNP | Ribonucleoprotein |
| RT | Reverse transcriptase |
| RV | Retrovirus |
| rAAV | Recombinant adeno-associated virus |
| SaCas9 | Staphylococcus aureus-derived Cas9 |
| SB | Sleeping Beauty |
| SFFV | Spleen focus-forming virus |
| sgRNA | Single-guide RNA |
| shRNA | Short hairpin RNA |
| siRNAs | Short interfering RNAs |
| SN | Substantia nigra |
| Sod1 | Superoxide dismutase 1 |
| SVZ | Subventricular zone |
| TadA | tRNA adenine deaminase |
| TBG | Thyroxine-binding globulin |
| Tg | Transgenic |
References
- Clark, J.F.; Dinsmore, C.J.; Soriano, P. A most formidable arsenal: Genetic technologies for building a better mouse. Genes Dev. 2020, 34, 1256–1286. [Google Scholar] [CrossRef]
- Sato, M.; Takabayashi, S.; Akasaka, E.; Nakamura, S. Recent Advances and Future Perspectives of In Vivo Targeted Delivery of Genome-Editing Reagents to Germ Cells, Embryos, and Fetuses in Mice. Cells 2020, 9, 799. [Google Scholar] [CrossRef]
- Takahashi, G.; Gurumurthy, C.B.; Wada, K.; Miura, H.; Sato, M.; Ohtsuka, M. GONAD: Genome-editing via Oviductal Nucleic Acids Delivery system: A novel microinjection independent genome engineering method in mice. Sci. Rep. 2015, 5, 11406. [Google Scholar] [CrossRef]
- Ohtsuka, M.; Sato, M.; Miura, H.; Takabayashi, S.; Matsuyama, M.; Koyano, T.; Arifin, N.; Nakamura, S.; Wada, K.; Gurumurthy, C.B. i-GONAD: A robust method for in situ germline genome engineering using CRISPR nucleases. Genome Biol. 2018, 19, 25. [Google Scholar] [CrossRef] [PubMed]
- National Academies of Sciences, Engineering, and Medicine; National Academy of Medicine; National Academy of Sciences; Committee on Human Gene Editing: Scientific, Medical, and Ethical Considerations. Human Genome Editing: Science, Ethics, and Governance; National Academies Press: Washington, DC, USA, 2017. [Google Scholar]
- Karda, R.; Rahim, A.A.; Wong, A.M.S.; Suff, N.; Diaz, J.A.; Perocheau, D.P.; Tijani, M.; Ng, J.; Baruteau, J.; Martin, N.P.; et al. Generation of light-producing somatic-transgenic mice using adeno-associated virus vectors. Sci. Rep. 2020, 10, 2121. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Inada, E.; Saitoh, I.; Sato, M. Recent Genome-Editing Approaches toward Post-Implanted Fetuses in Mice. BioTech 2023, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.X.; Loh, S.J.H.; Chan, W.K.; Soh, B.S. In Vivo Genome Editing as a Therapeutic Approach. Int. J. Mol. Sci. 2018, 19, 2721. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.M.; Jenkins, B.V.; O’Connor-Giles, K.M.; Wildonger, J. A CRISPR view of development. Genes Dev. 2014, 28, 1859–1872. [Google Scholar] [CrossRef]
- McVey, M.; Lee, S.E. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008, 24, 529–538. [Google Scholar] [CrossRef]
- Kim, S.I.; Matsumoto, T.; Kagawa, H.; Nakamura, M.; Hirohata, R.; Ueno, A.; Ohishi, M.; Sakuma, T.; Soga, T.; Yamamoto, T.; et al. Microhomology-assisted scarless genome editing in human iPSCs. Nat. Commun. 2018, 9, 939. [Google Scholar] [CrossRef]
- Komor, A.C.; Badran, A.H.; Liu, D.R. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 2017, 168, 20–36. [Google Scholar] [CrossRef] [PubMed]
- Nishida, K.; Arazoe, T.; Yachie, N.; Banno, S.; Kakimoto, M.; Tabata, M.; Mochizuki, M.; Miyabe, A.; Araki, M.; Hara, K.Y.; et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016, 353, aaf8729. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Kantor, A.; McClements, M.E.; MacLaren, R.E. CRISPR-Cas9 DNA Base-Editing and Prime-Editing. Int. J. Mol. Sci. 2020, 21, 6240. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Song, C.Q.; Jiang, T.; Richter, M.; Rhym, L.H.; Koblan, L.W.; Zafra, M.P.; Schatoff, E.M.; Doman, J.L.; Cao, Y.; Dow, L.E.; et al. Adenine base editing in an adult mouse model of tyrosinaemia. Nat. Biomed. Eng. 2020, 4, 125–130. [Google Scholar] [CrossRef]
- Song, C.Q.; Li, Y.; Mou, H.; Moore, J.; Park, A.; Pomyen, Y.; Hough, S.; Kennedy, Z.; Fischer, A.; Yin, H.; et al. Genome-Wide CRISPR Screen Identifies Regulators of Mitogen-Activated Protein Kinase as Suppressors of Liver Tumors in Mice. Gastroenterology 2017, 152, 1161–1173.e1. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, F.; Gao, G. CRISPR-Based Therapeutic Genome Editing: Strategies and In Vivo Delivery by AAV Vectors. Cell 2020, 181, 136–150. [Google Scholar] [CrossRef]
- Ryø, L.B.; Thomsen, E.A.; Mikkelsen, J.G. Production and Validation of Lentiviral Vectors for CRISPR/Cas9 Delivery. Methods Mol. Biol. 2019, 1961, 93–109. [Google Scholar]
- Swiech, L.; Heidenreich, M.; Banerjee, A.; Habib, N.; Li, Y.; Trombetta, J.; Sur, M.; Zhang, F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015, 33, 102–106. [Google Scholar] [CrossRef]
- Yang, S.; Chang, R.; Yang, H.; Zhao, T.; Hong, Y.; Kong, H.E.; Sun, X.; Qin, Z.; Jin, P.; Li, S.; et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Investig. 2017, 127, 2719–2724. [Google Scholar] [CrossRef]
- Lee, B.; Lee, K.; Panda, S.; Gonzales-Rojas, R.; Chong, A.; Bugay, V.; Park, H.M.; Brenner, R.; Murthy, N.; Lee, H.Y. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2018, 2, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Oh, J.; Shim, G.; Cho, B.; Chang, Y.; Kim, S.; Baek, S.; Kim, H.; Shin, J.; Choi, H.; et al. In vivo neuronal gene editing via CRISPR-Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer’s disease. Nat. Neurosci. 2019, 22, 524–528. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Ma, Y.; Li, Q.; Sun, Z.; Ma, L.; Wu, L.; Wang, L.; Zeng, L.; Shao, Y.; Chen, Y.; et al. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol. Med. 2016, 8, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Laoharawee, K.; DeKelver, R.C.; Podetz-Pedersen, K.M.; Rohde, M.; Sproul, S.; Nguyen, H.O.; Nguyen, T.; St Martin, S.J.; Ou, L.; Tom, S.; et al. Dose-Dependent Prevention of Metabolic and Neurologic Disease in Murine MPS II by ZFN-Mediated In Vivo Genome Editing. Mol. Ther. 2018, 26, 1127–1136. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, Z.T.; Xu, C.F.; Lu, Z.D.; Luo, Y.L.; Wang, J. Optimization of lipid-assisted nanoparticle for disturbing neutrophils-related inflammation. Biomaterials 2018, 172, 92–104. [Google Scholar] [CrossRef]
- Lee, Y.W.; Mout, R.; Luther, D.C.; Liu, Y.; Castellanos-García, L.; Burnside, A.S.; Ray, M.; Tonga, G.Y.; Hardie, J.; Nagaraj, H.; et al. In Vivo Editing of Macrophages through Systemic Delivery of CRISPR-Cas9-Ribonucleoprotein-Nanoparticle Nanoassemblies. Adv. Ther. 2019, 2, 1900041. [Google Scholar] [CrossRef]
- Stadtmauer, E.A.; Fraietta, J.A.; Davis, M.M.; Cohen, A.D.; Weber, K.L.; Lancaster, E.; Mangan, P.A.; Kulikovskaya, I.; Gupta, M.; Chen, F.; et al. CRISPR-engineered T cells in patients with refractory cancer. Science 2020, 367, eaba7365. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, L.; Xie, Y.; Wang, N.; Tang, R.; Zheng, W.; Jiang, X. Genome Editing for Cancer Therapy: Delivery of Cas9 Protein/sgRNA Plasmid via a Gold Nanocluster/Lipid Core-Shell Nanocarrier. Adv. Sci. 2017, 4, 1700175. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Li, F.; Wang, L.; Zhang, Z.K.; Wang, C.; He, B.; Li, J.; Chen, Z.; Shaikh, A.B.; Liu, J.; et al. Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma. Biomaterials 2017, 147, 68–85. [Google Scholar] [CrossRef]
- Zhang, H.; Qin, C.; An, C.; Zheng, X.; Wen, S.; Chen, W.; Liu, X.; Lv, Z.; Yang, P.; Xu, W.; et al. Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol. Cancer 2021, 20, 126. [Google Scholar] [CrossRef]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef]
- Bedbrook, C.N.; Deverman, B.E.; Gradinaru, V. Viral Strategies for Targeting the Central and Peripheral Nervous Systems. Annu. Rev. Neurosci. 2018, 41, 323–348. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tai, P.W.L.; Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019, 18, 358–378. [Google Scholar] [CrossRef]
- Haggerty, D.L.; Grecco, G.G.; Reeves, K.C.; Atwood, B. Adeno-Associated Viral Vectors in Neuroscience Research. Mol. Ther. Methods Clin. Dev. 2020, 17, 69–82. [Google Scholar] [CrossRef]
