Histone Methyltransferases G9a/Ehmt2 and GLP/Ehmt1 Are Associated with Cell Viability and Poorer Prognosis in Neuroblastoma and Ewing Sarcoma
Abstract
:1. Introduction
2. Results
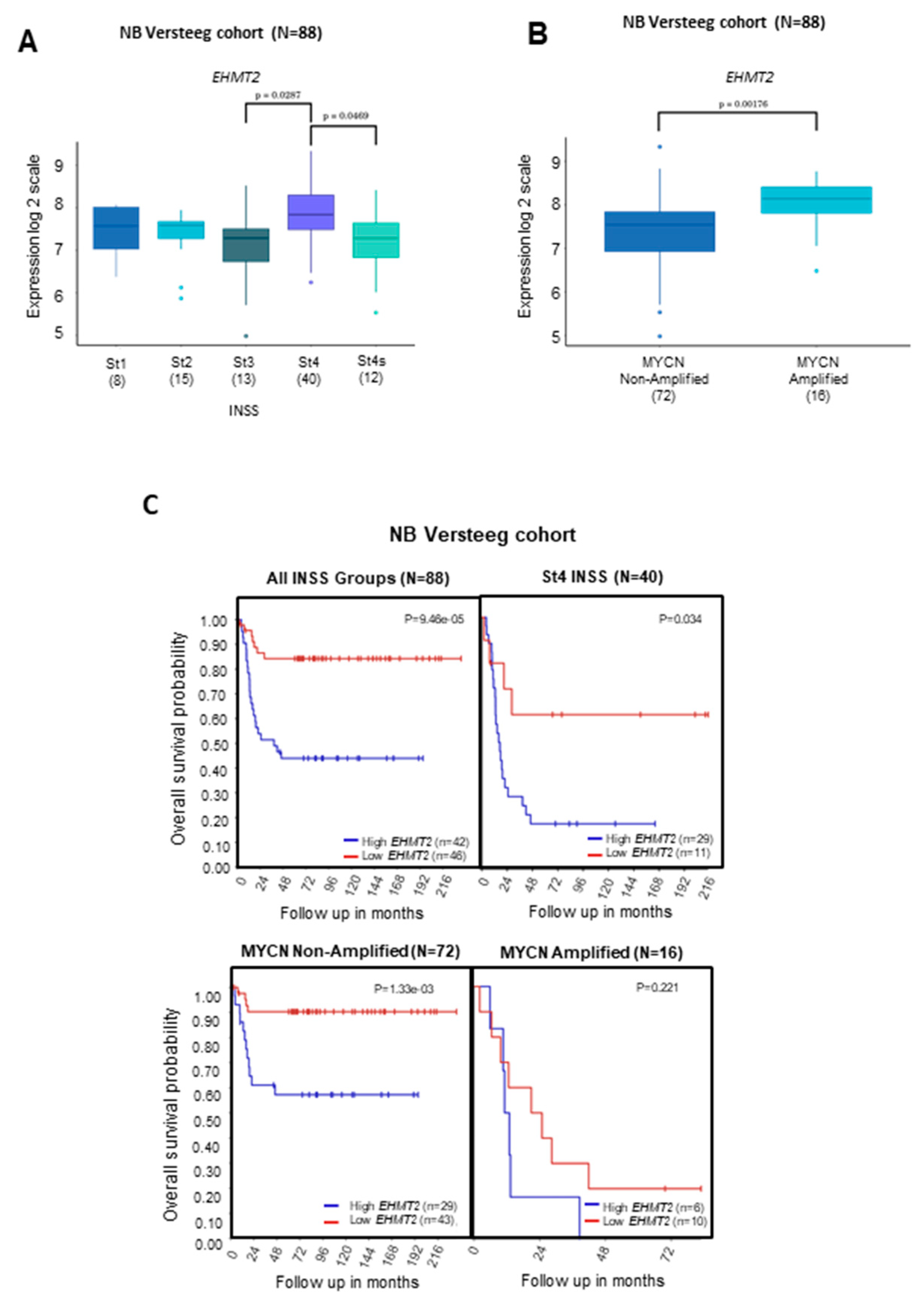
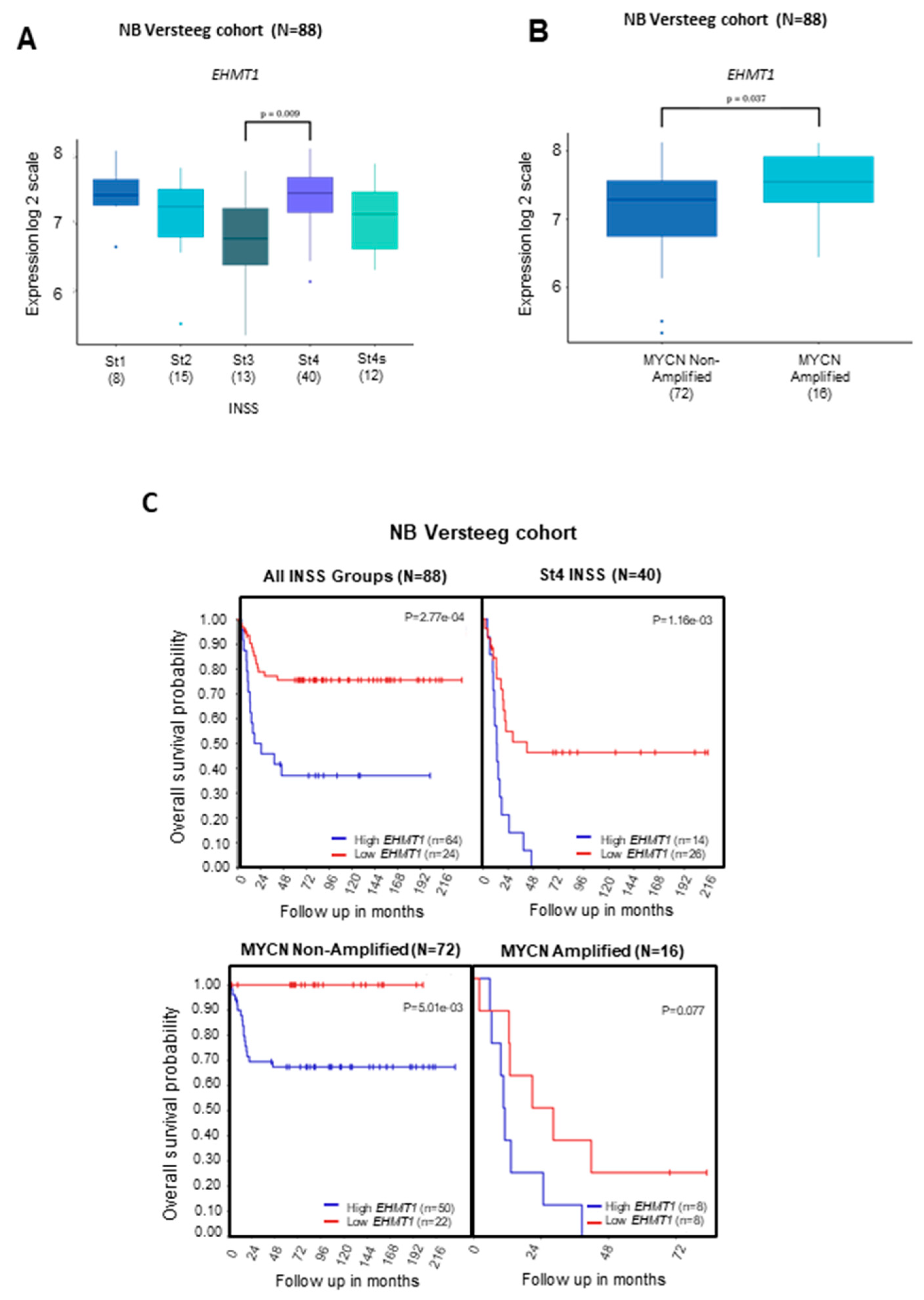
2.1. Transcriptional Levels of G9a/Ehmt2 and GLP/Ehmt1 on Survival Rates in Patients with NB
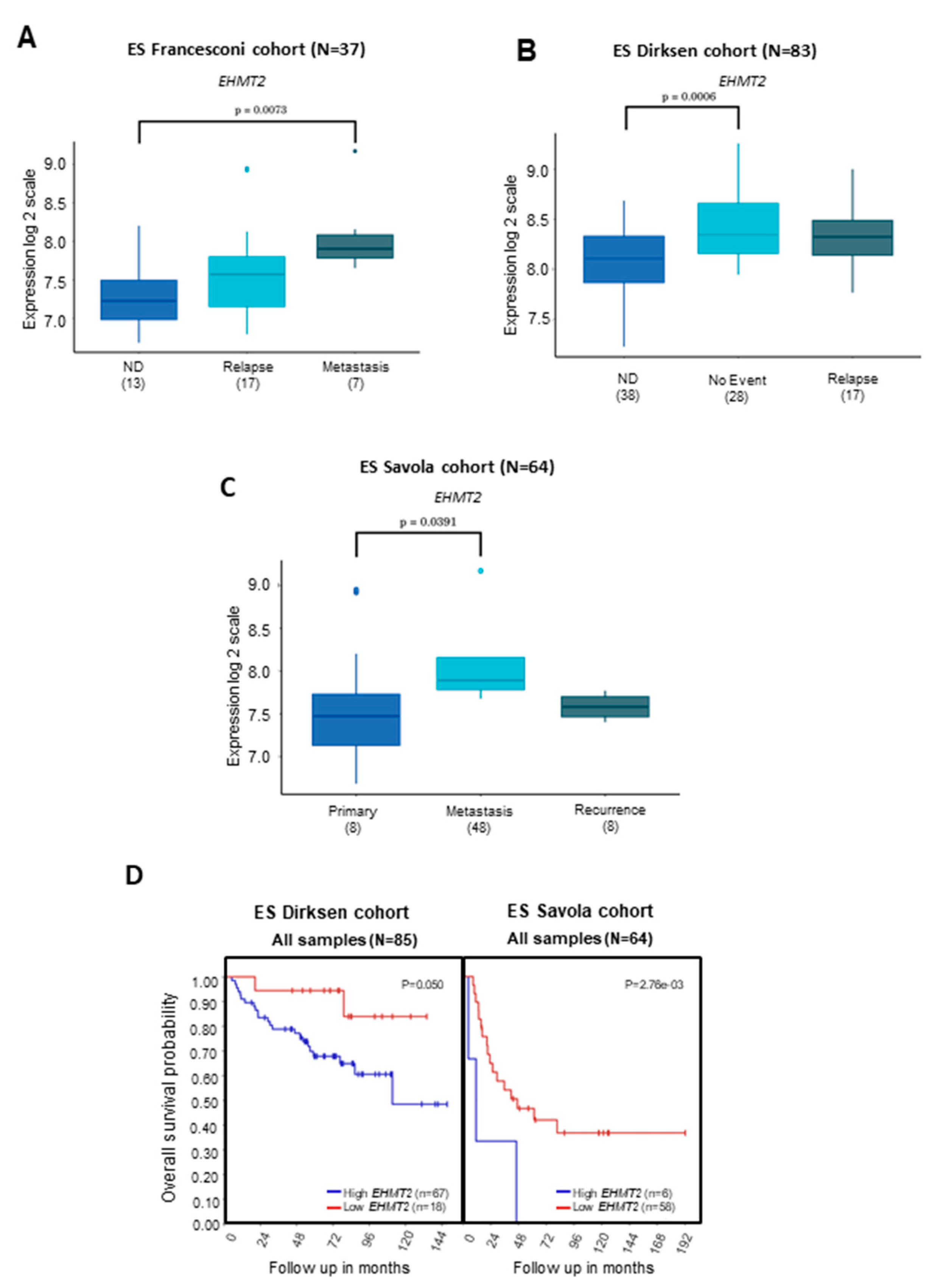
2.2. Transcriptional Levels of G9a/Ehmt2 and GLP/Ehmt1 on Survival Rates in Patients with ES
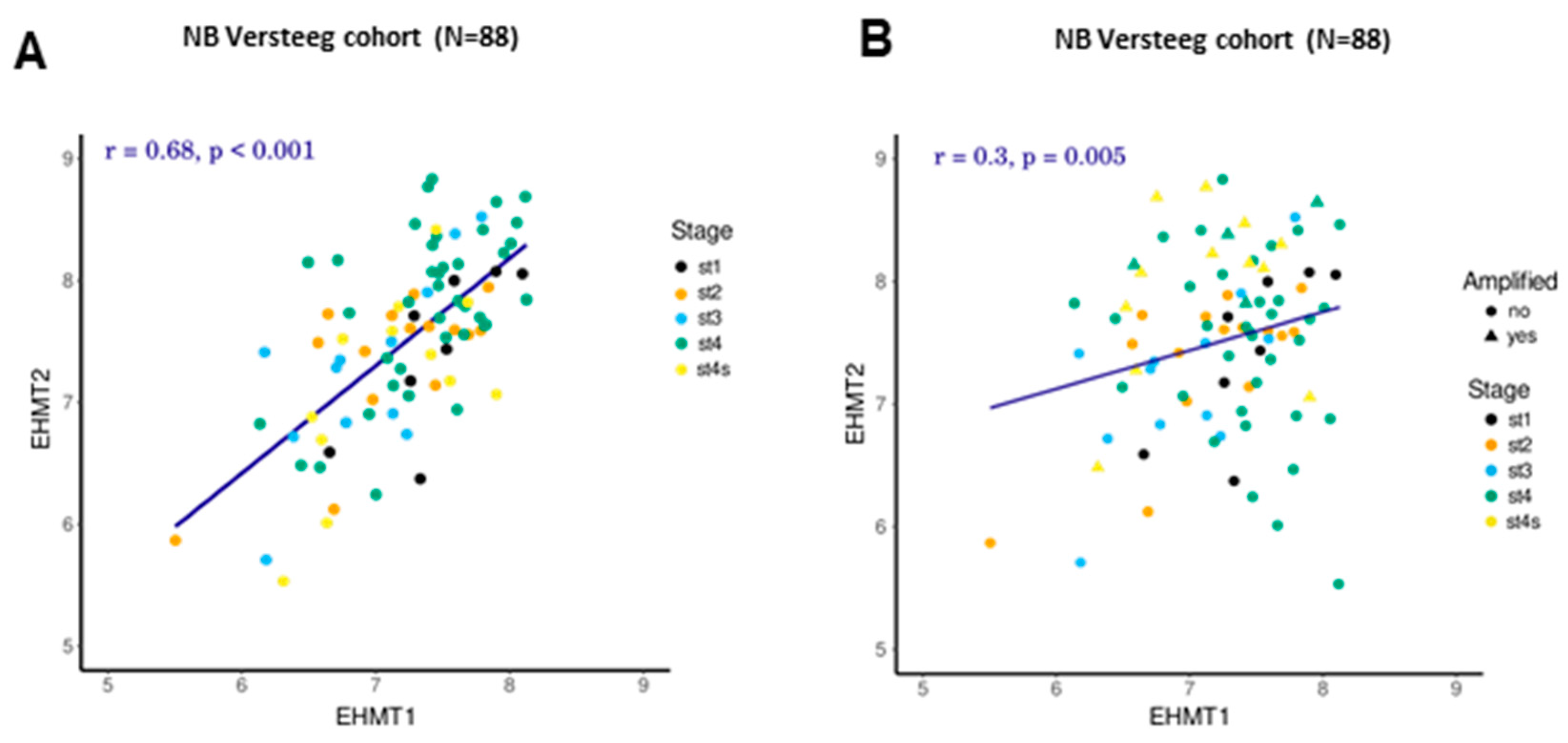
2.3. Correlation between Transcriptional Levels of G9a/Ehmt2 and GLP/Ehmt1 in Patients with NB
2.4. G9a/GLP Inhibition Reduces NB and ES Cell Viability
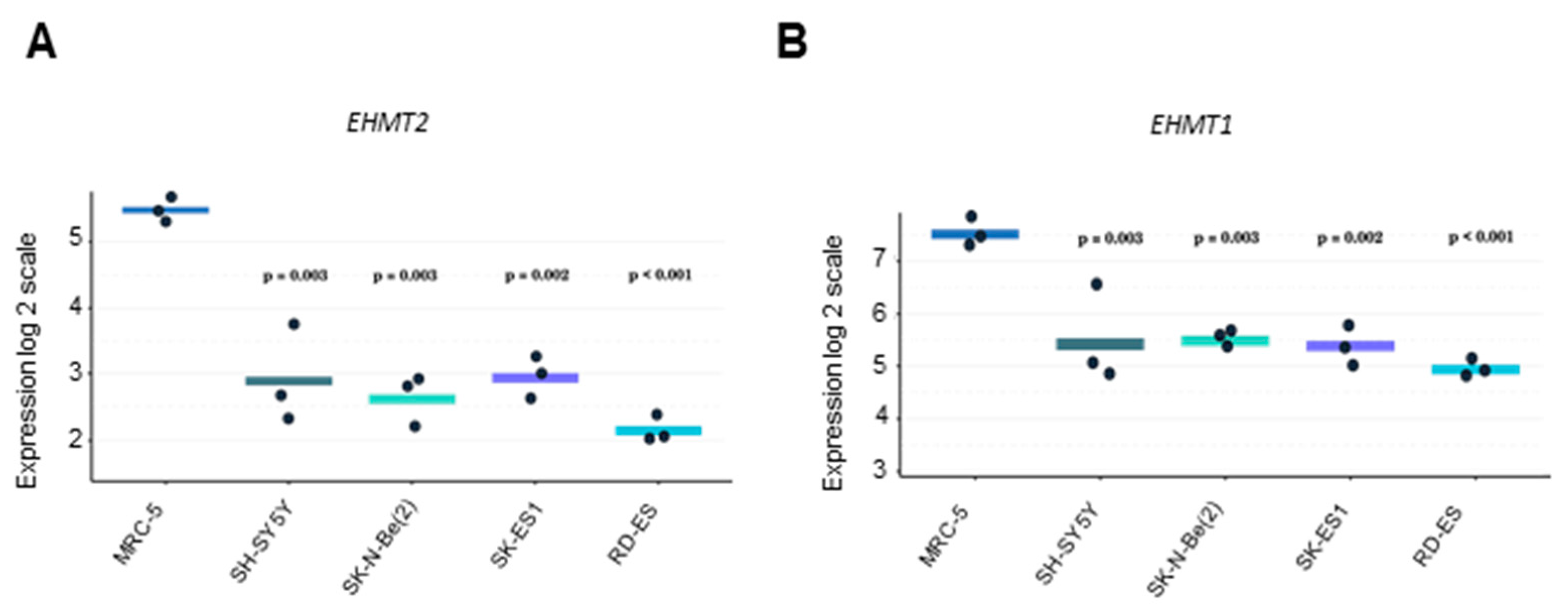
2.5. Expression of G9a/Ehmt2 and GLP/Ehmt1 mRNA Levels in NB, ES, and Non-Tumoral Human Cells
3. Discussion
4. Materials and Methods
4.1. Gene Expression
4.2. Cell Culture
4.3. Cell Viability and Drug Treatment
4.4. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
4.5. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lehnertz, B.; Ueda, Y.; Derijck, A.A.; Braunschweig, U.; Perez-Burgos, L.; Kubicek, S.; Chen, T.; Li, E.; Jenuwein, T.; Peters, A.H. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003, 13, 1192–1200. [Google Scholar] [CrossRef] [PubMed]
- Morera, L.; Lübbert, M.; Jung, M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin. Epigenet. 2016, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- Milner, C.M.; Campbell, R.D. The G9a gene in the human major histocompatibility complex encodes a novel protein containing ankyrin-like repeats. Biochem. J. 1993, 290 Pt 3, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.E.; Campbell, R.D.; Sanderson, C.M. Novel NG36/G9a gene products encoded within the human and mouse MHC class III