Neurolysin Knockout Mice in a Diet-Induced Obesity Model
Abstract
1. Introduction
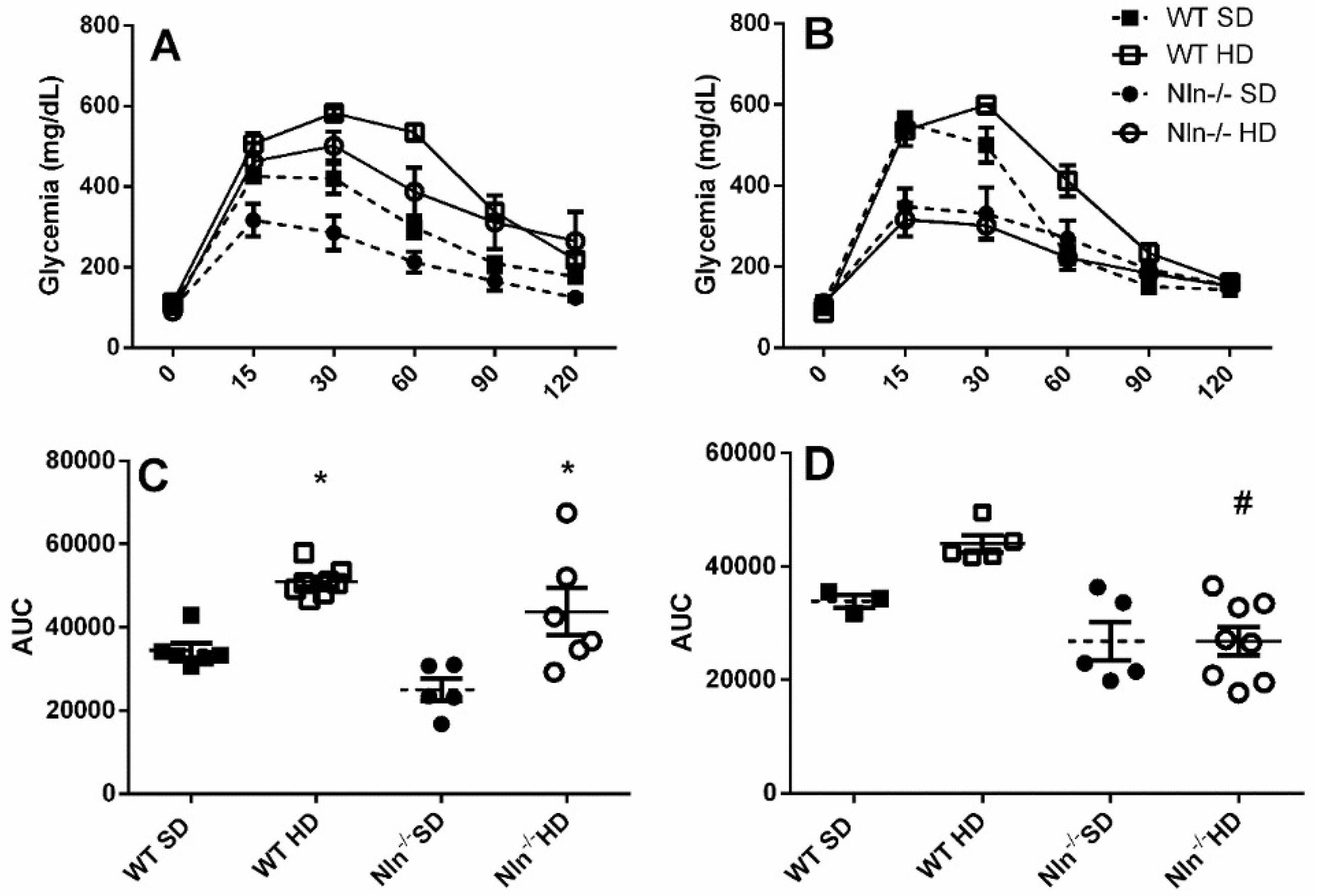
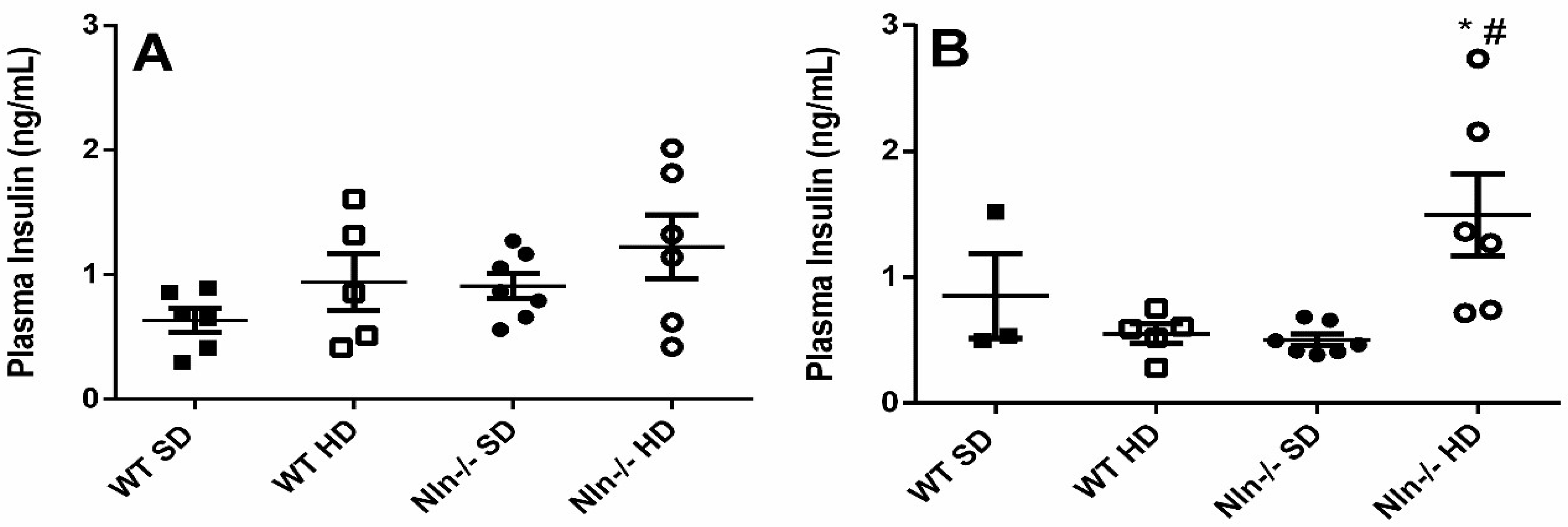
2. Results
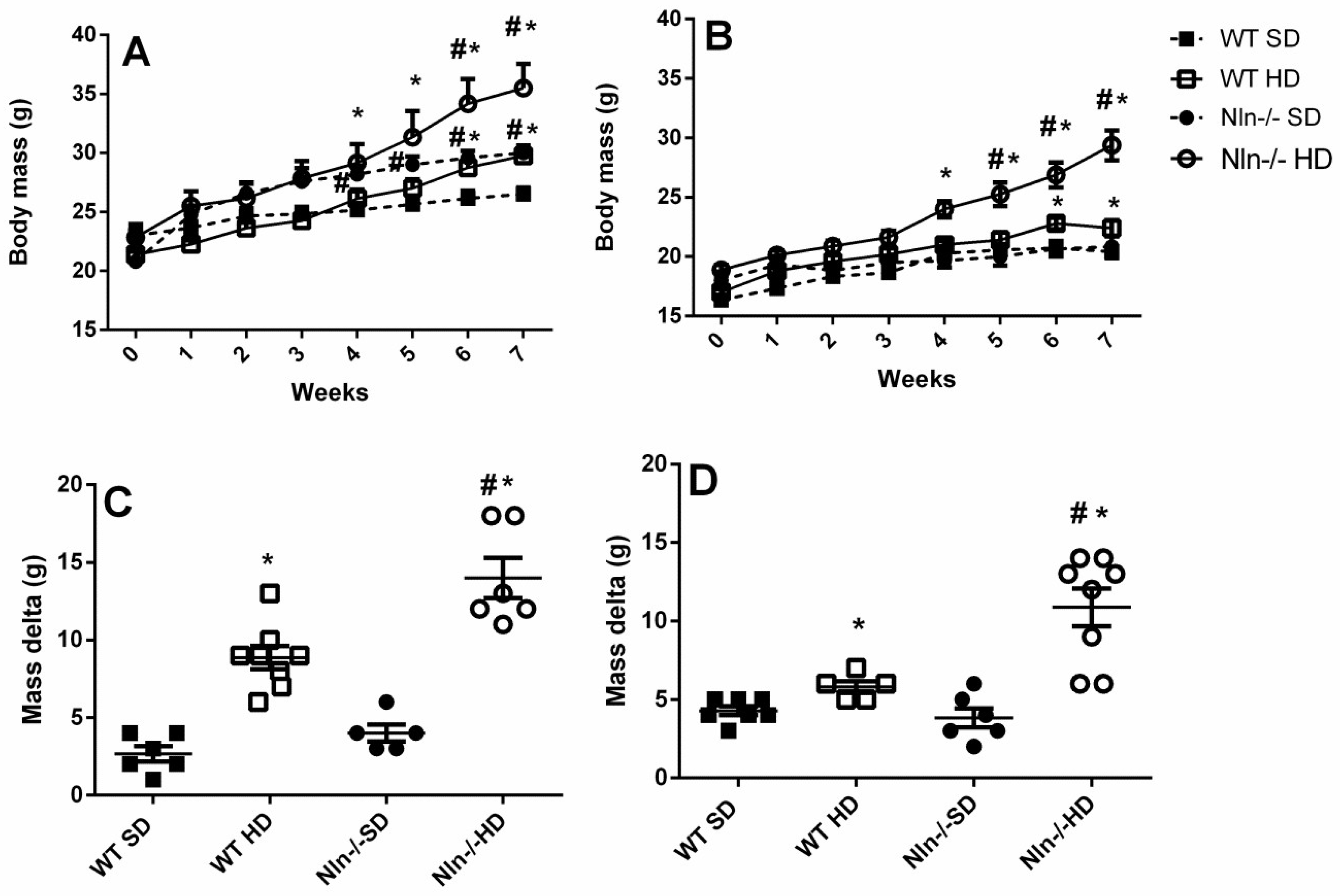
2.1. Body Mass, Food and Water Consumption
2.2. Analysis of Gene Expression
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Mice Genotyping
4.3. Diet-Induced Obesity
4.4. Measuring Animals Body Mass, Food, and Water Consumption
4.5. Glucose Tolerance Test (GTT) and Insulin Tolerance Test (ITT)
4.6. Plasma Insulin Measurements
4.7. Real-Time PCR (qRT-PCR)
4.8. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rioli, V.; Kato, A.; Portaro, F.C.; Cury, G.K.; te Kaat, K.; Vincent, B.; Checler, F.; Camargo, A.C.; Glucksman, M.J.; Roberts, J.L.; et al. Neuropeptide specificity and inhibition of recombinant isoforms of the endopeptidase 3.4.24.16 family: Comparison with the related recombinant endopeptidase 3.4.24.15. Biochem. Biophys. Res. Commun. 1998, 250, 5–11. [Google Scholar] [CrossRef]
- Checler, F.; Emson, P.C.; Vincent, J.P.; Kitabgi, P. Inactivation of neurotensin by rat brain synaptic membranes. Cleavage at the Pro10-Tyr11 bond by endopeptidase 24.11 (enkephalinase) and a peptidase different from proline-endopeptidase. J. Neurochem. 1984, 43, 1295–1301. [Google Scholar] [CrossRef]
- Dauch, P.; Vincent, J.P.; Checler, F. Molecular cloning and expression of rat brain endopeptidase 3.4.24.16. J. Biol. Chem. 1995, 270, 27266–27271. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Salvesen, G. (Eds.) Handbook of Proteolytic Enzymes, 3rd ed.; Academic Press: London, UK, 2013; p. iv. ISBN 9780123822192. [Google Scholar] [CrossRef]
- Checler, F.; Vincent, J.P.; Kitabgi, P. Purification and characterization of a novel neurotensin-degrading peptidase from rat brain synaptic membranes. J. Biol. Chem. 1986, 261, 11274–11281. [Google Scholar] [CrossRef]
- Fontenele-Neto, J.D.; Massarelli, E.E.; Gurgel Garrido, P.A.; Beaudet, A.; Ferro, E.S. Comparative fine structural distribution of endopeptidase 24.15 (EC3.4.24.15) and 24.16 (EC3.4.24.16) in rat brain. J. Comp. Neurol. 2001, 438, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Massarelli, E.E.; Casatti, C.A.; Kato, A.; Camargo, A.C.; Bauer, J.A.; Glucksman, M.J.; Roberts, J.L.; Hirose, S.; Ferro, E.S. Differential subcellular distribution of neurolysin (EC 3.4.24.16) and thimet oligopeptidase (EC 3.4.24.15) in the rat brain. Brain Res. 1999, 851, 261–265. [Google Scholar] [CrossRef]
- Vincent, B.; Beaudet, A.; Dauch, P.; Vincent, J.P.; Checler, F. Distinct properties of neuronal and astrocytic endopeptidase 3.4.24.16: A study on differentiation, subcellular distribution, and secretion processes. J. Neurosci. 1996, 16, 5049–5059. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Sugiura, N.; Saruta, Y.; Hosoiri, T.; Yasue, H.; Hirose, S. Targeting of endopeptidase 24.16 to different subcellular compartments by alternative promoter usage. J. Biol. Chem. 1997, 272, 15313–15322. [Google Scholar] [CrossRef]
- Lim, E.J.; Sampath, S.; Coll-Rodriguez, J.; Schmidt, J.; Ray, K.; Rodgers, D.W. Swapping the substrate specificities of the neuropeptidases neurolysin and thimet oligopeptidase. J. Biol. Chem. 2007, 282, 9722–9732. [Google Scholar] [CrossRef] [PubMed]
- Ray, K.; Hines, C.S.; Coll-Rodriguez, J.; Rodgers, D.W. Crystal structure of human thimet oligopeptidase provides insight into substrate recognition, regulation, and localization. J. Biol. Chem. 2004, 279, 20480–20489. [Google Scholar] [CrossRef]
- Ray, K.; Hines, C.S.; Rodgers, D.W. Mapping sequence differences between thimet oligopeptidase and neurolysin implicates key residues in substrate recognition. Protein Sci. 2002, 11, 2237–2246. [Google Scholar] [CrossRef]
- Oliveira, V.; Araujo, M.C.; Rioli, V.; de Camargo, A.C.; Tersariol, I.L.; Juliano, M.A.; Juliano, L.; Ferro, E.S. A structure-based site-directed mutagenesis study on the neurolysin (EC 3.4.24.16) and thimet oligopeptidase (EC 3.4.24.15) catalysis. FEBS Lett. 2003, 541, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Rioli, V.; Gozzo, F.C.; Heimann, A.S.; Linardi, A.; Krieger, J.E.; Shida, C.S.; Almeida, P.C.; Hyslop, S.; Eberlin, M.N.; Ferro, E.S. Novel natural peptide substrates for endopeptidase 24.15, neurolysin, and angiotensin-converting enzyme. J. Biol. Chem. 2003, 278, 8547–8555. [Google Scholar] [CrossRef] [PubMed]
- Barelli, H.; Vincent, J.P.; Checler, F. Rat kidney endopeptidase 24.16. Purification, physico-chemical characteristics and differential specificity towards opiates, tachykinins and neurotensin-related peptides. Eur. J. Biochem. 1993, 211, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Cavalcanti, D.M.L.P.; Castro, L.M.; Rosa Neto, J.C.; Seelaender, M.; Neves, R.X.; Oliveira, V.; Forti, F.L.; Iwai, L.K.; Gozzo, F.C.; Todiras, M.; et al. Neurolysin knockout mice generation and initial phenotype characterization. J. Biol. Chem. 2014, 289, 15426–15440. [Google Scholar] [CrossRef]
- Castro, L.M.; Cavalcanti, D.M.; Araujo, C.B.; Rioli, V.; Icimoto, M.Y.; Gozzo, F.C.; Juliano, M.; Juliano, L.; Oliveira, V.; Ferro, E.S. Peptidomic analysis of the neurolysin-knockout mouse brain. J. Proteom. 2014, 111, 238–248. [Google Scholar] [CrossRef]