- Dong, J.Y.; Fan, P.D.; Frizzell, R.A. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum. Gene Ther. 1996, 7, 2101–2112. [Google Scholar] [CrossRef] [PubMed]
- Boutin, S.; Monteilhet, V.; Veron, P.; Leborgne, C.; Benveniste, O.; Montus, M.F.; Masurier, C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: Implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010, 21, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Davidson, B.L.; Stein, C.S.; Heth, J.A.; Martins, I.; Kotin, R.M.; Derksen, T.A.; Zabner, J.; Ghodsi, A.; Chiorini, J.A. Recombinant adeno-associated virus type 2, 4, and 5 vectors: Transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA 2000, 97, 3428–3432. [Google Scholar] [CrossRef]
- Zincarelli, C.; Soltys, S.; Rengo, G.; Rabinowitz, J.E. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008, 16, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Duque, S.; Joussemet, B.; Riviere, C.; Marais, T.; Dubreil, L.; Douar, A.M.; Fyfe, J.; Moullier, P.; Colle, M.A.; Barkats, M. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol. Ther. 2009, 17, 1187–1196. [Google Scholar] [CrossRef]
- Fu, H.; Dirosario, J.; Killedar, S.; Zaraspe, K.; McCarty, D.M. Correction of neurological disease of mucopolysaccharidosis IIIB in adult mice by rAAV9 trans-blood-brain barrier gene delivery. Mol. Ther. 2011, 19, 1025–1033. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, B.; Mu, X.; Ahmed, S.S.; Su, Q.; He, R.; Wang, H.; Mueller, C.; Sena-Esteves, M.; Brown, R.; et al. Several rAAV vectors efficiently cross the blood-brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system. Mol. Ther. 2011, 19, 1440–1448. [Google Scholar] [CrossRef] [PubMed]
- Miyake, N.; Miyake, K.; Yamamoto, M.; Hirai, Y.; Shimada, T. Global gene transfer into the CNS across the BBB after neonatal systemic delivery of single-stranded AAV vectors. Brain Res. 2011, 1389, 19–26. [Google Scholar] [CrossRef]
- Deverman, B.E.; Pravdo, P.L.; Simpson, B.P.; Kumar, S.R.; Chan, K.Y.; Banerjee, A.; Wu, W.L.; Yang, B.; Huber, N.; Pasca, S.P.; et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016, 34, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Mellott, A.J.; Forrest, M.L.; Detamore, M.S. Physical non-viral gene delivery methods for tissue engineering. Ann. Biomed. Eng. 2013, 41, 446–468. [Google Scholar] [CrossRef] [PubMed]
- Wolff, J.A.; Malone, R.W.; Williams, P.; Chong, W.; Acsadi, G.; Jani, A.; Felgner, P.L. Direct gene transfer into mouse muscle in vivo. Science 1990, 247, 1465–1468. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.L.; Demeneix, B.A.; Quantin, B.; Coulombe, J.; Whalen, R.G. Plasmid DNA is superior to viral vectors for direct gene transfer into adult mouse skeletal muscle. Hum. Gene Ther. 1993, 4, 733–740. [Google Scholar] [CrossRef]
- Aihara, H.; Miyazaki, J. Gene transfer into muscle by electroporation in vivo. Nat. Biotechnol. 1998, 16, 867–870. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Boado, R.J.; Pardridge, W.M. Absence of toxicity of chronic weekly intravenous gene therapy with pegylated immunoliposomes. Pharm. Res. 2003, 20, 1779–1785. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Boado, R.J.; Pardridge, W.M. Lysosomal enzyme replacement of the brain with intravenous non-viral gene transfer. Pharm. Res. 2008, 25, 400–406. [Google Scholar] [CrossRef]
- Molnar, M.J.; Gilbert, R.; Lu, Y.; Liu, A.B.; Guo, A.; Larochelle, N.; Orlopp, K.; Lochmuller, H.; Petrof, B.J.; Nalbantoglu, J.; et al. Factors influencing the efficacy, longevity, and safety of electroporation-assisted plasmid-based gene transfer into mouse muscles. Mol. Ther. 2004, 10, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Mir, L.M.; Bureau, M.F.; Gehl, J.; Rangara, R.; Rouy, D.; Caillaud, J.M.; Delaere, P.; Branellec, D.; Schwartz, B.; Scherman, D. High-efficiency gene transfer into skeletal muscle mediated by electric pulses. Proc. Natl. Acad. Sci. USA 1999, 96, 4262–4267. [Google Scholar] [CrossRef]
- Gehl, J.; Sorensen, T.H.; Nielsen, K.; Raskmark, P.; Nielsen, S.L.; Skovsgaard, T.; Mir, L.M. In vivo electroporation of skeletal muscle: Threshold, efficacy and relation to electric field distribution. Biochim. Biophys. Acta. 1999, 1428, 233–240. [Google Scholar] [CrossRef]
- McMahon, J.M.; Signori, E.; Wells, K.E.; Fazio, V.M.; Wells, D.J. Optimisation of electrotransfer of plasmid into skeletal muscle by pretreatment with hyaluronidase—increased expression with reduced muscle damage. Gene Ther. 2001, 8, 1264–1270. [Google Scholar] [CrossRef]
- Liu, F.; Song, Y.; Liu, D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999, 6, 1258–1266. [Google Scholar] [CrossRef]
- Suda, T.; Yokoo, T.; Kanefuji, T.; Kamimura, K.; Zhang, G.; Liu, D. Hydrodynamic Delivery: Characteristics, Applications, and Technological Advances. Pharmaceutics 2023, 15, 1111. [Google Scholar] [CrossRef]
- Demorest, Z.L.; Low, W.C.; Freese, A.B.; Ohlfest, J.R. Non-Viral Gene Transfer Using the Sleeping Beauty Transposon for Long-Term Gene Expression in the CNS of Neonatal Mice. Mol. Ther. 2006, 13, S150. [Google Scholar] [CrossRef]
- Prabhakar, S.; Lule, S.; da Hora, C.C.; Breakefield, X.O.; Cheah, P.S. AAV9 transduction mediated by systemic delivery of vector via retro-orbital injection in newborn, neonatal and juvenile mice. Exp. Anim. 2021, 70, 450–458. [Google Scholar] [CrossRef]
- Kienstra, K.A.; Freysdottir, D.; Gonzales, N.M.; Hirschi, K.K. Murine neonatal intravascular injections: Modeling newborn disease. J. Am. Assoc. Lab. Anim. Sci. 2007, 46, 50–54. [Google Scholar]
- Ottoboni, L.; von Wunster, B.; Martino, G. Therapeutic Plasticity of Neural Stem Cells. Front. Neurol. 2020, 11, 148. [Google Scholar] [CrossRef]
- Kessler, P.D.; Podsakoff, G.M.; Chen, X.; McQuiston, S.A.; Colosi, P.C.; Matelis, L.A.; Kurtzman, G.J.; Byrne, B.J. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc. Natl. Acad. Sci. USA 1996, 93, 14082–14087. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Chou, B.; Fitzsimmons, B.; Miyanohara, A.; Shubayev, V.; Santucci, C.; Hefferan, M.; Marsala, M.; Hua, X.Y. In vivo gene knockdown in rat dorsal root ganglia mediated by self-complementary adeno-associated virus serotype 5 following intrathecal delivery. PLoS ONE 2012, 7, e32581. [Google Scholar] [CrossRef][Green Version]
- Machida, A.; Kuwahara, H.; Mayra, A.; Kubodera, T.; Hirai, T.; Sunaga, F.; Tajiri, M.; Hirai, Y.; Shimada, T.; Mizusawa, H.; et al. Intraperitoneal administration of AAV9-shRNA inhibits target gene expression in the dorsal root ganglia of neonatal mice. Mol. Pain 2013, 9, 36. [Google Scholar] [CrossRef]
- Titomirov, A.V.; Sukharev, S.; Kistanova, E. In vivo electroporation and stable transformation of skin cells of newborn mice by plasmid DNA. Biochim. Biophys. Acta. 1991, 1088, 131–134. [Google Scholar] [CrossRef]
- Daly, T.M.; Vogler, C.; Levy, B.; Haskins, M.E.; Sands, M.S. Neonatal gene transfer leads to widespread correction of pathology in a murine model of lysosomal storage disease. Proc. Natl. Acad. Sci. USA 1999, 96, 2296–2300. [Google Scholar] [CrossRef]
- Ponder, K.P.; Melniczek, J.R.; Xu, L.; Weil, M.A.; O’Malley, T.M.; O’Donnell, P.A.; Knox, V.W.; Aguirre, G.D.; Mazrier, H.; Ellinwood, N.M.; et al. Therapeutic neonatal hepatic gene therapy in mucopolysaccharidosis VII dogs. Proc. Natl. Acad. Sci. USA 2002, 99, 13102–13107. [Google Scholar] [CrossRef]
- Xu, L.; Mango, R.L.; Sands, M.S.; Haskins, M.E.; Ellinwood, N.M.; Ponder, K.P. Evaluation of pathological manifestations of disease in mucopolysaccharidosis VII mice after neonatal hepatic gene therapy. Mol. Ther. 2002, 6, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Kamata, Y.; Tanabe, A.; Kanaji, A.; Kosuga, M.; Fukuhara, Y.; Li, X.K.; Suzuki, S.; Yamada, M.; Azuma, N.; Okuyama, T. Long-term normalization in the central nervous system, ocular manifestations, and skeletal deformities by a single systemic adenovirus injection into neonatal mice with mucopolysaccharidosis VII. Gene Ther. 2003, 10, 406–414. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hartung, S.D.; Frandsen, J.L.; Pan, D.; Koniar, B.L.; Graupman, P.; Gunther, R.; Low, W.C.; Whitley, C.B.; McIvor, R.S. Correction of metabolic, craniofacial, and neurologic abnormalities in MPS I mice treated at birth with adeno-associated virus vector transducing the human alpha-L-iduronidase gene. Mol. Ther. 2004, 9, 866–875. [Google Scholar] [CrossRef]