regions. Mamm. Genome 2001, 12, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Skutt-Kakaria, K.; Davison, J.; Ou, Y.L.; Choi, E.; Malik, P.; Loeb, K.; Wood, B.; Georges, G.; Torok-Storb, B.; et al. G9a/GLP-dependent histone H3K9me2 patterning during human hematopoietic stem cell lineage commitment. Genes Dev. 2012, 26, 2499–2511. [Google Scholar] [CrossRef]
- Fong, K.W.; Zhao, J.C.; Lu, X.; Kim, J.; Piunti, A.; Shilatifard, A.; Yu, J. PALI1 promotes tumor growth through competitive recruitment of PRC2 to G9A-target chromatin for dual epigenetic silencing. Mol. Cell 2022, 82, 4611–4626.e7. [Google Scholar] [CrossRef]
- Vinson, D.A.; Stephens, K.E.; O’Meally, R.N.; Bhat, S.; Dancy, B.C.R.; Cole, R.N.; Yegnasubramanian, S.; Taverna, S.D. De novo methylation of histone H3K23 by the methyltransferases EHMT1/GLP and EHMT2/G9a. Epigenet. Chromatin 2022, 15, 36. [Google Scholar] [CrossRef]
- Chen, M.W.; Hua, K.T.; Kao, H.J.; Chi, C.C.; Wei, L.H.; Johansson, G.; Shiah, S.G.; Chen, P.S.; Jeng, Y.M.; Cheng, T.Y.; et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010, 70, 7830–7840. [Google Scholar] [CrossRef]
- Casciello, F.; Windloch, K.; Gannon, F.; Lee, J.S. Functional role of G9a histone methyltransferase in cancer. Front. Immunol. 2015, 6, 487. [Google Scholar] [CrossRef]
- Souza, B.K.; Freire, N.H.; Jaeger, M.; de Farias, C.B.; Brunetto, A.L.; Brunetto, A.T.; Roesler, R. EHMT2/G9a as an epigenetic target in pediatric and adult brain tumors. Int. J. Mol. Sci. 2021, 22, 11292. [Google Scholar] [CrossRef]