- Gomez, R.; Por, E.D.; Berg, K.A.; Clarke, W.P.; Glucksman, M.J.; Jeske, N.A. Metallopeptidase inhibition potentiates bradykinin-induced hyperalgesia. Pain 2011, 152, 1548–1554. [Google Scholar] [CrossRef]
- Jeske, N.A.; Berg, K.A.; Cousins, J.C.; Ferro, E.S.; Clarke, W.P.; Glucksman, M.J.; Roberts, J.L. Modulation of bradykinin signaling by EP24.15 and EP24.16 in cultured trigeminal ganglia. J. Neurochem. 2006, 97, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Norman, M.U.; Reeve, S.B.; Dive, V.; Smith, A.I.; Lew, R.A. Endopeptidases 3.4.24.15 and 24.16 in endothelial cells: Potential role in vasoactive peptide metabolism. Am. J. Physiol. Heart Circ. Physiol. 2003, 284, H1978–H1984. [Google Scholar] [CrossRef]
- Cotter, E.J.; von Offenberg Sweeney, N.; Coen, P.M.; Birney, Y.A.; Glucksman, M.J.; Cahill, P.A.; Cummins, P.M. Regulation of endopeptidases EC3.4.24.15 and EC3.4.24.16 in vascular endothelial cells by cyclic strain: Role of Gi protein signaling. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 457–463. [Google Scholar] [CrossRef]
- Barelli, H.; Fox-Threlkeld, J.; Dive, V.; Daniel, E.; Vincent, J.; Checler, F. Role of endopeptidase 3.4. 24.16 in the catabolism of neurotensin, in vivo, in the vascularly perfused dog ileum. Br. J. Pharmacol. 1994, 112, 127. [Google Scholar] [CrossRef]
- Dauch, P.; Vincent, J.P.; Checler, F. Specific inhibition of endopeptidase 24.16 by dipeptides. Eur. J. Biochem. 1991, 202, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Barchetta, I.; Baroni, M.G.; Melander, O.; Cavallo, M.G. New Insights in the Control of Fat Homeostasis: The Role of Neurotensin. Int. J. Mol. Sci. 2022, 23, 2209. [Google Scholar] [CrossRef]
- Ferro, E.S.; Gewehr, M.C.F.; Navon, A. Thimet Oligopeptidase Biochemical and Biological Significances: Past, Present, and Future Directions. Biomolecules 2020, 10, 1229. [Google Scholar] [CrossRef]
- Garrido, P.A.; Vandenbulcke, F.; Ramjaun, A.R.; Vincent, B.; Checler, F.; Ferro, E.; Beaudet, A. Confocal microscopy reveals thimet oligopeptidase (EC 3.4.24.15) and neurolysin (EC 3.4.24.16) in the classical secretory pathway. DNA Cell Biol. 1999, 18, 323–331. [Google Scholar] [CrossRef]
- Woulfe, J.; Checler, F.; Beaudet, A. Light and electron microscopic localization of the neutral metalloendopeptidase EC 3.4. 24.16 in the mesencephalon of the rat. Eur. J. Neurosci. 1992, 4, 1309–1319. [Google Scholar] [CrossRef]
- Gewehr, M.C.F.; Teixeira, A.A.S.; Santos, B.A.C.; Biondo, L.A.; Gozzo, F.C.; Cordibello, A.M.; Eichler, R.A.S.; Reckziegel, P.; Da Silva, R.N.O.; Dos Santos, N.B.; et al. The Relevance of Thimet Oligopeptidase in the Regulation of Energy Metabolism and Diet-Induced Obesity. Biomolecules 2020, 10, 321. [Google Scholar] [CrossRef]
- Santos, N.B.D.; Franco, R.D.; Camarini, R.; Munhoz, C.D.; Eichler, R.A.S.; Gewehr, M.C.F.; Reckziegel, P.; Llanos, R.P.; Dale, C.S.; Silva, V.; et al. Thimet Oligopeptidase (EC 3.4.24.15) Key Functions Suggested by Knockout Mice Phenotype Characterization. Biomolecules 2019, 9, 382. [Google Scholar] [CrossRef]
- Berti, D.A.; Morano, C.; Russo, L.C.; Castro, L.M.; Cunha, F.M.; Zhang, X.; Sironi, J.; Klitzke, C.F.; Ferro, E.S.; Fricker, L.D. Analysis of intracellular substrates and products of thimet oligopeptidase in human embryonic kidney 293 cells. J. Biol. Chem. 2009, 284, 14105–14116. [Google Scholar] [CrossRef]
- Ferro, E.S.; Tullai, J.W.; Glucksman, M.J.; Roberts, J.L. Secretion of metalloendopeptidase 24.15 (EC 3.4.24.15). DNA Cell Biol. 1999, 18, 781–789. [Google Scholar] [CrossRef]
- Ferro, E.S.; Tambourgi, D.V.; Gobersztejn, F.; Gomes, M.D.; Sucupira, M.; Armelin, M.C.; Kipnis, T.L.; Camargo, A.C. Secretion of a neuropeptide-metabolizing enzyme similar to endopeptidase 22.19 by glioma C6 cells. Biochem. Biophys. Res. Commun. 1993, 191, 275–281. [Google Scholar] [CrossRef]
- Jirácek, J.; Yiotakis, A.; Vincent, B.; Lecoq, A.; Nicolaou, A.; Checler, F.; Dive, V. Development of highly potent and selective phosphinic peptide inhibitors of zinc endopeptidase 24-15 using combinatorial chemistry. J. Biol. Chem. 1995, 270, 21701–21706. [Google Scholar] [CrossRef]
- Jirácek, J.; Yiotakis, A.; Vincent, B.; Checler, F.; Dive, V. Development of the first potent and selective inhibitor of the zinc endopeptidase neurolysin using a systematic approach based on combinatorial chemistry of phosphinic peptides. J. Biol. Chem. 1996, 271, 19606–19611. [Google Scholar] [CrossRef] [PubMed]
- Checler, F.; Barelli, H.; Dauch, P.; Dive, V.; Vincent, B.; Vincent, J.P. Neurolysin: Purification and assays. Methods Enzymol. 1995, 248, 593–614. [Google Scholar] [CrossRef]
- Shrimpton, C.N.; Glucksman, M.J.; Lew, R.A.; Tullai, J.W.; Margulies, E.H.; Roberts, J.L.; Smith, A.I. Thiol activation of endopeptidase EC 3.4.24.15. A novel mechanism for the regulation of catalytic activity. J. Biol. Chem. 1997, 272, 17395–17399. [Google Scholar] [CrossRef] [PubMed]