- Kobayashi, H.; Carbonaro, D.; Pepper, K.; Petersen, D.; Ge, S.; Jackson, H.; Shimada, H.; Moats, R.; Kohn, D.B. Neonatal gene therapy of MPS I mice by intravenous injection of a lentiviral vector. Mol. Ther. 2005, 11, 776–789. [Google Scholar] [CrossRef]
- Inagaki, K.; Fuess, S.; Storm, T.A.; Gibson, G.A.; McTiernan, C.F.; Kay, M.A.; Nakai, H. Robust systemic transduction with AAV9 vectors in mice: Efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 2006, 14, 45–53. [Google Scholar] [CrossRef]
- Pacak, C.A.; Mah, C.S.; Thattaliyath, B.D.; Conlon, T.J.; Lewis, M.A.; Cloutier, D.E.; Zolotukhin, I.; Tarantal, A.F.; Byrne, B.J. Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ. Res. 2006, 99, e3–e9. [Google Scholar] [CrossRef] [PubMed]
- Foust, K.D.; Poirier, A.; Pacak, C.A.; Mandel, R.J.; Flotte, T.R. Neonatal intraperitoneal or intravenous injections of recombinant adeno-associated virus type 8 transduce dorsal root ganglia and lower motor neurons. Hum. Gene Ther. 2008, 19, 61–70. [Google Scholar] [CrossRef]
- Hu, C.; Cela, R.G.; Suzuki, M.; Lee, B.; Lipshutz, G.S. Neonatal helper-dependent adenoviral vector gene therapy mediates correction of hemophilia A and tolerance to human factor VIII. Proc. Natl. Acad. Sci. USA 2011, 108, 2082–2087. [Google Scholar] [CrossRef] [PubMed]
- Rahim, A.A.; Wong, A.M.; Hoefer, K.; Buckley, S.M.; Mattar, C.N.; Cheng, S.H.; Chan, J.K.; Cooper, J.D.; Waddington, S.N. Intravenous administration of AAV2/9 to the fetal and neonatal mouse leads to differential targeting of CNS cell types and extensive transduction of the nervous system. Faseb J. 2011, 25, 3505–3518. [Google Scholar] [CrossRef]
- Hu, C.; Lipshutz, G.S. AAV-based neonatal gene therapy for hemophilia A: Long-term correction and avoidance of immune responses in mice. Gene Ther. 2012, 19, 1166–1176. [Google Scholar] [CrossRef] [PubMed]
- Dalkara, D.; Byrne, L.C.; Lee, T.; Hoffmann, N.V.; Schaffer, D.V.; Flannery, J.G. Enhanced gene delivery to the neonatal retina through systemic administration of tyrosine-mutated AAV9. Gene Ther. 2012, 19, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Bemelmans, A.P.; Duqué, S.; Rivière, C.; Astord, S.; Desrosiers, M.; Marais, T.; Sahel, J.A.; Voit, T.; Barkats, M. A single intravenous AAV9 injection mediates bilateral gene transfer to the adult mouse retina. PLoS ONE 2013, 8, e61618. [Google Scholar] [CrossRef] [PubMed]
- Gombash, S.E.; Cowley, C.J.; Fitzgerald, J.A.; Hall, J.C.; Mueller, C.; Christofi, F.L.; Foust, K.D. Intravenous AAV9 efficiently transduces myenteric neurons in neonate and juvenile mice. Front. Mol. Neurosci. 2014, 7, 81. [Google Scholar] [CrossRef]
- Mattar, C.N.; Wong, A.M.; Hoefer, K.; Alonso-Ferrero, M.E.; Buckley, S.M.; Howe, S.J.; Cooper, J.D.; Waddington, S.N.; Chan, J.K.; Rahim, A.A. Systemic gene delivery following intravenous administration of AAV9 to fetal and neonatal mice and late-gestation nonhuman primates. FASEB J. 2015, 29, 3876–3888. [Google Scholar] [CrossRef]
- Buckinx, R.; Van Remoortel, S.; Gijsbers, R.; Waddington, S.N.; Timmermans, J.P. Proof-of-concept: Neonatal intravenous injection of adeno-associated virus vectors results in successful transduction of myenteric and submucosal neurons in the mouse small and large intestine. Neurogastroenterol. Motil. 2016, 28, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Shibata, S.B.; Yoshimura, H.; Ranum, P.T.; Goodwin, A.T.; Smith, R.J.H. Intravenous rAAV2/9 injection for murine cochlear gene delivery. Sci. Rep. 2017, 7, 9609. [Google Scholar] [CrossRef] [PubMed]
- Counsell, J.R.; Karda, R.; Diaz, J.A.; Carey, L.; Wiktorowicz, T.; Buckley, S.M.K.; Ameri, S.; Ng, J.; Baruteau, J.; Almeida, F.; et al. Foamy Virus Vectors Transduce Visceral Organs and Hippocampal Structures following In Vivo Delivery to Neonatal Mice. Mol. Ther. Nucleic Acids 2018, 12, 626–634. [Google Scholar] [CrossRef]
- Gessler, D.J.; Tai, P.W.L.; Li, J.; Gao, G. Intravenous Infusion of AAV for Widespread Gene Delivery to the Nervous System. Methods Mol. Biol. 2019, 1950, 143–163. [Google Scholar] [PubMed]
- Passini, M.A.; Wolfe, J.H. Widespread gene delivery and structure-specific patterns of expression in the brain after intraventricular injections of neonatal mice with an adeno-associated virus vector. J. Virol. 2001, 75, 12382–12392. [Google Scholar] [CrossRef]
- Passini, M.A.; Watson, D.J.; Vite, C.H.; Landsburg, D.J.; Feigenbaum, A.L.; Wolfe, J.H. Intraventricular brain injection of adeno-associated virus type 1 (AAV1) in neonatal mice results in complementary patterns of neuronal transduction to AAV2 and total long-term correction of storage lesions in the brains of beta-glucuronidase-deficient mice. J. Virol. 2003, 77, 7034–7040. [Google Scholar]
- Boutin, C.; Diestel, S.; Desoeuvre, A.; Tiveron, M.C.; Cremer, H. Efficient in vivo electroporation of the postnatal rodent forebrain. PLoS ONE 2008, 3, e1883. [Google Scholar] [CrossRef]
- Chesler, A.T.; Le Pichon, C.E.; Brann, J.H.; Araneda, R.C.; Zou, D.J.; Firestein, S. Selective gene expression by postnatal electroporation during olfactory interneuron neurogenesis. PLoS ONE 2008, 3, e1517. [Google Scholar] [CrossRef]
- Pilpel, N.; Landeck, N.; Klugmann, M.; Seeburg, P.H.; Schwarz, M.K. Rapid, reproducible transduction of select forebrain regions by targeted recombinant virus injection into the neonatal mouse brain. J. Neurosci. Methods 2009, 182, 55–63. [Google Scholar] [CrossRef]
- Molotkov, D.A.; Yukin, A.Y.; Afzalov, R.A.; Khiroug, L.S. Gene delivery to postnatal rat brain by non-ventricular plasmid injection and electroporation. J. Vis. Exp. 2010, 43, 2244. [Google Scholar]
- Fernández, M.E.; Croce, S.; Boutin, C.; Cremer, H.; Raineteau, O. Targeted electroporation of defined lateral ventricular walls: A novel and rapid method to study fate specification during postnatal forebrain neurogenesis. Neural Dev. 2011, 6, 13. [Google Scholar] [CrossRef]
- Kim, J.Y.; Ash, R.T.; Ceballos-Diaz, C.; Levites, Y.; Golde, T.E.; Smirnakis, S.M.; Jankowsky, J.L. Viral transduction of the neonatal brain delivers controllable genetic mosaicism for visualising and manipulating neuronal circuits in vivo. Eur. J. Neurosci. 2013, 37, 1203–1220. [Google Scholar] [CrossRef]
- Chakrabarty, P.; Rosario, A.; Cruz, P.; Siemienski, Z.; Ceballos-Diaz, C.; Crosby, K.; Jansen, K.; Borchelt, D.R.; Kim, J.Y.; Jankowsky, J.L.; et al. Capsid serotype and timing of injection determines AAV transduction in the neonatal mice brain. PLoS ONE 2013, 8, e67680. [Google Scholar] [CrossRef]
- Lattanzi, A.; Salvagno, C.; Maderna, C.; Benedicenti, F.; Morena, F.; Kulik, W.; Naldini, L.; Montini, E.; Martino, S.; Gritti, A. Therapeutic benefit of lentiviral-mediated neonatal intracerebral gene therapy in a mouse model of globoid cell leukodystrophy. Hum. Mol. Genet. 2014, 23, 3250–3268. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Morishita, R.; Iwamoto, I.; Nagata, K. Establishment of an in vivo electroporation method into postnatal newborn neurons in the dentate gyrus. Hippocampus 2014, 24, 1449–1457. [Google Scholar] [CrossRef]
- Kim, J.Y.; Grunke, S.D.; Levites, Y.; Golde, T.E.; Jankowsky, J.L. Intracerebroventricular viral injection of the neonatal mouse brain for persistent and widespread neuronal transduction. J. Vis. Exp. 2014, 91, 51863. [Google Scholar]
- Hammond, S.L.; Leek, A.N.; Richman, E.H.; Tjalkens, R.B. Cellular selectivity of AAV serotypes for gene delivery in neurons and astrocytes by neonatal intracerebroventricular injection. PLoS ONE 2017, 12, e0188830. [Google Scholar] [CrossRef] [PubMed]