- Tachibana, M.; Ueda, J.; Fukuda, M.; Takeda, N.; Ohta, T.; Iwanari, H.; Sakihama, T.; Kodama, T.; Hamakubo, T.; Shinkai, Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005, 19, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Termanis, A.; Özkan, B.; Bao, X.X.; Culley, J.; de Lima Alves, F.; Rappsilber, J.; Ramsahoye, B.; Stancheva, I. G9a/GLP complex maintains imprinted DNA methylation in embryonic stem cells. Cell Rep. 2016, 15, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, N.A.; Kallweit, L.M.; Trnka, M.J.; Clemmer, C.L.; Al-Sady, B. Heterodimerization of H3K9 histone methyltransferases G9a and GLP activates methyl reading and writing capabilities. J. Biol. Chem. 2021, 297, 101276. [Google Scholar] [CrossRef] [PubMed]
- Pinto, N.R.; Applebaum, M.A.; Volchenboum, S.L.; Matthay, K.K.; London, W.B.; Ambros, P.F.; Nakagawara, A.; Berthold, F.; Schleiermacher, G.; Park, J.R.; et al. Advances in risk classification and treatment strategies for neuroblastoma. J. Clin. Oncol. 2015, 33, 3008–3017. [Google Scholar] [CrossRef]
- Yu, A.L.; Gilman, A.L.; Ozkaynak, M.F.; Naranjo, A.; Diccianni, M.B.; Gan, J.; Hank, J.A.; Batova, A.; London, W.B.; Tenney, S.C.; et al. Long-term follow-up of a phase III study of ch14.18 (dinutuximab) + cytokine immunotherapy in children with high-risk neuroblastoma: COG Study ANBL0032. Clin. Cancer Res. 2021, 27, 2179–2189. [Google Scholar] [CrossRef] [PubMed]
- Grünewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; de Álava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Ver. Dis. Primers 2018, 4, 5. [Google Scholar] [CrossRef]
- Eaton, B.R.; Claude, L.; Indelicato, D.J.; Vatner, R.; Yeh, B.; Schwarz, R.; Laack, N. Ewing sarcoma. Pediatr. Blood Cancer 2021, 68 (Suppl. S2), e28355. [Google Scholar] [CrossRef]
- Riggi, N.; Suvà, M.L.; Stamenkovic, I. Ewing’s sarcoma. N. Engl. J. Med. 2021, 384, 154–164. [Google Scholar] [CrossRef]
- Lawlor, E.R.; Thiele, C.J. Epigenetic changes in pediatric solid tumors: Promising new targets. Clin. Cancer Res. 2012, 18, 2768–2779. [Google Scholar] [CrossRef]