- Malvezzi, A.; Higa, P.M.; Amaral, A.T.-d.; Silva, G.M.; Gozzo, F.C.; Ferro, E.S.; Castro, L.M.; de Rezende, L.; Monteiro, G.; Demasi, M. The cysteine-rich protein thimet oligopeptidase as a model of the structural requirements for S-glutathiolation and oxidative oligomerization. PLoS ONE 2012, 7, e39408. [Google Scholar] [CrossRef] [PubMed]
- Neves, R.L.; Marem, A.; Carmona, B.; Arata, J.G.; Cyrillo Ramos, M.P.; Justo, G.Z.; Machado de Melo, F.H.; Oliveira, V.; Icimoto, M.Y. Expression of thimet oligopeptidase (THOP) modulated by oxidative stress in human multidrug resistant (MDR) leukemia cells. Biochimie 2023, 212, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Heymsfield, S.B.; Wadden, T.A. Mechanisms, pathophysiology, and management of obesity. N. Eng. J. Med. 2017, 376, 254–266. [Google Scholar] [CrossRef]
- Blüher, M. Obesity: Global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019, 15, 288–298. [Google Scholar] [CrossRef]
- da Costa Louzada, M.L.; Baraldi, L.G.; Steele, E.M.; Martins, A.P.B.; Canella, D.S.; Moubarac, J.-C.; Levy, R.B.; Cannon, G.; Afshin, A.; Imamura, F. Consumption of ultra-processed foods and obesity in Brazilian adolescents and adults. Prev. Med. 2015, 81, 9–15. [Google Scholar] [CrossRef]
- de Mendonça, M.; Rocha, K.C.; de Sousa, É.; Pereira, B.M.; Oyama, L.M.; Rodrigues, A.C. Aerobic exercise training regulates serum extracellular vesicle miRNAs linked to obesity to promote their beneficial effects in mice. Am. J. Physiol. -Endocrinol. Metab. 2020, 319, E579–E591. [Google Scholar] [CrossRef] [PubMed]
- Gewehr, M.C.F.; Silverio, R.; Rosa-Neto, J.C.; Lira, F.S.; Reckziegel, P.; Ferro, E.S. Peptides from Natural or Rationally Designed Sources Can Be Used in Overweight, Obesity, and Type 2 Diabetes Therapies. Molecules 2020, 25, 1093. [Google Scholar] [CrossRef]
- Wronska, A.; Kmiec, Z. Structural and biochemical characteristics of various white adipose tissue depots. Acta Physiol. 2012, 205, 194–208. [Google Scholar] [CrossRef] [PubMed]
- de Moura e Dias, M.; dos Reis, S.A.; da Conceição, L.L.; Sediyama, C.M.N.d.O.; Pereira, S.S.; de Oliveira, L.L.; Gouveia Peluzio, M.d.C.; Martinez, J.A.; Milagro, F.I. Diet-induced obesity in animal models: Points to consider and influence on metabolic markers. Diabetol. Metab. Syndr. 2021, 13, 32. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, I.A.; Luck, K.; Spirohn, K.; Wang, Y.; Pollis, C.; Schlabach, S.; Bian, W.; Kim, D.K.; Kishore, N.; Hao, T.; et al. Network-based prediction of protein interactions. Nat. Commun. 2019, 10, 1240. [Google Scholar] [CrossRef]
- Menche, J.; Sharma, A.; Kitsak, M.; Ghiassian, S.D.; Vidal, M.; Loscalzo, J.; Barabasi, A.L. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 2015, 347, 1257601. [Google Scholar] [CrossRef]
- Sahni, N.; Yi, S.; Zhong, Q.; Jailkhani, N.; Charloteaux, B.; Cusick, M.E.; Vidal, M. Edgotype: A fundamental link between genotype and phenotype. Curr. Opin. Genet. Dev. 2013, 23, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Prieur, X.; Mok, C.Y.; Velagapudi, V.R.; Nunez, V.; Fuentes, L.; Montaner, D.; Ishikawa, K.; Camacho, A.; Barbarroja, N.; O’Rahilly, S.; et al. Differential lipid partitioning between adipocytes and tissue macrophages modulates macrophage lipotoxicity and M2/M1 polarization in obese mice. Diabetes 2011, 60, 797–809. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.; Cusick, M.E.; Barabasi, A.L. Interactome networks and human disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef]
- Sethi, J.K.; Vidal-Puig, A.J. Targeting fat to prevent diabetes. Cell Metab. 2007, 5, 323–325. [Google Scholar] [CrossRef][Green Version]
- Goh, K.-I.; Cusick, M.E.; Valle, D.; Childs, B.; Vidal, M.; Barabási, A.-L. The human disease network. Proc. Natl. Acad. Sci. USA 2007, 104, 8685–8690. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Menche, J.; Huang, C.C.; Ort, T.; Zhou, X.; Kitsak, M.; Sahni, N.; Thibault, D.; Voung, L.; Guo, F.; et al. A disease module in the interactome explains disease heterogeneity, drug response and captures novel pathways and genes in asthma. Hum. Mol. Genet. 2015, 24, 3005–3020. [Google Scholar] [CrossRef]
- Pawson, T.; Nash, P. Protein-protein interactions define specificity in signal transduction. Genes. Dev. 2000, 14, 1027–1047. [Google Scholar] [CrossRef]
- Pawson, T.; Scott, J.D. Signaling through scaffold, anchoring, and adaptor proteins. Science 1997, 278, 2075–2080. [Google Scholar] [CrossRef]
- Ferro, E.S.; Hyslop, S.; Camargo, A.C. Intracellullar peptides as putative natural regulators of protein interactions. J. Neurochem. 2004, 91, 769–777. [Google Scholar] [CrossRef]
- Dasgupta, S.; Yang, C.; Castro, L.M.; Tashima, A.K.; Ferro, E.S.; Moir, R.D.; Willis, I.M.; Fricker, L.D. Analysis of the Yeast Peptidome and Comparison with the Human Peptidome. PLoS ONE 2016, 11, e0163312. [Google Scholar] [CrossRef]
- Dasgupta, S.; Fishman, M.; Mahallati, H.; Castro, L.; Tashima, A.; Ferro, E.; Fricker, L. Reduced Levels of Proteasome Products in a Mouse Striatal Cell Model of Huntington’s Disease. PLoS ONE 2015, 10, e0145333. [Google Scholar] [CrossRef]