- Torregrosa, T.; Lehman, S.; Hana, S.; Marsh, G.; Xu, S.; Koszka, K.; Mastrangelo, N.; McCampbell, A.; Henderson, C.E.; Lo, S.C. Use of CRISPR/Cas9-mediated disruption of CNS cell type genes to profile transduction of AAV by neonatal intracerebroventricular delivery in mice. Gene Ther. 2021, 28, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhu, T.; Qiao, C.; Zhou, L.; Wang, B.; Zhang, J.; Chen, C.; Li, J.; Xiao, X. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat. Biotechnol. 2005, 23, 321–328. [Google Scholar] [CrossRef]
- VandenDriessche, T.; Vanslembrouck, V.; Goovaerts, I.; Zwinnen, H.; Vanderhaeghen, M.L.; Collen, D.; Chuah, M.K. Long-term expression of human coagulation factor VIII and correction of hemophilia A after in vivo retroviral gene transfer in factor VIII-deficient mice. Proc. Natl. Acad. Sci. USA 1999, 96, 10379–10384. [Google Scholar] [CrossRef]
- Matsuda, T.; Cepko, C.L. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. USA 2004, 101, 16–22. [Google Scholar] [CrossRef]
- Matsuda, T.; Cepko, C.L. Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. USA 2007, 104, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Fu, Q.; Zhou, Y.; Wang, J.; Liu, Y.; Duan, X.; Jia, S.; Peng, J.; Gao, B.; Du, J.; et al. High levels of gene expression in the hepatocytes of adult mice, neonatal mice and tree shrews via retro-orbital sinus hydrodynamic injections of naked plasmid DNA. J. Control. Release 2012, 161, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, S.; Sakurai, F.; Shimizu, K.; Ohashi, K.; Nakamura, S.; Tachibana, M.; Mizuguchi, H. Evaluation of transduction properties of an adenovirus vector in neonatal mice. Biomed Res. Int. 2015, 2015, 685374. [Google Scholar] [CrossRef] [PubMed]
- Daly, T.M.; Okuyama, T.; Vogler, C.; Haskins, M.E.; Muzyczka, N.; Sands, M.S. Neonatal intramuscular injection with recombinant adeno-associated virus results in prolonged beta-glucuronidase expression in situ and correction of liver pathology in mucopolysaccharidosis type VII mice. Hum. Gene Ther. 1999, 10, 85–94. [Google Scholar] [CrossRef]
- Signori, E.; Rinaldi, M.; Fioretti, D.; Iurescia, S.; Seripa, D.; Perrone, G.; Norata, G.D.; Catapano, A.L.; Fazio, V.M. ApoE gene delivery inhibits severe hypercholesterolemia in newborn ApoE-KO mice. Biochem. Biophys. Res. Commun. 2007, 361, 543–548. [Google Scholar] [CrossRef]
- Sly, W.S.; Quinton, B.A.; McAlister, W.H.; Rimoin, D.L. Beta glucuronidase deficiency: Report of clinical, radiologic, and biochemical features of a new mucopolysaccharidosis. J. Pediatr. 1973, 82, 249–257. [Google Scholar] [CrossRef]
- Sands, M.S.; Vogler, C.; Kyle, J.W.; Grubb, J.H.; Levy, B.; Galvin, N.; Sly, W.S.; Birkenmeier, E.H. Enzyme replacement therapy for murine mucopolysaccharidosis type VII. J. Clin. Investig. 1994, 93, 2324–2331. [Google Scholar] [CrossRef]
- Mallard, C.; Ek, C.J.; Vexler, Z.S. The myth of the immature barrier systems in the developing brain: Role in perinatal brain injury. J. Physiol. 2018, 596, 5655–5664. [Google Scholar] [CrossRef] [PubMed]
- Ohmi, K.; Greenberg, D.S.; Rajavel, K.S.; Ryazantsev, S.; Li, H.H.; Neufeld, E.F. Activated microglia in cortex of mouse models of mucopolysaccharidoses I and IIIB. Proc. Natl. Acad. Sci. USA 2003, 100, 1902–1907. [Google Scholar] [CrossRef]
- Fisher, K.J.; Jooss, K.; Alston, J.; Yang, Y.; Haecker, S.E.; High, K.; Pathak, R.; Raper, S.E.; Wilson, J.M. Recombinant adeno-associated virus for muscle directed gene therapy. Nat. Med. 1997, 3, 306–312. [Google Scholar] [CrossRef]
- Gregorevic, P.; Blankinship, M.J.; Allen, J.M.; Crawford, R.W.; Meuse, L.; Miller, D.G.; Russell, D.W.; Chamberlain, J.S. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat. Med. 2004, 10, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Mor, G.; Yamshchikov, G.; Sedegah, M.; Takeno, M.; Wang, R.; Houghten, R.A.; Hoffman, S.; Klinman, D.M. Induction of neonatal tolerance by plasmid DNA vaccination of mice. J. Clin. Investig. 1996, 98, 2700–2705. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, A.; Ma, S.; Langellotto, F.; Gao, G.; Punzo, C. Retinal gene delivery by rAAV and DNA electroporation. Curr. Protoc. Microbiol. 2013, 28, 14D.4.1–14D.4.32. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Yamamura, K.; Miyazaki, J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991, 108, 193–199. [Google Scholar] [PubMed]
- Petrs-Silva, H.; Dinculescu, A.; Li, Q.; Deng, W.T.; Pang, J.J.; Min, S.H.; Chiodo, V.; Neeley, A.W.; Govindasamy, L.; Bennett, A.; et al. Novel properties of tyrosine-mutant AAV2 vectors in the mouse retina. Mol. Ther. 2011, 19, 293–301. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, L.; Bell, P.; McMenamin, D.; He, Z.; White, J.; Yu, H.; Xu, C.; Morizono, H.; Musunuru, K.; et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016, 34, 334–338. [Google Scholar] [CrossRef]
- Ohmori, T.; Nagao, Y.; Mizukami, H.; Sakata, A.; Muramatsu, S.I.; Ozawa, K.; Tominaga, S.I.; Hanazono, Y.; Nishimura, S.; Nureki, O.; et al. CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Sci. Rep. 2017, 7, 4159. [Google Scholar] [CrossRef]
- Wang, L.; Yang, Y.; Breton, C.A.; White, J.; Zhang, J.; Che, Y.; Saveliev, A.; McMenamin, D.; He, Z.; Latshaw, C.; et al. CRISPR/Cas9-mediated in vivo gene targeting corrects hemostasis in newborn and adult factor IX-knockout mice. Blood 2019, 133, 2745–2752. [Google Scholar] [CrossRef]
- Lisjak, M.; De Caneva, A.; Marais, T.; Barbon, E.; Biferi, M.G.; Porro, F.; Barzel, A.; Zentilin, L.; Kay, M.A.; Mingozzi, F.; et al. Promoterless Gene Targeting Approach Combined to CRISPR/Cas9 Efficiently Corrects Hemophilia B Phenotype in Neonatal Mice. Front. Genome Ed. 2022, 4, 785698. [Google Scholar] [CrossRef]
- Hana, S.; Peterson, M.; McLaughlin, H.; Marshall, E.; Fabian, A.J.; McKissick, O.; Koszka, K.; Marsh, G.; Craft, M.; Xu, S.; et al. Highly efficient neuronal gene knockout in vivo by CRISPR-Cas9 via neonatal intracerebroventricular injection of AAV in mice. Gene Ther. 2021, 28, 646–658. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Amoasii, L.; Mireault, A.A.; McAnally, J.R.; Li, H.; Sanchez-Ortiz, E.; Bhattacharyya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 2016, 351, 400–403. [Google Scholar] [CrossRef]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Castellanos Rivera, R.M.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.W.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef]
- Li, D.; Yue, Y.; Duan, D. Marginal level dystrophin expression improves clinical outcome in a strain of dystrophin/utrophin double knockout mice. PLoS ONE 2010, 5, e15286. [Google Scholar] [CrossRef] [PubMed]
- van Putten, M.; Hulsker, M.; Young, C.; Nadarajah, V.D.; Heemskerk, H.; van der Weerd, L.; t Hoen, P.A.; van Ommen, G.J.; Aartsma-Rus, A.M. Low dystrophin levels increase survival and improve muscle pathology and function in dystrophin/utrophin double-knockout mice. FASEB J. 2013, 27, 2484–2495. [Google Scholar] [CrossRef]
- Foust, K.D.; Nurre, E.; Montgomery, C.L.; Hernandez, A.; Chan, C.M.; Kaspar, B.K. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 2009, 27, 59–65. [Google Scholar] [CrossRef]
- Chan, K.Y.; Jang, M.J.; Yoo, B.B.; Greenbaum, A.; Ravi, N.; Wu, W.L.; Sánchez-Guardado, L.; Lois, C.; Mazmanian, S.K.; Deverman, B.E.; et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017, 20, 1172–1179. [Google Scholar] [CrossRef]
- Hu, Y.; Zhou, Y.; Zhang, M.; Ge, W.; Li, Y.; Yang, L.; Wei, G.; Han, L.; Wang, H.; Yu, S.; et al. CRISPR/Cas9-Engineered Universal CD19/CD22 Dual-Targeted CAR-T Cell Therapy for Relapsed/Refractory B-cell Acute Lymphoblastic Leukemia. Clin. Cancer Res. 2021, 27, 2764–2772. [Google Scholar] [CrossRef]
- Fu, B.; Liao, J.; Chen, S.; Li, W.; Wang, Q.; Hu, J.; Yang, F.; Hsiao, S.; Jiang, Y.; Wang, L.; et al. CRISPR-Cas9-mediated gene editing of the BCL11A enhancer for pediatric β(0)/β(0) transfusion-dependent β-thalassemia. Nat. Med. 2022, 28, 1573–1580. [Google Scholar] [CrossRef]
- Ottaviano, G.; Georgiadis, C.; Gkazi, S.A.; Syed, F.; Zhan, H.; Etuk, A.; Preece, R.; Chu, J.; Kubat, A.; Adams, S.; et al. Phase 1 clinical trial of CRISPR-engineered CAR19 universal T cells for treatment of children with refractory B cell leukemia. Sci. Transl. Med. 2022, 14, eabq3010. [Google Scholar] [CrossRef]