- Riggi, N.; Knoechel, B.; Gillespie, S.M.; Rheinbay, E.; Boulay, G.; Suvà, M.L.; Rossetti, N.E.; Boonseng, W.E.; Oksuz, O.; Cook, E.B.; et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014, 26, 668–681. [Google Scholar] [CrossRef]
- Almeida, V.R.; Vieira, I.A.; Buendia, M.; Brunetto, A.T.; Gregianin, L.J.; Brunetto, A.L.; Klamt, F.; de Farias, C.B.; Abujamra, A.L.; Lopez, P.L.D.C.; et al. Combined treatments with a retinoid receptor agonist and epigenetic modulators in human neuroblastoma cells. Mol. Neurobiol. 2017, 54, 7610–7619. [Google Scholar] [CrossRef] [PubMed]
- Veschi, V.; Liu, Z.; Voss, T.C.; Ozbun, L.; Gryder, B.; Yan, C.; Hu, Y.; Ma, A.; Jin, J.; Mazur, S.J.; et al. Epigenetic siRNA and chemical screens identify SETD8 inhibition as a therapeutic strategy for p53 activation in high-risk neuroblastoma. Cancer Cell 2017, 31, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Souza, B.K.; da Costa Lopez, P.L.; Menegotto, P.R.; Vieira, I.A.; Kersting, N.; Abujamra, A.L.; Brunetto, A.T.; Brunetto, A.L.; Gregianin, L.; de Farias, C.B.; et al. Targeting histone deacetylase activity to arrest cell growth and promote neural differentiation in Ewing sarcoma. Mol. Neurobiol. 2018, 55, 7242–7258. [Google Scholar] [CrossRef] [PubMed]
- Souza, B.K.; Freire, N.H.; Jaeger, M.; de Farias, C.B.; Brunetto, A.L.; Brunetto, A.T.; Roesler, R. G9a/EHMT2 is a potential prognostic biomarker and molecular target in SHH medulloblastoma. Neuromol. Med. 2022, 24, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, J.J.; Domingo-Fernández, R.; Ebus, M.E.; Lindner, S.; Koster, J.; Drabek, K.; Mestdagh, P.; van Sluis, P.; Valentijn, L.J.; van Nes, J.; et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 2012, 44, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Scotlandi, K.; Remondini, D.; Castellani, G.; Manara, M.C.; Nardi, F.; Cantiani, L.; Francesconi, M.; Mercuri, M.; Caccuri, A.M.; Serra, M.; et al. Overcoming resistance to conventional drugs in Ewing sarcoma and identification of molecular predictors of outcome. J. Clin. Oncol. 2009, 27, 2209–2216. [Google Scholar] [CrossRef]
- Savola, S.; Klami, A.; Myllykangas, S.; Manara, C.; Scotlandi, K.; Picci, P.; Knuutila, S.; Vakkila, J. High expression of complement component 5 (C5) at tumor site associates with superior survival in Ewing’s sarcoma family of tumour patients. ISRN Oncol. 2011, 2011, 168712. [Google Scholar] [CrossRef]
- Selvanathan, S.P.; Graham, G.T.; Erkizan, H.V.; Dirksen, U.; Natarajan, T.G.; Dakic, A.; Yu, S.; Liu, X.; Paulsen, M.T.; Ljungman, M.E.; et al. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc. Natl. Acad. Sci. USA 2015, 112, E1307–E1316. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Seeger, R.C.; Schwab, M.; Varmus, H.E.; Bishop, J.M. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 1984, 224, 1121–1124. [Google Scholar] [CrossRef]
- Peifer, M.; Hertwig, F.; Roels, F.; Dreidax, D.; Gartlgruber, M.; Menon, R.; Krämer, A.; Roncaioli, J.L.; Sand, F.; Heuckmann, J.M.; et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 2015, 526, 700–704. [Google Scholar] [CrossRef]
- Pugh, T.J.; Morozova, O.; Attiyeh, E.F.; Asgharzadeh, S.; Wei, J.S.; Auclair, D.; Carter, S.L.; Cibulskis, K.; Hanna, M.; Kiezun, A.; et al. The genetic landscape of high-risk neuroblastoma. Nat. Genet. 2013, 45, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Bellamy, J.; Szemes, M.; Melegh, Z.; Dallosso, A.; Kollareddy, M.; Catchpoole, D.; Malik, K. Increased efficacy of histone methyltransferase G9a inhibitors against MYCN-amplified neuroblastoma. Front. Oncol. 2020, 10, 818. [Google Scholar] [CrossRef]