- Dasgupta, S.; Castro, L.M.; Dulman, R.; Yang, C.; Schmidt, M.; Ferro, E.S.; Fricker, L.D. Proteasome inhibitors alter levels of intracellular peptides in HEK293T and SH-SY5Y cells. PLoS ONE 2014, 9, e103604. [Google Scholar] [CrossRef] [PubMed]
- Ferro, E.S.; Rioli, V.; Castro, L.M.; Fricker, L.D. Intracellular peptides: From discovery to function. EuPA Open Proteom. 2014, 3, 143–151. [Google Scholar] [CrossRef]
- Gelman, J.S.; Sironi, J.; Berezniuk, I.; Dasgupta, S.; Castro, L.M.; Gozzo, F.C.; Ferro, E.S.; Fricker, L.D. Alterations of the Intracellular Peptidome in Response to the Proteasome Inhibitor Bortezomib. PLoS ONE 2013, 8, e53263. [Google Scholar] [CrossRef]
- Cunha, F.M.; Berti, D.A.; Ferreira, Z.S.; Klitzke, C.F.; Markus, R.P.; Ferro, E.S. Intracellular peptides as natural regulators of cell signaling. J. Biol. Chem. 2008, 283, 24448–24459. [Google Scholar] [CrossRef]
- Machado, M.F.; Cunha, F.M.; Berti, D.A.; Heimann, A.S.; Klitzke, C.F.; Rioli, V.; Oliveira, V.; Ferro, E.S. Substrate phosphorylation affects degradation and interaction to endopeptidase 24.15, neurolysin, and angiotensin-converting enzyme. Biochem. Biophys. Res. Commun. 2006, 339, 520–525. [Google Scholar] [CrossRef]
- Russo, L.C.; Castro, L.M.; Gozzo, F.C.; Ferro, E.S. Inhibition of thimet oligopeptidase by siRNA alters specific intracellular peptides and potentiates isoproterenol signal transduction. FEBS Lett. 2012, 586, 3287–3292. [Google Scholar] [CrossRef] [PubMed]
- Gewehr, M.C.; Navon, A.; Ferro, E.S. Intracellular peptides as drug prototypes. In Peptide and Peptidomimetic Therapeutics; Elsevier: Amsterdam, The Netherlands, 2022; pp. 255–289. [Google Scholar]
- de Araujo, C.B.; Heimann, A.S.; Remer, R.A.; Russo, L.C.; Colquhoun, A.; Forti, F.L.; Ferro, E.S. Intracellular Peptides in Cell Biology and Pharmacology. Biomolecules 2019, 9, 150. [Google Scholar] [CrossRef]
- Russo, L.C.; Asega, A.F.; Castro, L.M.; Negraes, P.D.; Cruz, L.; Gozzo, F.C.; Ulrich, H.; Camargo, A.C.; Rioli, V.; Ferro, E.S. Natural intracellular peptides can modulate the interactions of mouse brain proteins and thimet oligopeptidase with 14-3-3epsilon and calmodulin. Proteomics 2012, 12, 2641–2655. [Google Scholar] [CrossRef]
- Parada, C.A.; de Oliveira, I.P.; Gewehr, M.C.F.; Machado-Neto, J.A.; Lima, K.; Eichler, R.A.S.; Lopes, L.R.; Bechara, L.R.G.; Ferreira, J.C.B.; Festuccia, W.T.; et al. Effect of FKBP12-Derived Intracellular Peptides on Rapamycin-Induced FKBP-FRB Interaction and Autophagy. Cells 2022, 11, 385. [Google Scholar] [CrossRef]
- Masi, L.N.; Martins, A.R.; Crisma, A.R.; do Amaral, C.L.; Davanso, M.R.; Serdan, T.D.A.; da Cunha de Sá, R.D.C.; Cruz, M.M.; Alonso-Vale, M.I.C.; Torres, R.P.; et al. Combination of a high-fat diet with sweetened condensed milk exacerbates inflammation and insulin resistance induced by each separately in mice. Sci. Rep. 2017, 7, 3937. [Google Scholar] [CrossRef] [PubMed]
- Birch, G.G.; Mwangelwa, O.M. Colorimetric determination of sugars in sweetened condensed milk products. J. Sci. Food Agric. 1974, 25, 1355–1362. [Google Scholar] [CrossRef]
- Fan, X.; Yao, H.; Liu, X.; Shi, Q.; Lv, L.; Li, P.; Wang, R.; Tang, T.; Qi, K. High-Fat Diet Alters the Expression of Reference Genes in Male Mice. Front. Nutr. 2020, 7. [Google Scholar] [CrossRef]
- Guaita-Cespedes, M.; Grillo-Risco, R.; Hidalgo, M.R.; Fernández-Veledo, S.; Burks, D.J.; de la Iglesia-Vayá, M.; Galán, A.; Garcia-Garcia, F. Deciphering the sex bias in housekeeping gene expression in adipose tissue: A comprehensive meta-analysis of transcriptomic studies. Biol. Sex. Differ. 2023, 14, 20. [Google Scholar] [CrossRef]
- Dallner, O.S.; Marinis, J.M.; Lu, Y.H.; Birsoy, K.; Werner, E.; Fayzikhodjaeva, G.; Dill, B.D.; Molina, H.; Moscati, A.; Kutalik, Z.; et al. Dysregulation of a long noncoding RNA reduces leptin leading to a leptin-responsive form of obesity. Nat. Med. 2019, 25, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Gruzdeva, O.; Borodkina, D.; Uchasova, E.; Dyleva, Y.; Barbarash, O. Leptin resistance: Underlying mechanisms and diagnosis. Diabetes Metab. Syndr. Obes. 2019, 12, 191–198. [Google Scholar] [CrossRef]
- Ruiz-Medina, B.E.; Cadena-Medina, D.A.; Esparza, E.; Arrieta, A.J.; Kirken, R.A. Isoproterenol-induced beta-2 adrenergic receptor activation negatively regulates interleukin-2 signaling. Biochem. J. 2018, 475, 2907–2923. [Google Scholar] [CrossRef]
- Cui, J.; Ding, Y.; Chen, S.; Zhu, X.; Wu, Y.; Zhang, M.; Zhao, Y.; Li, T.R.; Sun, L.V.; Zhao, S.; et al. Disruption of Gpr45 causes reduced hypothalamic POMC expression and obesity. J. Clin. Invest. 2016, 126, 3192–3206. [Google Scholar] [CrossRef]
- Li, J.; Wu, H.; Liu, Y.; Yang, L. High fat diet induced obesity model using four strainsof mice: Kunming, C57BL/6, BALB/c and ICR. Exp. Anim. 2020, 69, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, M.K.; Hallahan, N.L.; Brown, S.H.; Liu, M.; Mitchell, T.W.; Cooney, G.J.; Turner, N. Mouse strain-dependent variation in obesity and glucose homeostasis in response to high-fat feeding. Diabetologia 2013, 56, 1129–1139. [Google Scholar] [CrossRef]