- Sharma, A.; Boelens, J.J.; Cancio, M.; Hankins, J.S.; Bhad, P.; Azizy, M.; Lewandowski, A.; Zhao, X.; Chitnis, S.; Peddinti, R.; et al. CRISPR-Cas9 Editing of the HBG1 and HBG2 Promoters to Treat Sickle Cell Disease. N. Engl. J. Med. 2023, 389, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, S.; Kesari, K.K.; Rachamalla, M.; Mani, S.; Ashraf, G.M.; Jha, S.K.; Kumar, P.; Ambasta, R.K.; Dureja, H.; Devkota, H.P.; et al. CRISPR/Cas9 gene editing: New hope for Alzheimer’s disease therapeutics. J. Adv. Res. 2022, 40, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Harish, V.; Mohd, S.; Singh, S.K.; Tewari, D.; Tatiparthi, R.; Harshita; Vishwas, S.; Sutrapu, S.; Dua, K.; et al. Role of CRISPR/Cas9 in the treatment of Duchenne muscular dystrophy and its delivery strategies. Life Sci. 2023, 330, 122003. [Google Scholar] [CrossRef] [PubMed]
- Pavathuparambil Abdul Manaph, N.; Al-Hawwas, M.; Liu, L.; Liu, D.; Hayball, J.; Zhou, X.F. Facial vein injection of human cells in severe combined immunodeficiency (SCID) neonatal mice. MethodsX 2018, 5, 1281–1286. [Google Scholar] [CrossRef]
- Wang, D.; Mou, H.; Li, S.; Li, Y.; Hough, S.; Tran, K.; Li, J.; Yin, H.; Anderson, D.G.; Sontheimer, E.J.; et al. Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses. Hum. Gene Ther. 2015, 26, 432–442. [Google Scholar] [CrossRef]
- Hinderer, C.; Bell, P.; Louboutin, J.P.; Zhu, Y.; Yu, H.; Lin, G.; Choa, R.; Gurda, B.L.; Bagel, J.; O’Donnell, P.; et al. Neonatal Systemic AAV Induces Tolerance to CNS Gene Therapy in MPS I Dogs and Nonhuman Primates. Mol. Ther. 2015, 23, 1298–1307. [Google Scholar] [CrossRef]
- Dorschner, R.A.; Lin, K.H.; Murakami, M.; Gallo, R.L. Neonatal skin in mice and humans expresses increased levels of antimicrobial peptides: Innate immunity during development of the adaptive response. Pediatr. Res. 2003, 53, 566–572. [Google Scholar] [CrossRef] [PubMed]
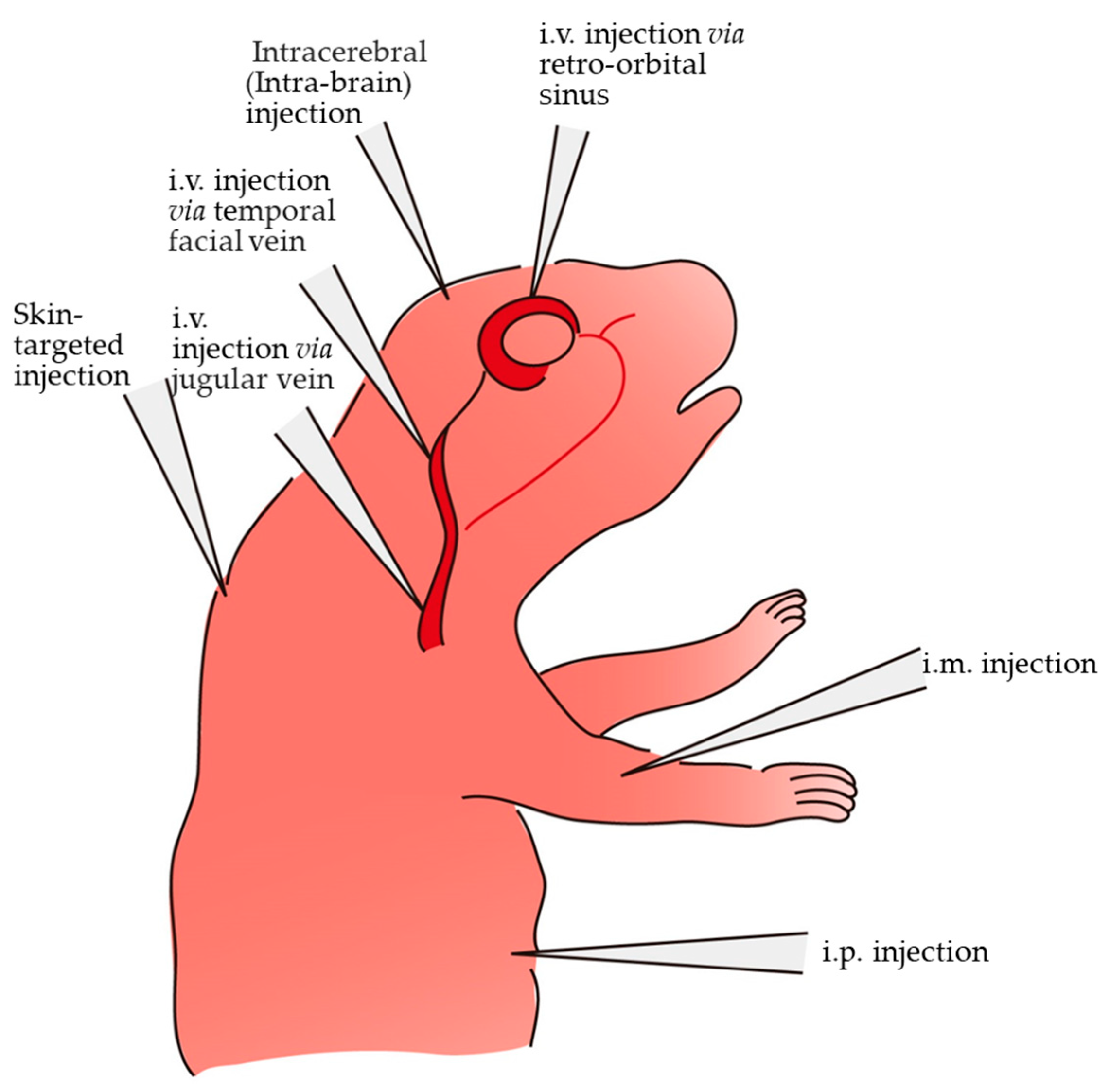

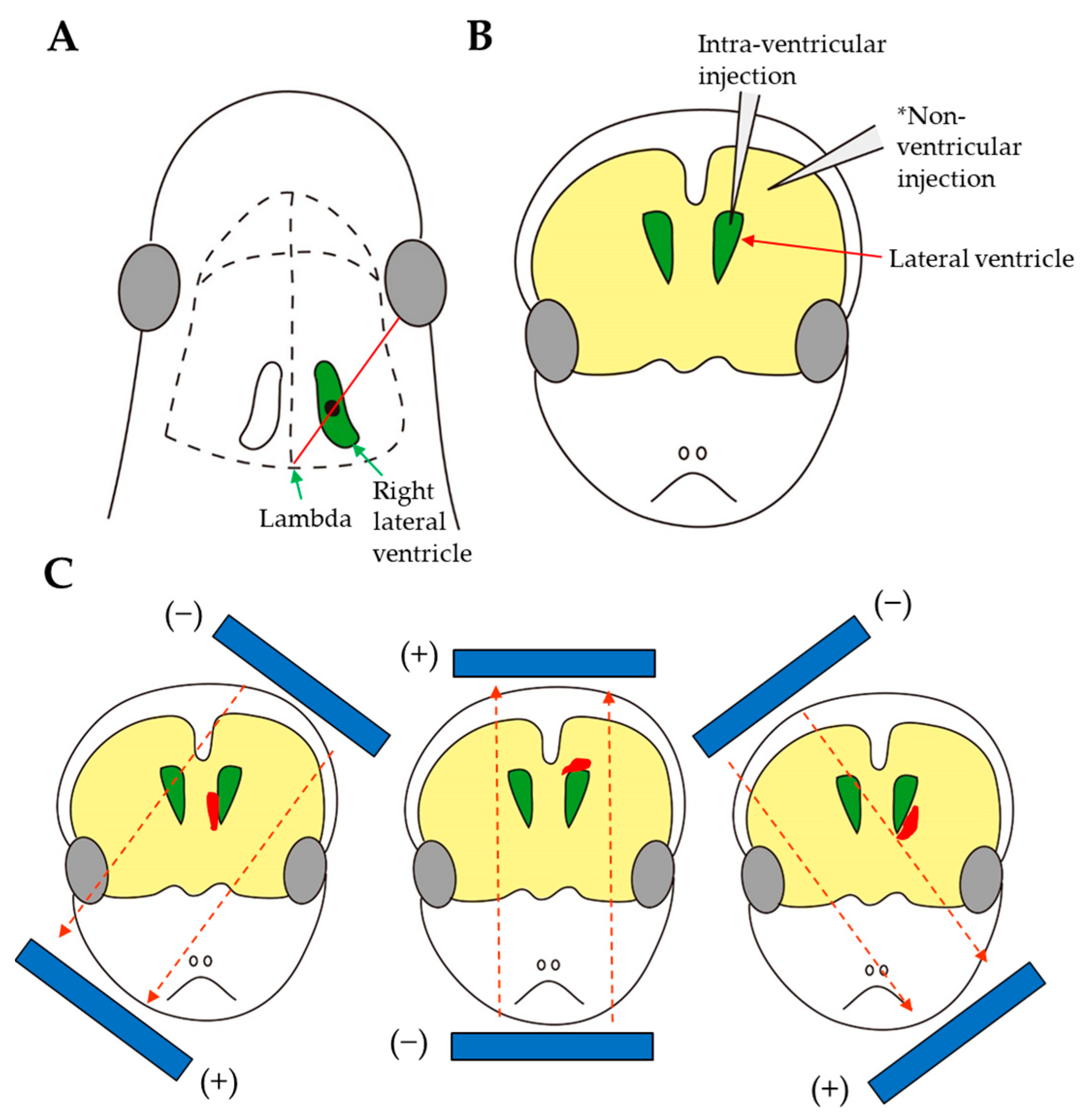
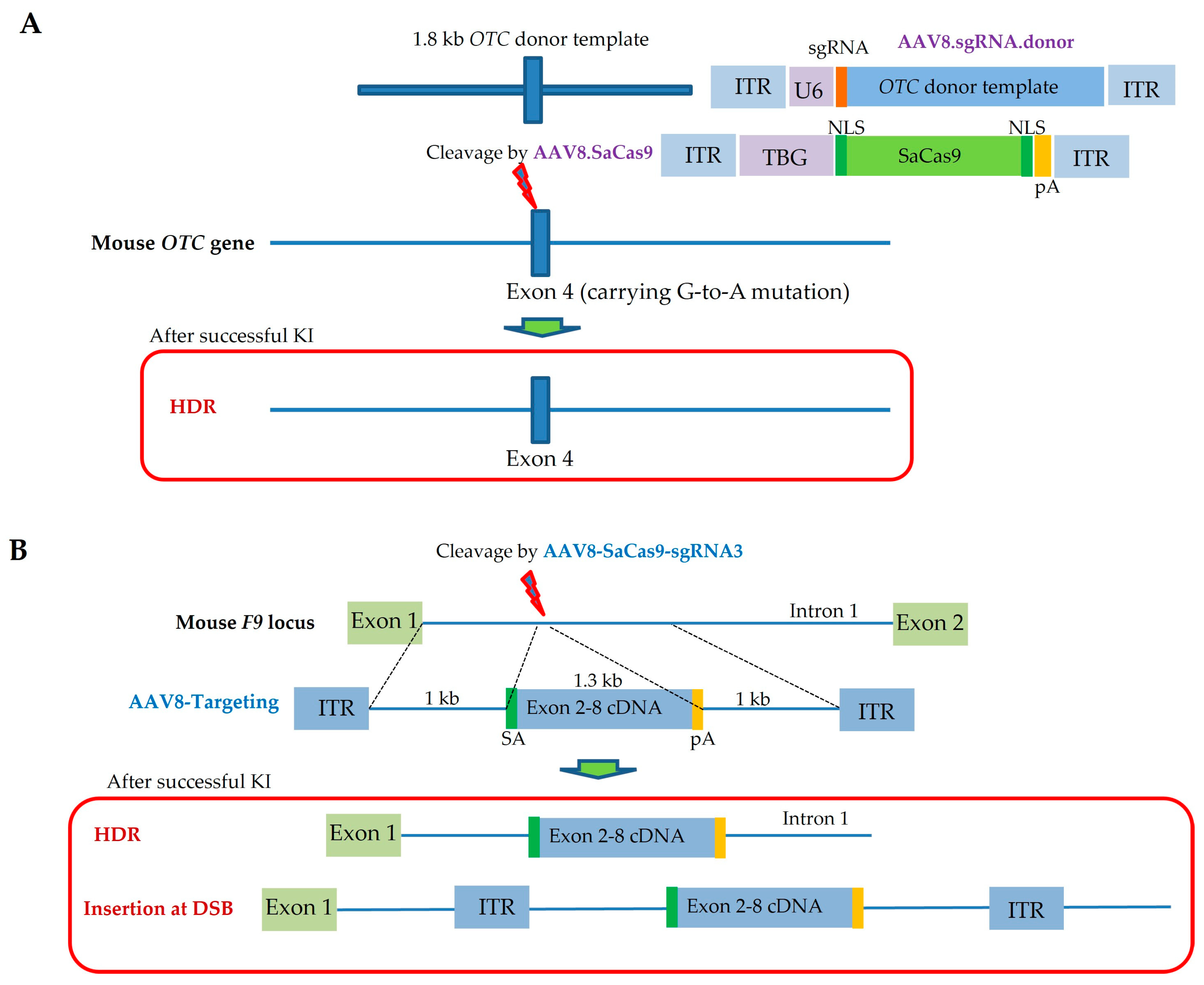
| Route and Method of Gene Delivery | Type of Nucleic Acid(s) Introduced | Outcome | Target Gene or Gene of Interest (GOI) Introduced | References |
|---|---|---|---|---|
| Facial-vein-mediated injection (also called i.v. injection) | Recombinant adeno-associated virus (rAAV) | Single injection of rAAV carrying human beta-glucuronidase (GUSB) cDNA into mucopolysaccharidosis type VII (MPS VII) model mice resulted in the expression of GUSB in most organs for 16 weeks and decreased or complete prevention of lysosomal storage; particularly, cells in the CNS were cleared of disease, suggesting viral infection beyond the blood–brain barrier (BBB) | GUSB | Daly et al., 1999 [66] |
| Facial-vein-mediated injection (also called i.v. injection) | Retroviral vector (RV) | Neonatal injection of RV carrying canine GUSB cDNA into dogs with MPS VII resulted in clonal expansion of hepatocytes and secretion of active GUSB into serum, along with clinical improvements; this is the first successful application of gene therapy in preventing a lysosomal storage disease in a large animal | GUSB | Ponder et al. 2002 [67] |
| Facial-vein-mediated injection (also called i.v. injection) | RV | Neonatal injection of RV carrying canine GUSB cDNA into MPS VII mice resulted in transduction of 6 to 35% of hepatocytes, which secreted GUSB into the blood; the secreted enzyme was taken up by other tissues and reduced the histopathological evidence of lysosomal storage in the liver, spleen, kidneys, small intestine, neurons, and glial cells | GUSB | Xu et al., 2002 [68] |
| Facial-vein-mediated injection (also called i.v. injection) | Adenoviral vector (AD) | Neonatal single injection of AD carrying human GUSB cDNA into MPS VII mice resulted in the recovery of more than 20% of GUSB activity in the brain, leading to the prevention of lysosomal storage, and lacking characteristic facial skeletal deformities associated with bone deformity, mental retardation, corneal clouding, and retinal degeneration | GUSB | Kamata et al., 2003 [69] |
| Facial-vein-mediated injection (also called i.v. injection) | rAAV2 | Neonatal i.v. injection of rAAV2 carrying human iduronidase (IDUA) cDNA was performed using newborns with mucopolysaccharidosis type I (MPS I) to determine the potential for the gene delivery approach; high levels of IDUA activity were present in the treated animals and persisted for the 5-month duration of the study; successful correction of metabolic, craniofacial, and neurological abnormalities in MPS I mice was achieved | IDUA | Hartung et al., 2004 [70] |
| Facial-vein-mediated injection (also called i.v. injection) | Lentiviral vector (LV) | Single injection of LV into MPS I model mice resulted in the expression of IDUA activity, decreased glycosaminoglycan (GAG) storage, prevention of skeletal abnormalities, a more normal gross appearance, and improved survival; most strikingly, transduction of neurons at high levels was prominent | IDUA | Kobayashi et al., 2005 [71] |
| Peripheral (tail vein) i.v. injection | rAAV2/8 rAAV2/9 | Both rAAV2/8 and rAAV2/9 vectors caused substantial transduction in the heart, skeletal muscle, and pancreas; importantly, rAAV2/9 transduced myocardium 5–10-fold higher than rAAV2/8, resulting in over 80% cardiomyocyte transduction, and suggesting that rAAV2/9 is superior to rAAV2/8 specifically for cardiac gene delivery | F9 nlslacZ lacZ | Inagaki et al., 2006 [72] |
| Facial-vein-mediated injection (also called i.v. injection) | Sleeping Beauty (SB) transposon | Newborn mice were first i.p. injected with mannitol, and 1 h later luciferase-transposon DNA conjugated with polyethylenimine (PEI) was injected into the lateral ventricle; the treated animals showed significantly higher luciferase expression one month after gene delivery, suggesting chromosomal integration of the luciferase transgene and the usefulness of mannitol pretreatment combined with transposon-mediated gene transfer for long-term gene expression in the mammalian brain | Luciferase | Demorest et al., 2006 [58] |
| Facial-vein-mediated injection (also called i.v. injection) | rAAV2/1 rAAV2/8 rAAV2/9 | When comparing rAAV2/1 with rAAV2/8 and the newer rAAV2/9 vectors for the transduction of skeletal muscle, both rAAV2/8 and rAAV2/9 were able to transduce myocardium at approximately 20- and 200-fold (respectively) higher levels than rAAV2/1; thus, rAAV2/9 is more readily able to cross the vasculature and leads to preferential cardiac transduction in vivo, efficiently transducing cardiac tissue | lacZ | Pacak et al., 2006 [73] |
| Facial-vein- or external jugular-vein-mediated injection (also called i.v. injection) | - | Two techniques (injection via the external jugular vein, and via the superficial temporal vein) for i.v. injection in neonatal mice from birth to 6 days of age were described; both allow the injection of a variety of substances, including cells, medications, toxins, and cytokines, and both permit serial injections | - | Kienstra et al., 2007 [60] |
| Facial-vein-mediated or intraperitoneal (i.p.) injection | rAAV8 | Neonatal administration of rAAV8 by i.v. or i.p. injection was performed to test whether lower motor neurons (LMNs) are transduced, indicating that dorsal root ganglion transduction occurs across all timepoints and injection routes | GFP | Foust et al. 2008 [74] |
| Facial-vein-mediated injection (also called i.v. injection) | Helper-dependent adenoviral vector (AD) | Intravenous (i.v.) injection of AV expressing human factor 8 (F8) to neonatal hemophilia A (HA)-knockout (KO) mice resulted in the production of high levels of F8, and its expression lasted until >1 y of age, which is associated with correction of HA and tolerance to human F8 | F8 | Hu et al., 2011 [75] |