- Coward, W.R.; Brand, O.J.; Pasini, A.; Jenkins, G.; Knox, A.J.; Pang, L. Interplay between EZH2 and G9a regulates CXCL10 gene repression in idiopathic pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2018, 58, 449–460. [Google Scholar] [CrossRef]
- Ligresti, G.; Caporarello, N.; Meridew, J.A.; Jones, D.L.; Tan, Q.; Choi, K.M.; Haak, A.J.; Aravamudhan, A.; Roden, A.C.; Prakash, Y.S.; et al. CBX5/G9a/H3K9me-mediated gene repression is essential to fibroblast activation during lung fibrosis. JCI Insight 2019, 5, e127111. [Google Scholar] [CrossRef]
- Minakawa, T.; Kanki, Y.; Nakamura, K.; Yamashita, J.K. Protein kinase A accelerates the rate of early stage differentiation of pluripotent stem cells. Biochem. Biophys. Res. Commun. 2020, 524, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Zylicz, J.J.; Borensztein, M.; Wong, F.C.; Huang, Y.; Lee, C.; Dietmann, S.; Surani, M.A. G9a regulates temporal preimplantation developmental program and lineage segregation in blastocyst. eLife 2018, 7, e33361. [Google Scholar] [CrossRef] [PubMed]
- García-Domínguez, D.J.; Hajji, N.; López-Alemany, R.; Sánchez-Molina, S.; Figuerola-Bou, E.; Morón Civanto, F.J.; Rello-Varona, S.; Andrés-León, E.; Benito, A.; Keun, H.C.; et al. Selective histone methyltransferase G9a inhibition reduces metastatic development of Ewing sarcoma through the epigenetic regulation of NEU1. Oncogene 2022, 41, 2638–2650. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Xie, X.; Xu, Y.; Huang, G. Identification of driver genes associated with chemotherapy resistance of Ewing’s sarcoma. Onco Targets Ther. 2018, 11, 6947–6956. [Google Scholar] [CrossRef]