- Nishikawa, S.; Yasoshima, A.; Doi, K.; Nakayama, H.; Uetsuka, K. Involvement of sex, strain and age factors in high fat diet-induced obesity in C57BL/6J and BALB/cA mice. Exp. Anim. 2007, 56, 263–272. [Google Scholar] [CrossRef]
- Blaak, E.E.; Saris, W.H. Postprandial thermogenesis and substrate utilization after ingestion of different dietary carbohydrates. Metabolism 1996, 45, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.A.; Navas-Carretero, S.; Saris, W.H.; Astrup, A. Personalized weight loss strategies-the role of macronutrient distribution. Nat. Rev. Endocrinol. 2014, 10, 749–760. [Google Scholar] [CrossRef]
- Levin, B.E.; Dunn-Meynell, A.A. Defense of body weight depends on dietary composition and palatability in rats with diet-induced obesity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 282, R46–R54. [Google Scholar] [CrossRef]
- Hall, K.D. A review of the carbohydrate-insulin model of obesity. Eur. J. Clin. Nutr. 2018, 72, 183. [Google Scholar] [CrossRef]
- Beetch, M.; Harandi-Zadeh, S.; Shen, K.; Lubecka, K.; Kitts, D.D.; O’Hagan, H.M.; Stefanska, B. Dietary antioxidants remodel DNA methylation patterns in chronic disease. Br. J. Pharmacol. 2020, 177, 1382–1408. [Google Scholar] [CrossRef]
- Yang, Z.; Kubant, R.; Cho, C.E.; Kranenburg, E.; Beaudry, J.; Bottiglieri, T.; Anderson, G.H. Micronutrients in High-Fat Diet Modify Insulin Resistance and Its Regulatory Genes in Adult Male Mice. Mol. Nutr. Food Res. 2023, 67, e2300199. [Google Scholar] [CrossRef]
- Zhang, Y.; Proenca, R.; Maffei, M.; Barone, M.; Leopold, L.; Friedman, J.M. Positional cloning of the mouse obese gene and its human homologue. Nature 1994, 372, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Campfield, L.A.; Smith, F.J.; Burn, P. The OB protein (leptin) pathway--a link between adipose tissue mass and central neural networks. Horm. Metab. Res. 1996, 28, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Hamann, A.; Matthaei, S. Regulation of energy balance by leptin. Exp. Clin. Endocrinol. Diabetes 1996, 104, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.M.; Halaas, J.L. Leptin and the regulation of body weight in mammals. Nature 1998, 395, 763–770. [Google Scholar] [CrossRef]
- Fischer, A.W.; Cannon, B.; Nedergaard, J. Leptin: Is It Thermogenic? Endocr. Rev. 2020, 41, 232–260. [Google Scholar] [CrossRef]
- Fischer, A.W.; Hoefig, C.S.; Abreu-Vieira, G.; de Jong, J.M.A.; Petrovic, N.; Mittag, J.; Cannon, B.; Nedergaard, J. Leptin Raises Defended Body Temperature without Activating Thermogenesis. Cell Rep. 2016, 14, 1621–1631. [Google Scholar] [CrossRef] [PubMed]
- Cannon, B.; Nedergaard, J. Brown adipose tissue: Function and physiological significance. Physiol. Rev. 2004, 84, 277–359. [Google Scholar] [CrossRef] [PubMed]
- Castro, É.; Vieira, T.S.; Oliveira, T.E.; Ortiz-Silva, M.; Andrade, M.L.; Tomazelli, C.A.; Peixoto, A.S.; Sobrinho, C.R.; Moreno, M.F.; Gilio, G.R.; et al. Adipocyte-specific mTORC2 deficiency impairs BAT and iWAT thermogenic capacity without affecting glucose uptake and energy expenditure in cold-acclimated mice. Am. J. Physiol. Endocrinol. Metab. 2021, 321, E592–E605. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, L.F.G.; Castro, É.; Eichler, R.; Moreno, M.F.; de Sousa, É.; Jardim, G.F.R.; Peixoto, Á.S.; Moraes, M.N.; Castrucci, A.M.L.; Nedergaard, J.; et al. Cold acclimation and pioglitazone combined increase thermogenic capacity of brown and white adipose tissues but this does not translate into higher energy expenditure in mice. Am. J. Physiol. Endocrinol. Metab. 2023, 324, E358–E373. [Google Scholar] [CrossRef] [PubMed]
- Silvério, R.; Barth, R.; Heimann, A.S.; Reckziegel, P.; Dos Santos, G.J.; Romero-Zerbo, S.Y.; Bermúdez-Silva, F.J.; Rafacho, A.; Ferro, E.S. Pep19 Has a Positive Effect on Insulin Sensitivity and Ameliorates Both Hepatic and Adipose Tissue Phenotype of Diet-Induced Obese Mice. Int. J. Mol. Sci. 2022, 23, 4082. [Google Scholar] [CrossRef]
- Reckziegel, P.; Festuccia, W.T.; Britto, L.R.G.; Jang, K.L.L.; Romao, C.M.; Heimann, J.C.; Fogaca, M.V.; Rodrigues, N.S.; Silva, N.R.; Guimaraes, F.S.; et al. A novel peptide that improves metabolic parameters without adverse central nervous system effects. Sci. Rep. 2017, 7, 14781. [Google Scholar] [CrossRef]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef]
- Lönnqvist, F.; Nordfors, L.; Jansson, M.; Thörne, A.; Schalling, M.; Arner, P. Leptin secretion from adipose tissue in women. Relationship to plasma levels and gene expression. J. Clin. Invest. 1997, 99, 2398–2404. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.0031. [Google Scholar] [CrossRef]
- Gonzalez-Carter, D.; Goode, A.; Fiammengo, R.; Dunlop, I.E.; Dexter, D.T.; Porter, A. Inhibition of Leptin–ObR interaction does not prevent leptin translocation across a human blood–brain barrier model. J. Neuroendocrinol. 2016, 28. [Google Scholar] [CrossRef]
- Sahu, A.; Carraway, R.E.; Wang, Y.P. Evidence that neurotensin mediates the central effect of leptin on food intake in rat. Brain Res. 2001, 888, 343–347. [Google Scholar] [CrossRef]