| Facial-vein-mediated injection (also called i.v. injection) | rAAV2/9 | Neonatal administration of rAAV2/9 produced global delivery to the central (i.e., brain, spinal cord, and all layers of the retina) and peripheral nervous system (i.e., myenteric plexus and innervating nerves), which can provide a therapeutic strategy for the treatment of early lethal genetic diseases, such as Gaucher disease | GFP | Rahim et al. 2011 [76] |
| Facial-vein-mediated injection (also called i.v. injection) | rAAVrh10 | Neonatal co-injection of two AAVrh10s (in which one had a heavy chain and the other a light chain for F8) into mice with HA resulted in long-term correction and avoidance of immune responses in the AAV-F8-treated mice | F8 | Hu and Lipshutz 2012 [77] |
| i.v.-mediated injection | rAAV9 carrying mutating capsid surface tyrosines | A double-tyrosine mutant of rAAV9 significantly enhanced gene delivery to the CNS and retina, and that gene expression could be restricted to rod photoreceptor cells by incorporating a rhodopsin promoter, which may provide a new methodology for the development of retinal gene therapies or the creation of animal models of neurodegenerative diseases | GFP | Dalkara et al., 2012 [78] |
| Facial-vein-mediated injection (also called i.v. injection) | Self-complementary (sc) rAAV9 | Neonatal i.v. injection of rAAV9 resulted in gene transfer to all layers of the retina (including retinal pigment epithelium cells, photoreceptors, bipolar cells, Müller cells, and retinal ganglion cells) in adult mice; in particular, the cells on the inner side of the retina were transduced with the highest efficiency, suggesting that this vector serotype is able to cross mature blood–eye barriers | GFP | Bemelmans et al., 2013 [79] |
| Facial-vein-mediated injection (also called i.v. injection) | rAAV2/1 rAAV2/5 rAAV2/6 rAAV2/8 rAAV2/9 | Intravenous (i.v.) injection of AAV9 resulted in the transduction of 25–57% of enteric nervous system (ENS) myenteric neurons; AAV9 transduction in enteric glia was very low compared to CNS astrocytes; AAV8 resulted in comparable transduction in neonatal mice to AAV9, while AAV1, -5, and -6 were less efficient, suggesting that systemic AAV9 has a high affinity for peripheral neural tissue and will be useful for future therapeutic development and basic studies of the ENS | GFP | Gombash et al., 2014 [80] |
| Facial-vein-mediated injection (also called i.v. injection) | Single-stranded (ss) and scAAV9 | Extensive GFP expression was observed in organs throughout the body, with the epithelial and muscle cells being particularly well transduced, suggesting that AAV9 can potentially be used for clinical systemic gene therapy protocols | GFP | Mattar et al., 2015 [81] |
| Facial-vein-mediated injection (also called i.v. injection) | rAAV2/8 rAAV2/9 | Both rAAV2/8 and rAAV2/9 showed equal potential in transducing the ENS, with 25–30% of the neurons expressing EGFP; all enteric neuron subtypes, but not glia, expressed the reporter protein; these findings will be used for novel preclinical applications aimed at manipulating and imaging the ENS in the short term, and for gene therapy in the longer term | EGFP | Buckinx et al., 2016 [82] |
| Facial-vein-mediated injection (also called i.v. injection) | rAAV2/9 | Resulted in binaural transduction of inner hair cells, spiral ganglion neurons, and vestibular hair cells; transduction efficiency increased in a dose-dependent manner; inner hair cells were transduced in an apex-to-base gradient, with transduction reaching 96% in the apical turn; intravenous delivery of rAAV2/9 represents a novel and atraumatic technique for inner-ear transgene delivery in early postnatal mice | EGFP | Shibata et al., 2017 [83] |
| Facial-vein-mediated injection (also called i.v. injection) or intracerebral injection | Foamy virus (PFV) | Systemic PFV vector delivery to neonatal mice gave transgene expression in the heart, xiphisternum, liver, pancreas, and gut, whereas intracranial administration produced brain expression; transgene expression was highly localized to the hippocampal architecture, despite vector delivery being administered to the lateral ventricle; PFV can be used for neonatal gene delivery targeted to hippocampal neurons, for gene therapy of neurological disorders | EGFP | Counsell et al., 2018 [84] |
| Facial-vein-mediated injection (also called i.v. injection) | AAVs | Intravenous (i.v.) administration can achieve widespread delivery of rAAVs to the CNS, which are considered to be promising therapeutic tools for treating genetic defects of the CNS, due to their excellent safety profile and ability to cross the BBB | - | Gessler et al., 2019 [85] |
| Facial-vein-mediated injection (also called i.v. injection) | rAAV8 | Intravenous (i.v.) administration can achieve widespread gene expression in the central and peripheral nervous system, liver, kidneys, and skeletal muscle; i.v. injection of rAAV8 carrying a spleen focus-forming virus (SFFV) promoter and nuclear factor-κB (NF-κB) binding sequence for bioluminescence and biosensor evaluation resulted in a 10-fold increase in luc expression after single administration of lipopolysaccharide (LPS), and whole-body bioluminescence persisted for up to 240 days | GFP luc | Karda et al., 2020 [6] |
| Intracerebral injection | rAAV2 | Gene transfer throughout the CNS was achieved without germline transmission, and gene expression lasted for at least 1 year | GUSB | Passini and Wolfe 2001 [86] |
| Intracerebral injection | rAAV1 rAAV2 rAAV5 | The 0gene delivery pattern after neonatal intracerebral injection was assessed using different rAAV serotypes; consequently, rAAV5 showed very limited brain transduction, even though it has different transduction patterns than rAAV2; in contrast, rAAV1 vectors showed robust widespread transduction; in the majority of structures, rAAV1 transduced many more cells than rAAV2 | GUSB | Passini et al., 2003 [87] |
| Intracerebral injection and subsequent in vivo electroporation (EP) | Plasmid DNA | Intraventricular injection of DNA followed by EP induced strong expression of transgenes in the radial glia, neuronal precursors and neurons of the olfactory system; overexpression of the cell-cycle inhibitor p21 resulted in interference with the proliferation of neural stem cells | hNCAM-140 p21 | Boutin et al., 2008 [88] |
| Intracerebral injection and subsequent in vivo EP | Plasmid DNA | Neonatal intracerebral injection was performed to determine how neuroprogenitors in the subventricular zone (SVZ) give rise to neuroblasts that migrate along the rostral migratory stream (RMS); labeling was found in all classes of interneurons in the olfactory bulb (OB), persisted to adulthood, and had no adverse effects | EGFP | Chesler et al., 2008 [89] |
| Intracerebral injection | rAAV8 LV | Region-specific recombination of a “stop-floxed” Rosa26 reporter allele was achieved upon targeted injection of rAAV vectors expressing Cre-recombinase (Cre); utilizing LV, efficient transduction of neuroprogenitors in the SVZ occurred and, as a result, approximately 20% of labeled migrating neuroblasts were generated along the RMS into the OB | Cre | Pilpel et al., 2009 [90] |
| Intracerebral injection and subsequent in vivo EP | Plasmid DNA | Neonatal non-ventricular injection into deep cortical layers or the striatum region, followed by EP, was performed to create a local expression pattern in the area of interest and in situ transfection of non-migratory cell types, e.g., cortical astrocytes, which may be used for two-photon in vivo imaging | EGFP | Molotkov et al., 2010 [91] |
| Intracerebral injection and subsequent in vivo EP | Plasmid DNA | Improvement of the EP technique allowing for targeted transgene delivery to specific walls of the lateral ventricle, accurately and reproducibly, was performed to trace perinatal neural stem cells, which successfully enabled fate mapping of the progeny of RGCs | EGFP | Fernandez et al., 2011 [92] |
| Intracerebral injection | rAAV8 | Neonatal intra-cerebral injection of viral vectors into the hindbrain enables postnatal dendritic maturation in cerebellar Purkinje neurons for in vivo imaging of mature Purkinje neurons at a resolution sufficient for complete analytical reconstruction | YFP tdTomato iCre tTA | Kim et al., 2013 [93] |