- Ke, X.X.; Zhang, D.; Zhu, S.; Xia, Q.; Xiang, Z.; Cui, H. Inhibition of H3K9 methyltransferase G9a repressed cell proliferation and induced autophagy in neuroblastoma cells. PLoS ONE 2014, 9, e106962. [Google Scholar] [CrossRef]
- Lu, Z.; Tian, Y.; Salwen, H.R.; Chlenski, A.; Godley, L.A.; Raj, J.U.; Yang, Q. Histone-lysine methyltransferase EHMT2 is involved in proliferation, apoptosis, cell invasion, and DNA methylation of human neuroblastoma cells. Anticancer Drugs 2013, 24, 484–493. [Google Scholar] [CrossRef]
- Bachman, K.E.; Park, B.H.; Rhee, I.; Rajagopalan, H.; Herman, J.G.; Baylin, S.B.; Kinzler, K.W.; Vogelstein, B. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell 2003, 3, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, R.J.; Klimecki, W.T.; Lau, S.S.; Feinstein, Y.; Futscher, B.W. 5-Aza-2’-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene 2007, 26, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Barsyte-Lovejoy, D.; Li, F.; Xiong, Y.; Korboukh, V.; Huang, X.P.; Allali-Hassani, A.; Janzen, W.P.; Roth, B.L.; Frye, S.V.; et al. Discovery of an in vivo chemical probe of the lysine methyltransferases G9a and GLP. J. Med. Chem. 2013, 56, 8931–8942. [Google Scholar] [CrossRef] [PubMed]


| Time (h) | SH-SY5Y | SK-N-Be(2) | SK-ES 1 | ED-ES |
|---|---|---|---|---|
| 24 | 4.5 ± 0.08 | 4.5 ± 0.09 | 1.9 ± 0.07 | 2.6 ± 0.09 |
| 48 | 6.0 ± 0.07 | 4.2 ± 0.07 | 4.6 ± 0.06 | 2.3 ± 0.06 |
| 72 | 5.3 ± 0.06 | 2.4 ± 0.07 | 7.5 ± 0.07 | 2.0 ± 0.10 |
| 96 | 3.3 ± 0.08 | 2.0 ± 0.10 | 7.4 ± 0.08 | 2.6 ± 0.07 |
| Gene | Primers |
|---|---|
| Ehmt1 | primer sense 5′-GCT GCT GGG AGA AGA GAC AC-3′ primer anti-sense 5′-AGC ATT TGC ATG ACT GCT GG-3′ |
| G9a/Ehmt2 | primer sense 5′-GAG GTC TAC TGC ATA GAT GCC-3′ primer anti-sense 5′-CAG ACG GTC CTG CTC CAG GGC-3′ |