- Tsujino, N.; Yamanaka, A.; Ichiki, K.; Muraki, Y.; Kilduff, T.S.; Yagami, K.; Takahashi, S.; Goto, K.; Sakurai, T. Cholecystokinin activates orexin/hypocretin neurons through the cholecystokinin A receptor. J. Neurosci. 2005, 25, 7459–7469. [Google Scholar] [CrossRef]
- Checler, F.; Ferro, E.S. Neurolysin: From Initial Detection to Latest Advances. Neurochem. Res. 2018, 43, 2017–2024. [Google Scholar] [CrossRef]
- Song, E.S.; Rodgers, D.W.; Hersh, L.B. Insulin-degrading enzyme is not secreted from cultured cells. Sci. Rep. 2018, 8, 2335. [Google Scholar] [CrossRef] [PubMed]
- Berti, D.A.; Russo, L.C.; Castro, L.M.; Cruz, L.; Gozzo, F.C.; Heimann, J.C.; Lima, F.B.; Oliveira, A.C.; Andreotti, S.; Prada, P.O.; et al. Identification of intracellular peptides in rat adipose tissue: Insights into insulin resistance. Proteomics 2012, 12, 2668–2681. [Google Scholar] [CrossRef]
- Heimann, A.S.; Favarato, M.H.; Gozzo, F.C.; Rioli, V.; Carreno, F.R.; Eberlin, M.N.; Ferro, E.S.; Krege, J.H.; Krieger, J.E. ACE gene titration in mice uncovers a new mechanism for ACE on the control of body weight. Physiol. Genom. 2005, 20, 173–182. [Google Scholar] [CrossRef]
- Iwanaga, T.; Nio-Kobayashi, J. Cellular expression of CD26/dipeptidyl peptidase IV. Biomed. Res. 2021, 42, 229–237. [Google Scholar] [CrossRef]
- Lentzen, H.; Reinsch, I.; Linke, J. Angiotensin-converting enzyme, enkephalinase A and aminopeptidases in the breakdown of enkephalin--studies in cell cultures. Clin. Exp. Hypertens. A 1984, 6, 1829–1832. [Google Scholar] [CrossRef] [PubMed]
- Shimasaki, T.; Masaki, T.; Mitsutomi, K.; Ueno, D.; Gotoh, K.; Chiba, S.; Kakuma, T.; Yoshimatsu, H. The dipeptidyl peptidase-4 inhibitor des-fluoro-sitagliptin regulates brown adipose tissue uncoupling protein levels in mice with diet-induced obesity. PLoS ONE 2013, 8, e63626. [Google Scholar] [CrossRef]
- Chae, Y.N.; Kim, T.H.; Kim, M.K.; Shin, C.Y.; Jung, I.H.; Sohn, Y.S.; Son, M.H. Beneficial Effects of Evogliptin, a Novel Dipeptidyl Peptidase 4 Inhibitor, on Adiposity with Increased Ppargc1a in White Adipose Tissue in Obese Mice. PLoS ONE 2015, 10, e0144064. [Google Scholar] [CrossRef] [PubMed]
- João, A.L.; Reis, F.; Fernandes, R. The incretin system ABCs in obesity and diabetes - novel therapeutic strategies for weight loss and beyond. Obes. Rev. 2016, 17, 553–572. [Google Scholar] [CrossRef]
- Popoviciu, M.S.; Păduraru, L.; Yahya, G.; Metwally, K.; Cavalu, S. Emerging Role of GLP-1 Agonists in Obesity: A Comprehensive Review of Randomised Controlled Trials. Int. J. Mol. Sci. 2023, 24, 10449. [Google Scholar] [CrossRef]
- Kim, H.S.; Jung, C.H. Oral Semaglutide, the First Ingestible Glucagon-Like Peptide-1 Receptor Agonist: Could It Be a Magic Bullet for Type 2 Diabetes? Int. J. Mol. Sci. 2021, 22, 9936. [Google Scholar] [CrossRef]
- Rakhat, Y.; Wang, L.; Han, W.; Rustemova, A.; Kulzhanova, N.; Yamada, Y.; Yabe, D.; Seino, Y.; Yada, T. Oral Semaglutide under Human Protocols and Doses Regulates Food Intake, Body Weight, and Glycemia in Diet-Induced Obese Mice. Nutrients 2023, 15, 3765. [Google Scholar] [CrossRef]
- Queathem, E.D.; Welly, R.J.; Clart, L.M.; Rowles, C.C.; Timmons, H.; Fitzgerald, M.; Eichen, P.A.; Lubahn, D.B.; Vieira-Potter, V.J. White Adipose Tissue Depots Respond to Chronic Beta-3 Adrenergic Receptor Activation in a Sexually Dimorphic and Depot Divergent Manner. Cells 2021, 10, 3453. [Google Scholar] [CrossRef]
- Kim, S.-N.; Jung, Y.-S.; Kwon, H.-J.; Seong, J.K.; Granneman, J.G.; Lee, Y.-H. Sex differences in sympathetic innervation and browning of white adipose tissue of mice. Biol. Sex. Differ. 2016, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Black, M.H.; Watanabe, R.M.; Trigo, E.; Takayanagi, M.; Lawrence, J.M.; Buchanan, T.A.; Xiang, A.H. High-fat diet is associated with obesity-mediated insulin resistance and β-cell dysfunction in Mexican Americans. J. Nutr. 2013, 143, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Camporez, J.P.G.; Jornayvaz, F.R.; Lee, H.-Y.; Kanda, S.; Guigni, B.A.; Kahn, M.; Samuel, V.T.; Carvalho, C.R.O.; Petersen, K.F.; Jurczak, M.J.; et al. Cellular Mechanism by Which Estradiol Protects Female Ovariectomized Mice From High-Fat Diet-Induced Hepatic and Muscle Insulin Resistance. Endocrinology 2013, 154, 1021–1028. [Google Scholar] [CrossRef]
- Silva, R.N.O.; Llanos, R.P.; Eichler, R.A.S.; Oliveira, T.B.; Gozzo, F.C.; Festuccia, W.T.; Ferro, E.S. New Intracellular Peptide Derived from Hemoglobin Alpha Chain Induces Glucose Uptake and Reduces Blood Glycemia. Pharmaceutics 2021, 13, 2175. [Google Scholar] [CrossRef]
- Heimann, A.S.; Gupta, A.; Gomes, I.; Rayees, R.; Schlessinger, A.; Ferro, E.S.; Unterwald, E.M.; Devi, L.A. Generation of G protein-coupled receptor antibodies differentially sensitive to conformational states. PLoS ONE 2017, 12, e0187306. [Google Scholar] [CrossRef]
- Gupta, A.; Heimann, A.S.; Gomes, I.; Devi, L.A. Antibodies against G-protein coupled receptors: Novel uses in screening and drug development. Comb. Chem. High. Throughput Screen. 2008, 11, 463–467. [Google Scholar] [CrossRef] [PubMed]

| Consumption: | WT SD | WT HD | Nln-/- SD | Nln-/- HD |
|---|---|---|---|---|
| Food (g/day/animal) | 5.02 ± 0.27 | 1.87 ± 0.25 * | 5.9 ± 0.46 | 2.57 ± 0.10 *.# |