| Intracerebral injection | rAAV2/1 rAAV2/2 rAAV2/5 rAAV2/7 rAAV2/8 rAAV2/9 | The effects of the timing of the injection on the rAAV tropism and biodistribution of six commonly used rAAVs after neonatal intracerebral gene delivery was assessed; consequently, rAAV2/8 and 2/9 resulted in the most widespread distribution in the brain; injection on neonatal day P0 resulted in mostly neuronal transduction, whereas administration at later periods of development resulted in more non-neuronal transduction; rAAV2/5 showed widespread transduction of astrocytes irrespective of the time of injection; none of the serotypes tested showed any microglial transduction | EGFP | Chakrabarty et al., 2013 [94] |
| Intracerebral injection | LV | Neonatal intracerebral injection of LV carrying the β-galactocerebrosidase (GALC) gene in the external capsule of twitcher mice, a severe model of globoid cell leukodystrophy, resulted in the restoration of GALC activity in the whole CNS of treated mice as early as 8 days post-injection; this approach will be useful for neonatal LV-mediated intracerebral gene therapy | GALC GFP | Lattanzi et al., 2014 [95] |
| Intracerebral injection and subsequent in vivo EP | Plasmid DNA | Neonatal intracerebral injection of plasmid and subsequent in vivo EP were performed to analyze the RMS and postnatal OB neurogenesis; consequently, GFP-positive cells in the dentate gyrus (DG) were observed to extend branched dendrites and long axons into the molecular layer and the hilus; the expression of GFP in these neurons was sustained for at least 9 months | GFP | Ito et al., 2014 [96] |
| Intracerebral injection | rAAV8 | Intraventricular injection of rAAV8 within the first 24 h after birth resulted in widespread transduction of neurons throughout the brain; expression began within days of the injection and persisted for the lifetime of the animal; this versatile manipulation enables studies ranging from early postnatal brain development to aging and degeneration in the adult | YFP tdTomato | Kim et al., 2014 [97] |
| Intracerebral injection | rAAV2/1 rAAVDJ8 rAAV9 | Tropism of rAAV2/1, rAAVDJ8, and rAAV9 throughout different brain regions and cell types was assessed through neonatal intracerebral injection; rAAV2/1 infections were more prevalent in the cortical layers but penetrated to the midbrain less than rAAVDJ8 and rAAV9; rAAVDJ8 displayed more tropism in astrocytes compared to rAAV9 in the substantia nigra (SN) region | GFP | Hammond et al., 2017 [98] |
| Intracerebral injection | rAAV9 AAV-PHP.B AAV-PHP.eB | Neonatal intra-brain injection of rAAV9, AAV-PHP.B, and AAV-PHP.eB carrying CRISPR reagents was used to examine cell-type-specific gene ablation; consequently, AAV-PHP.B variants exhibited marked disruption of neuron-related genes, but only modest disruption of the astrocyte- or oligodendrocyte-specific genes was observed by all three AAV variants, which could facilitate profiling of AAV cellular tropism in the murine CNS | NeuN GFAP MOG | Torregrosa et al., 2021 [99] |
| Intraperitoneal (i.p.) injection | rAAV1 rAAV2 rAAV5 rAAV6 rAAV7 rAAV8 | Neonatal single injection of various serotypes of rAAVs via i.p. or i.v. routes was performed; rAAV8 was the most efficient vector for crossing the blood vessel barrier to attain systemic gene transfer in both skeletal and cardiac muscles; rAAV1 and rAAV6, which demonstrated robust infection in skeletal muscle cells, were less effective in crossing the blood vessel barrier; gene expression persisted in the muscle and heart but diminished in the liver, which showed rapid cell division; this approach will be useful for muscle-directed systemic gene therapy | GFP | Wang et al., 2005 [100] |
| i.p. injection | Single-stranded AAV9 (ssAAV9) carrying short hairpin RNA (shRNA) | Neonatal i.p. injection of shRNA-ssAAV9 resulted in ~80% reduction in target mRNA in the DRG, along with 75% suppression of the protein; the suppression effect lasted for more than 3 months; this approach may be helpful for elucidating the mechanisms of pain and sensory ganglionopathies | Sod1 | Machida et al., 2013 [64] |
| i.p. and i.v. injection | rAAV8 | Neonatal i.p. or i.v. injection of rAAV8 through entry into the nervous system was performed to target lower motor neurons (LMNs) for future gene therapy against spinal muscular atrophy and amyotrophic lateral sclerosis; consequently, spinal cords were positively transduced; furthermore, fibers in the dorsal horns and columns were labeled, indicating dorsal root ganglion transduction with these techniques | GFP | Foust et al., 2008 [74] |
| i.v. injection via the retro-orbital sinus | RV | Neonatal retro-orbital-sinus-mediated injection of an RV vector carrying F8 cDNA into F8-deficient mice resulted in successful correction of HA | F8 | Vanden Driessche et al., 1999 [101] |
| Injection into the subretinal space and subsequent in vivo EP | Plasmid DNA | Neonatal injection of plasmid DNA into the subretinal space of mice, and subsequent in vivo EP, resulted in successful transfection of retinal cells; transfection of plasmid DNA carrying RNAi resulted in a knockdown phenotype of a target gene’s expression, which was similar to the phenotype shown in KO mice | GFP | Matsuda and Cepko 2004 [102] |
| Injection into the subretinal space and subsequent in vivo EP | Plasmid DNA | Conditional temporal and spatial regulation of gene expression in the retinas of postnatal rats was assessed using Cre/loxP-mediated inducible expression vectors and 4-hydroxytamoxifen treatment, which enables conditional activation of Cre recombinase; transgene expression was successfully induced in a cell-type- and time-specific manner | CreERT2 DsRed GFP | Matsuda and Cepko 2007 [103] |
| Hydrodynamics-based gene delivery (HGD) via the retro-orbital sinus | Plasmid DNA | High levels of gene expression in the hepatocytes of neonatal mice were achieved, which will provide a way to perform gene delivery to animals that are difficult to inject via the tail vein; it will also be beneficial for exploring gene function and treating genetic disease | Luciferase | Yan et al., 2012 [104] |
| i.v. injection via the retro-orbital sinus | First-generation AD vector | The highest AD vector genome copy numbers and transgene expression were found in the neonatal liver; the neonatal heart exhibited the second-highest levels of transgene expression among the organs examined; no apparent hepatotoxicity was observed in neonatal mice; these findings may be helpful for performing gene therapy using AD vectors in neonates | Luciferase | Iizuka et al., 2015 [105] |
| i.v. injection via the retro-orbital sinus | rAAV9 | Showing the benefits of i.v. injection via the retro-orbital sinus as an effective route of rAAV9-mediated gene delivery in neonates | Cre | Prabhakar et al., 2021 [59] |
| Intramuscular (i.m.) injection | rAAV | Neonatal i.m. injection of an rAAV vector carrying human GUSB cDNA into MPS VII mice resulted in high-level intramuscular GUSB expression as early as 2 weeks of age, and for at least 16 weeks; GUSB activity was detected in both the liver and spleen at later timepoints, indicating that rAAV vectors can successfully infect neonatal muscle and persist through the rapid growth phase following birth | GUSB | Daly et al., 1999 [106] |
| i.m. injection | Plasmid DNA | Neonatal intramuscular injection of plasmid DNA into hypercholesterolemic mice (ApoE KO mice) resulted in a reduction in the incidence of severe hypercholesterolemia; notably, when naked DNA was administrated early, no immune response was generated against the human APOE3, allowing for repeated administrations; this approach will be useful for treating many genetic childhood diseases where early administration is required to prevent developmental damage | APOE3 | Signori et al., 2007 [107] |