| ACTB | primer sense 5′-AAC TGG AAC GGT GAA GGT G-3′ primer anti-sense 5′-AGA GAA GTG GGG TGG CTT TT-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, B.K.; Freire, N.H.; Monteiro, T.S.; Herlinger, A.L.; Jaeger, M.; Dalmolin, M.G.S.; de Farias, C.B.; Gregianin, L.; Brunetto, A.T.; Brunetto, A.L.; et al. Histone Methyltransferases G9a/Ehmt2 and GLP/Ehmt1 Are Associated with Cell Viability and Poorer Prognosis in Neuroblastoma and Ewing Sarcoma. Int. J. Mol. Sci. 2023, 24, 15242. https://doi.org/10.3390/ijms242015242
Souza BK, Freire NH, Monteiro TS, Herlinger AL, Jaeger M, Dalmolin MGS, de Farias CB, Gregianin L, Brunetto AT, Brunetto AL, et al. Histone Methyltransferases G9a/Ehmt2 and GLP/Ehmt1 Are Associated with Cell Viability and Poorer Prognosis in Neuroblastoma and Ewing Sarcoma. International Journal of Molecular Sciences. 2023; 24(20):15242. https://doi.org/10.3390/ijms242015242
Chicago/Turabian StyleSouza, Barbara Kunzler, Natalia Hogetop Freire, Thiago Santos Monteiro, Alice Laschuk Herlinger, Mariane Jaeger, Matheus G. S. Dalmolin, Caroline Brunetto de Farias, Lauro Gregianin, André T. Brunetto, Algemir L. Brunetto, and et al. 2023. "Histone Methyltransferases G9a/Ehmt2 and GLP/Ehmt1 Are Associated with Cell Viability and Poorer Prognosis in Neuroblastoma and Ewing Sarcoma" International Journal of Molecular Sciences 24, no. 20: 15242. https://doi.org/10.3390/ijms242015242
APA StyleSouza, B. K., Freire, N. H., Monteiro, T. S., Herlinger, A. L., Jaeger, M., Dalmolin, M. G. S., de Farias, C. B., Gregianin, L., Brunetto, A. T., Brunetto, A. L., Thiele, C. J., & Roesler, R. (2023). Histone Methyltransferases G9a/Ehmt2 and GLP/Ehmt1 Are Associated with Cell Viability and Poorer Prognosis in Neuroblastoma and Ewing Sarcoma. International Journal of Molecular Sciences, 24(20), 15242. https://doi.org/10.3390/ijms242015242