| Water (mL/day/animal) | 1.3 ± 0.15 | 1.5 ± 0.05 | 1.5 ± 0.09 | 1.3 ± 0.16 |
| Calories (Kcal/day/animal) | 19.08 ± 0.95 | 15.98 ± 2.07 | 22.43 ± 2.24 | 21.97 ± 1.05 |
| Consumption: | WT SD | WT HD | Nln-/- SD | Nln-/- HD |
|---|---|---|---|---|
| Food (g/day/animal) | 3.91 ± 0.59 | 1.39 ± 0.15 * | 4.38 ± 0.20 | 2.09 ± 0.16 *. # |
| Water (mL/day/animal) | 1.6 ± 0.1 | 2.0 ± 0.2 * | 1.7 ± 0.2 | 1.0 ± 0.1 # |
| Calories (Kcal/day/animal) | 14.3 ± 1.56 | 11.88 ± 1.20 | 16.6 ± 0.83 | 17.86 ± 0.90 |
| Tissues | WT SD | WT HD | Nln-/- SD | Nln-/- HD |
|---|---|---|---|---|
| Liver | 4.48 ± 0.09 | 2.95 ± 0.07 * | 4.70 ± 0.16 | 2.90 ± 0.10 * |
| Inguinal Fat | 0.80 ± 0.06 | 2.24 ± 0.30 * | 0.76 ± 0.06 | 2.98 ± 0.29 * |
| Gonadal Fat | 1.31 ± 0.12 | 3.67 ± 0.47 * | 0.92 ± 0.09 | 4.37 ± 0.25 * |
| Retroperitoneal Fat | 0.38 ± 0.08 | 1.34 ± 0.13 * | 0.33 ± 0.06 | 2.36 ± 0.13 *,# |
| Gastrocnemius muscle | 1.15 ± 0.02 | 0.96 ± 0.03 * | 1.07 ± 0.04 | 0.89 ± 0.08* |
| Tissues | WT SD | WT HD | Nln-/- SD | Nln-/- HD |
|---|---|---|---|---|
| Liver | 4.22 ± 0.35 | 3.45 ± 0.09 * | 4.4 ± 0.18 | 3.32 ± 0.09 * |
| Inguinal Fat | 1.01 ± 0.11 | 1.92 ± 0.13 * | 0.86 ± 0.07 | 3.26 ± 0.19 *,# |
| Gonadal Fat | 1.10 ± 0.17 | 2.76 ± 0.26 * | 0.61 ± 0.11 | 4.29 ± 0.41 *,# |
| Retroperitoneal Fat | 0.39 ± 0.11 | 1.16 ± 0.11 * | 0.27 ± 0.06 | 2.55 ± 0.23 *,# |
| Gastrocnemius muscle | 1.10 ± 0.02 | 1.05 ± 0.02 | 1.13 ± 0.08 | 0.94 ± 0.04 * |
| Gene | WT SD | WT HD | Nln-/- SD | Nln-/- HD |
|---|---|---|---|---|
| PPAR-alpha | 1.04 ± 0.22 | 54.23 ± 6.66 * | 5.46 ± 0.85 # | 47.71 ± 6.54 * |
| PPAR-gamma | 1.02 ± 0.05 | 0.82 ± 0.12 | 0.51 ± 0.01 # | 0.43 ± 0.10 # |
| FAS | 1.01 ± 0.04 | 0.8 ± 0.15 | 0.98 ± 0.17 | 1.04 ± 0.17 |
| LPL | 1.02 ± 0.18 | 7.64 ± 0.92 * | 4.4 ± 1.29 # | 7.13 ± 1.54 * |
| FABP4 | 1.02 ± 0.15 | 1.84 ± 0.27 | 1.48 ± 0.29 | 2.94 ± 0.78 * |
| Leptin | 1.02 ± 0.14 | 16.16 ± 3.83 * | 1.54 ± 0.49 | 46.16 ± 6.89 *,# |
| CD206 | 1.02 ± 0.04 | 3.63 ± 1.33 * | 1.86 ± 0.30 | 4.07 ± 0.42 * |
| CD11C | 1.05 ± 0.26 | 7.28 ± 2.46 * | 1.43 ± 0.6 | 14.75 ± 6.05 * |
| PGC-1 alpha | 1.02 ± 0.07 | 0.74 ± 0.05 | 0.8 ± 0.08 | 0.79 ± 0.20 |
| F4-80 | 1.02 ± 0.07 | 2.87 ± 1.26 * | 1.0 ± 0.14 | 3.98 ± 0.63 * |
| CD36 | 1.00 ± 0.08 | 2.33 ± 0.57 | 1.88 ± 0.41 | 2.07 ± 0.48 |
| ADBR3 | 1.02 ± 0.18 | 0.59 ± 0.08 * | 0.48 ± 0.13 # | 0.32 ± 0.09 # |
| NEP | 1.00 ± 0.01 | 1.22 ± 0.09 | 1.37 ± 0.27 | 1.73 ± 0.27 |
| POP | 1.00 ± 0.06 | 1.33 ± 0.30 | 1.03 ± 0.16 | 1.33 ± 0.13 |
| DPP4 | 1.00 ± 0.03 | 1.72 ± 0.27 | 1.70 ± 0.28 | 1.98 ± 0.50 |
| IDE | 1.01 ± 0.09 | 6.46 ± 0.93 * | 4.59 ± 0.97 # | 8.33 ± 0.36 * |
| ACE1 | 1.03 ± 0.21 | 2.30 ± 0.57 * | 2.06 ± 0.30 # | 2.89 ± 0.39 * |
| β5-Prot | 1.01 ± 0.13 | 2.72 ± 0.31 * | 2.02 ± 0.41 # | 4.03 ± 0.40 *,# |
| Gene | WT SD | WT HD | Nln-/- SD | Nln-/- HD |
|---|---|---|---|---|
| PPAR-alpha | 1.24 ± 0.60 | 29.7 ± 3.71 * | 5.28 ± 1.58 # | 25.30 ± 5.59 * |
| PPAR-gamma | 1.16 ± 0.41 | 0.86 ± 0.18 | 0.51 ± 0.11 # | 0.39 ± 0.06 # |
| FAS | 1.10 ± 0.33 | 0.43 ± 0.04 | 0.69 ± 0.06 | 0.55 ± 0.11 |
| LPL | 1.09 ± 0.29 | 23.76 ± 3.41 * | 14.45 ± 3.50 # | 29.17 ± 6.84 * |
| FABP4 | 1.03 ± 0.17 | 1.52 ± 0.28 | 1.39 ± 0.21 | 2.25 ± 0.65 * |
| Leptin | 1.06 ± 0.23 | 9.09 ± 2.66 * | 2.21 ± 0.91 | 24.11 ± 4.69 *,# |
| CD206 | 1.04 ± 0.23 | 2.99 ± 1.26 * | 2.03 ± 0.43 | 3.04 ± 0.05 * |
| CD11C | 1.03 ± 0.17 | 6.18 ± 2.28 * | 2.20 ± 1.37 | 12.63 ± 5.55 * |
| PGC-1 alpha | 1.03 ± 0.18 | 0.58 ± 0.01 | 1.04 ± 0.23 | 0.63 ± 0.21 |
| F4-80 | 1.07 ± 0.27 | 3.67 ± 1.81 * | 1.62 ± 0.17 | 4.87 ± 1.18 * |
| CD36 | 1.04 ± 0.21 | 1.83 ± 0.34 | 1.28 ± 0.38 | 1.57 ± 0.41 |
| ADBR3 | 1.21 ± 0.55 | 0.69 ± 0.04 * | 0.67 ± 0.07 # | 0.34 ± 0.05 # |
| NEP | 1.06 ± 0.26 | 0.99 ± 0.13 | 1.34 ± 0.19 | 1.28 ± 0.04 |
| POP | 1.12 ± 0.35 | 1.30 ± 0.35 | 1.23 ± 0.12 | 1.21 ± 0.15 |
| DPP4 | 1.08 ± 0.29 | 1.48 ± 0.27 | 2.16 ± 0.56 | 1.52 ± 0.19 |
| IDE | 1.11 ± 0.31 | 3.53 ± 0.48 * | 2.93 ± 0.43 # | 4.31 ± 0.36 * |
| ACE1 | 1.00 ± 0.11 | 1.76 ± 0.51 * | 2.09 ± 0.51 # | 2.01 ± 0.02 |
| β5-Prot | 1.00 ± 0.10 | 1.82 ± 0.21 * | 1.57 ± 0.09 # | 2.51 ± 0.07 *,# |
| Gene | WT SD | WT HD | Nln-/- SD | Nln-/- HD |
|---|---|---|---|---|
| PPAR-alpha | 2.75 ± 0.17 | 12.31 ± 3.74 * | 6.25 ± 1.95 # | 16.13 ± 1.17 * |
| PPAR-gamma | 0.43 ± 0.10 | 0.77 ± 0.15 | 0.63 ± 0.11 | 0.60 ± 0.06 |
| FAS | 0.73 ± 0.07 | 0.36 ± 0.02 * | 0.98 ± 0.08 | 0.72 ± 0.10 # |
| LPL | 1.07 ± 0.08 | 4.19 ± 0.55 * | 0.56 ± 0.1 | 4.86 ± 0.37 * |
| FABP4 | 0.72 ± 0.04 | 1.61 ± 0.29 * | 0.55 ± 0.14 | 1.27 ± 0.12 * |
| Leptin | 2.91 ± 0.84 | 8.02 ± 1.29 * | 2.94 ± 0.63 | 25.44 ± 7.42 *,# |
| CD206 | 0.60 ± 0.02 | 1.98 ± 0.28 * | 0.71 ± 0.14 | 2.28 ± 0.29 * |
| CD11C | 0.84 ± 0.15 | 2.09 ± 0.34 * | 0.62 ± 0.13 | 4.42 ± 0.68 * |
| PGC-1 alpha | 0.93 ± 0.12 | 0.57 ± 0.12 | 0.46 ± 0.11# | 0.57 ± 0.07 |
| F4-80 | 0.26 ± 0.03 | 1.25 ± 0.08 * | 0.45 ± 0.07 | 1.80 ± 0.23 * |
| CD36 | 0.69 ± 0.05 | 2.18 ± 0.42 * | 0.57 ± 0.14 | 2.45 ± 0.55 * |
| ADBR3 | 0.39 ± 0.07 | 0.91 ± 0.19 * | 0.40 ± 0.08 | 0.36 ± 0.06 # |
| NEP | 0.79 ± 0.10 | 1.14 ± 0.16 | 0.90 ± 0.11 | 1.29 ± 0.11 |