| Injection into the skin and subsequent in vivo EP | Plasmid DNA | Subcutaneous injection of two plasmid DNAs (one with a neomycin-resistance gene (neo) and the other with an immortalizing gene) into the skin cells of newborn mice (at P1-3), followed by subsequent in vivo EP, resulted in the generation of stably transfected fibroblasts | neo | Titomirov et al., 1991 [65] |
| Route and Method of Gene Delivery | Type of Nucleic Acid(s) Introduced | Outcome | Target Gene or Gene of Interest (GOI) Introduced | References |
|---|---|---|---|---|
| Facial-vein-mediated injection (also called i.v. injection) | Dual rAAV (one with the Cas9 gene and the other with gRNA/donor DNA) | Neonatal i.v. injection of dual AAVs into OTC-deficient mice was performed to correct metabolic liver disease caused by lethal hyperammonemia; consequently, 10% of hepatocytes were restored, and reduced survival after feeding with a chow diet was avoided | OTC | Yang et al., 2016 [118] |
| Facial-vein-mediated injection (also called i.v. injection) | Dual rAAV8 (one with SaCas9/sgRNA and the other with donor DNA) | Neonatal i.v. injection of an SaCas9/sgRNA-expressing rAAV8 vector into wild-type mice resulted in mutations in the F9 gene in hepatocytes, sufficiently developing HB; also, it was possible to generate HDR-based correction of the mutated F9 gene in HB model mice; this approach will provide a flexible approach to induce DSB-mediated mutations in target genes in hepatocytes, and also to cure congenital hemorrhagic disease | F9 | Ohmori et al., 2017 [119] |
| Facial-vein-mediated injection (also called i.v. injection) | Dual rAAV8 (one with SaCas9 and the other with gRNA/donor DNA containing F9 cDNA carrying a hyperactive F9 Padua mutation | Neonatal i.v. injection of dual rAAVs into F9 KO mice resulted in expression of F9 at a normal level over 8 months, suggesting the use of the CRISPR approach to achieve lifelong expression of therapeutic proteins | F9 | Wang et al., 2019 [120] |
| Facial-vein-mediated injection (also called i.v. injection) | rAAV8 carrying donor DNA and rAAV8 carrying SaCas9 and sgRNA | Neonatal i.v. injection of dual rAAV8 vectors into F9 KO mice resulted in the stable expression of human F9, reaching up to 150% of the human levels, showing a clotting capacity comparable to wild-type animals, and demonstrating the rescue of the disease phenotype | F9 | Lisjak et al., 2022 [121] |
| Intracerebral injection | AAV-PHP.B carrying sgRNA | Neonatal intracerebral injection of AAV-PHP.B carrying sgRNA into Cas9 mice resulted in a 99.4% rate of biallelic indels in the transduced cells, leading to a more than 70% reduction in the quantity of total NeuN proteins in the cortex, hippocampus, and spinal cord | NeuN | Hana et al., 2021 [122] |
| i.p. injection | Dual rAAV9 carrying CRISPR components | To correct Duchenne muscular dystrophy (DMD) by skipping mutant dystrophin exons in postnatal muscle tissue in vivo, rAAV9 carrying CRISPR/Cas9 components was i.p. injected into neonatal mdx mice (P1), a model of DMD; as a result, dystrophin protein expression was observed in cardiac and skeletal muscle to varying degrees, and expression increased from 3 to 12 weeks after te injection | Dystrophin | Long et al., 2016 [123] |
| i.m. injection | Dual rAAV8 carrying CRISPR components | Neonatal i.m. injection of rAAV8 carrying CRISPR components into an mdx mouse model of DMD was performed to improve muscle function; removal of mutated exon 23 from the dystrophin gene resulted in expression of the modified dystrophin gene, partial recovery of functional dystrophin protein in skeletal myofibers and cardiac muscle, and significant enhancement of muscle force, suggesting that this approach is useful as a potential therapy to treat DMD | Dystrophin | Nelson et al., 2016 [124] |
| i.m. injection | Dual rAAV9 carrying CRISPR components | To test whether the removal of one or more exons from the mutated transcript in DMD-related dystrophin could produce an in-frame mRNA and a truncated but still functional protein, dual rAAVs carrying CRISPR/Cas9 components were subjected to i.m. injection into neonatal mdx model mice; as a result, in the treated mice, restoration of dystrophin expression was observed in myofibers, cardiomyocytes, and muscle stem cells | Dystrophin | Tabebordbar et al., 2016 [125] |
| Procedure or Property | Newborns | Adults |
|---|---|---|
| Anesthesia | Relatively easy, because newborn pups are prone to hypothermia after being placed on a chilled paper towel on top of ice for 30–60 s; therefore, there is no need for an anesthetic agent, but the surgical procedure must be performed within ~30 min | Requires administration of an anesthetic agent or isoflurane; surgical procedure can be performed for more than 1 h |
| Immunological tolerance | The immune system of newborn pups at P0 to P7 (whose condition is also called “immune immaturity”) is not well established; therefore, it is possible to transplant xenogenic cells such as human-derived cells | The immune system is already tightly established; therefore, it is impossible to transplant xenogenic cells such as human-derived cells |
| Intravenous (i.v.) injection | Relatively easy; a small amount of solution (up to 50 μL) can be injected via the facial vein; therefore, the reagents required can be reduced at a minimal level; however, the appearance of the facial vein is temporal, as it disappears up to 6 days after birth; tail vein injection becomes possible from P15 onward; therefore, it may be theoretically impossible to perform i.v. injection during the periods of P7 to P14 | Relatively easy; relatively large amounts of solution (~1 mL) are generally required; therefore, the reagents used appear to be costly |
| Gene delivery beyond the blood–brain barrier (BBB) after i.v. injection | Due to the incomplete development of the BBB at this stage, small molecules such as recombinant adeno-associated viruses (rAAVs) injected exogenously can easily penetrate into cells of the central nervous system (CNS) via the BBB | Large molecules cannot be transferred inside a brain, since the established BBB does not allow penetration beyond the BBB |
| Intracerebral injection | Relatively easy; requires peeling of the skin and skull with fine forceps prior to the insertion of a capillary pipette under a dissecting microscope | It requires drilling of a skull prior to the insertion of the capillary pipette, which is laborious and time-consuming |
| Retro-orbital-sinus-mediated injection | Relatively easy | Relatively easy; repeated treatment is possible |
| Manipulation of internal organ | Relatively difficult, because organs are too small and fragile; therefore, their handling must be carried out under observation using a dissecting microscope, requiring special caution upon surgical treatment and skin closure after surgery, because the skin itself is thin and labile | Relatively easy; manipulation of internal organs can be performed during their exposure onto the skin; also, skin closure is easy after surgery |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakamura, S.; Morohoshi, K.; Inada, E.; Sato, Y.; Watanabe, S.; Saitoh, I.; Sato, M. Recent Advances in In Vivo Somatic Cell Gene Modification in Newborn Pups. Int. J. Mol. Sci. 2023, 24, 15301. https://doi.org/10.3390/ijms242015301
Nakamura S, Morohoshi K, Inada E, Sato Y, Watanabe S, Saitoh I, Sato M. Recent Advances in In Vivo Somatic Cell Gene Modification in Newborn Pups. International Journal of Molecular Sciences. 2023; 24(20):15301. https://doi.org/10.3390/ijms242015301
Chicago/Turabian StyleNakamura, Shingo, Kazunori Morohoshi, Emi Inada, Yoko Sato, Satoshi Watanabe, Issei Saitoh, and Masahiro Sato. 2023. "Recent Advances in In Vivo Somatic Cell Gene Modification in Newborn Pups" International Journal of Molecular Sciences 24, no. 20: 15301. https://doi.org/10.3390/ijms242015301
APA StyleNakamura, S., Morohoshi, K., Inada, E., Sato, Y., Watanabe, S., Saitoh, I., & Sato, M. (2023). Recent Advances in In Vivo Somatic Cell Gene Modification in Newborn Pups. International Journal of Molecular Sciences, 24(20), 15301. https://doi.org/10.3390/ijms242015301