| POP | 0.89 ± 0.16 | 0.96 ± 0.12 | 0.96 ± 0.10 | 1.34 ± 0.13 |
| DPP4 | 0.82 ± 0.21 | 1.35 ± 0.27 * | 0.87 ± 0.12 | 1.45 ± 0.20 * |
| IDE | 4.85 ± 1.14 | 5.98 ± 0.65 | 4.23 ± 0.90 | 6.96 ± 0.93 |
| ACE1 | 0.62 ± 0.09 | 1.15 ± 0.25 * | 0.60 ± 0.10 | 1.45 ± 0.17 * |
| β5-Prot | 1.16 ± 0.11 | 2.16 ± 0.17 | 1.92 ± 0.26 | 3.32 ± 0.44 |
| Gene | WT SD | WT HD | Nln-/- SD | Nln-/- HD |
|---|---|---|---|---|
| PPAR-alpha | 3.29 ± 1.21 | 9.66 ± 0.38 * | 5.67 ± 1.75 | 17.35 ± 1.48 * |
| PPAR-gamma | 0.97 ± 0.29 | 1.16 ± 0.26 | 0.66± 0.14 | 1.04 ± 0.23 |
| FAS | 0.90 ± 0.05 | 0.33 ± 0.05 * | 1.13 ± 0.34 | 0.71 ± 0.12 # |
| LPL | 11.91 ± 2.98 | 24.08 ± 2.29 * | 3.94 ± 1.87 | 26.48 ± 6.07 * |
| FABP4 | 1.30 ± 0.55 | 1.97 ± 0.34 | 0.94 ± 0.21 | 1.53 ± 0.22 * |
| Leptin | 2.60 ± 0.71 | 7.34 ± 0.45 * | 2.70 ± 0.59 | 16.71 ± 3.36 *,# |
| CD206 | 0.97 ± 0.09 | 2.31 ± 0.25 * | 0.81 ± 0.24 | 3.03 ± 0.63 * |
| CD11C | 1.56 ± 0.13 | 2.61 ± 0.42 * | 0.85 ± 0.16 | 6.06 ± 1.31 * |
| PGC-1 alpha | 1.53 ± 0.11 | 0.98 ± 0.19 | 0.69 ± 0.16# | 1.27 ± 0.47 |
| F4-80 | 0.86 ± 0.13 | 2.05 ± 0.30 * | 0.70 ± 0.17 | 3.09 ± 0.38 * |
| CD36 | 1.51 ± 0.28 | 2.66 ± 0.47 * | 0.63 ± 0.19 | 3.35 ± 0.84 * |
| ADBR3 | 1.18 ± 0.60 | 1.57 ± 0.31 | 0.51 ± 0.07 | 0.61 ± 0.11 # |
| NEP | 1.20 ± 0.18 | 1.33 ± 0.20 | 1.36 ± 0.17 | 1.91 ± 0.42 |
| POP | 1.49 ± 0.38 | 1.33 ± 0.19 | 1.48 ± 0.07 | 2.36 ± 0.67 |
| DPP4 | 0.99 ± 0.19 | 1.70 ± 0.34 * | 1.00 ± 0.09 | 2.19 ± 0.68 * |
| IDE | 4.93 ± 0.85 | 4.56 ± 0.86 | 5.60 ± 0.79 | 7.22 ± 2.47 |
| ACE1 | 0.95 ± 0.06 | 1.38 ± 0.30 * | 0.99 ± 0.01 | 1.88 ± 0.51 * |
| β5-Prot | 2.13 ± 0.47 | 2.05 ± 0.21 | 1.49 ± 0.27 | 3.25 ± 0.72 |
| Gene | Sequence | Amplicon (bp) | Access Number |
|---|---|---|---|
| Peroxisome proliferator activated receptor gamma (PPAR-gamma) | Fwd: ATCTTAACTGCCGGATCC Rev: CAAACCTGATGGCATTGTGAG | 102 | NM_001127330.2 |
| Peroxisome proliferator activated receptor alpha (PPAR-alpha) | Fwd: TGCAATTCGCTTTGGAA Rev: CTTGCCCAGAGATTTGAGGT | 118 | NM_011144.6 |
| Fatty acid synthase (FAS) | Fwd: GATTCGGTGTCTGCTGTC Rev: CATGCTTTAGCACCTGCTGT | 95 | NM_007988.3 |
| Lipoprotein lipase (LPL) | Fwd: GTCTGGCCACTGGACAAA Rev: CCCACTTTCAAACACCCAAA | 122 | NM_008509.2 |
| Leptin | Fwd: CCAGGATGACACCAAAACCCT Rev: TGAAGTCCAAGCCAGTGACC | 107 | NM_008493.3 |
| CD36 molecule (CD36) | Fwd: GATTGGTTGAGACCCCG Rev: GCTCCACACATTTCAGAAGGC | 174 | NM_001159558.1 |
| Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC 1 alpha) | Fwd: AAGGGCCAAACAGAGAG Rev: AGTAAATCACACGGCGCTCTT | 63 | NM_008904.3 |
| Mannose receptor (CD206) | Fwd: TGTGTTCAGCTATTGGACGC Rev: CGGAATTTCTGGGATTCAGCTTC | 133 | NM_008625.2 |
| Integrin alpha X (CD11C) | Fwd: CTGGATAGCCTTTCTTCTGCTG Rev: GCACACTGTGTCCGAACTCA | 113 | NM_021334.3 |
| Adhesion G protein-coupled receptor E1 (F4/80) | Fwd: AACATGCAACCTGCCACAAC Rev: TTCACAGGATTCGTCCAGGC | 110 | NM_010130.4 |
| Fatty acid-binding protein 4 (FABP4) | Fwd: CGCAGACGACAGGAAGGT Rev: TTCCATCCCACTTCTGCAC | 77 | NM_024406.3 |
| Ribosomal protein L19 (RPL19) | Fwd: CAATGCCAACTCCCGTCA Rev: GTGTTTTTCCGGCAACGAG | 102 | NM_009078.2 |
| Adrenergic receptor, beta 3 (ADBR3) | Fwd: ACCCTGATGATCGACATGTTCC Rev: GCCATAGTGAGGAGACAGGG | 129 | NM_013462.3 |
| Neprilysin (NEP) | Fwd: CCTGAACTTTGCCCAGGTGT Rev: GCGGCAATGAAAGGCATCTG | 148 | NM_001289462.1 |
| Angiotensin I converting enzyme (ACE1) | Fwd: ACCCTAGGACCTGCCAATCT Rev: CGTGAGGAAGCCAGGATGTT | 164 | NM_207624.5 |
| Prolyl oligopeptidase (POP) | Fwd: GGGTGCTCCGACACTAAACA Rev: GACGGGTACTGGATGTCGTC | 98 | NM_011156.3 |
| Insulin degrading enzyme (IDE) | Fwd: GTCCATGTTCTTGCCAGGGA Rev: TTCACGAGGGGAAACAGTGG | 161 | NM_031156.3 |
| Dipeptidyl peptidase 4 (DPP4) | Fwd: GACGGCAGAGGAAGTGGTT Rev: CGCTTGCTATCCACAAATCCC | 134 | NM_010074.3 |
| Proteasome subunit beta 5 (Prot-β5) | Fwd: CCAAACTGCTCGCTAACATGG Rev: GTTCCCCTCGCTGTCTACG | 119 | NM_011186.1 |
| Ribosomal protein L19 (RPL19) | Fwd: CAATGCCAACTCCCGTCA Rev: GTGTTTTTCCGGCAACGAG | 102 | NM_009078.2 |
| Cyclophilin A | Fwd: TATCTGCACTGCCAAGACTGAGT Rev: CTTCTTGCTGGTCTTGCCATTCC | 127 | NM_008907.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caprioli, B.; Eichler, R.A.S.; Silva, R.N.O.; Martucci, L.F.; Reckziegel, P.; Ferro, E.S. Neurolysin Knockout Mice in a Diet-Induced Obesity Model. Int. J. Mol. Sci. 2023, 24, 15190. https://doi.org/10.3390/ijms242015190
Caprioli B, Eichler RAS, Silva RNO, Martucci LF, Reckziegel P, Ferro ES. Neurolysin Knockout Mice in a Diet-Induced Obesity Model. International Journal of Molecular Sciences. 2023; 24(20):15190. https://doi.org/10.3390/ijms242015190
Chicago/Turabian StyleCaprioli, Bruna, Rosangela A. S. Eichler, Renée N. O. Silva, Luiz Felipe Martucci, Patricia Reckziegel, and Emer S. Ferro. 2023. "Neurolysin Knockout Mice in a Diet-Induced Obesity Model" International Journal of Molecular Sciences 24, no. 20: 15190. https://doi.org/10.3390/ijms242015190
APA StyleCaprioli, B., Eichler, R. A. S., Silva, R. N. O., Martucci, L. F., Reckziegel, P., & Ferro, E. S. (2023). Neurolysin Knockout Mice in a Diet-Induced Obesity Model. International Journal of Molecular Sciences, 24(20), 15190. https://doi.org/10.3390/ijms242015190






