Spatiotemporal Patterns of Five Small Heat Shock Protein Genes in Hyphantria cunea in Response to Thermal Stress
Abstract
:1. Introduction
2. Results
2.1. Molecular Cloning and Sequence Analysis
2.2. Multiple Sequence Alignments
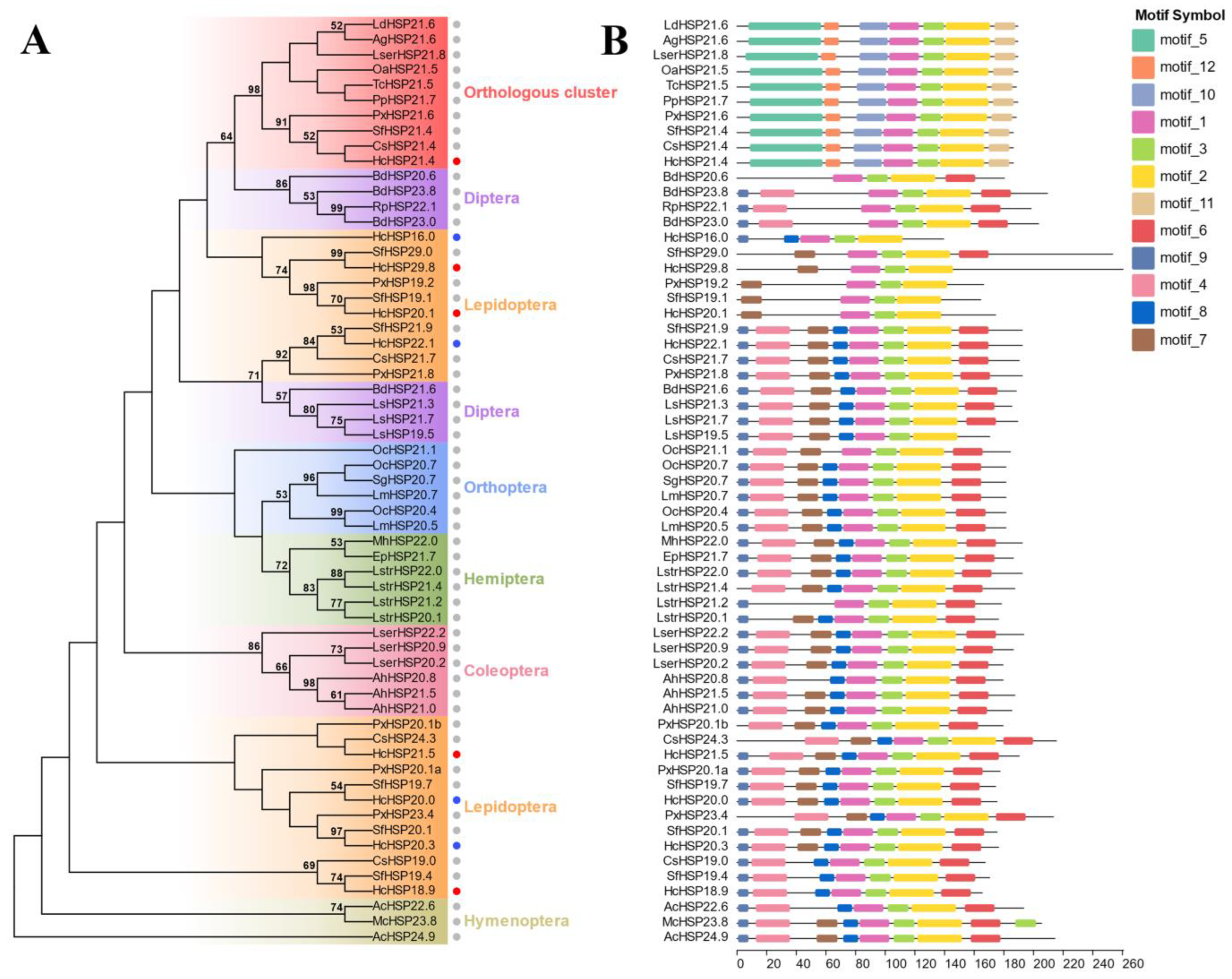
2.3. Phylogenetic Analysis
2.4. Temporal and Spatial Expression Patterns of HcHSPs under Normal Conditions
2.5. Heat-Induced Expression of HcHSPs on the Temporal and Spatial Scales
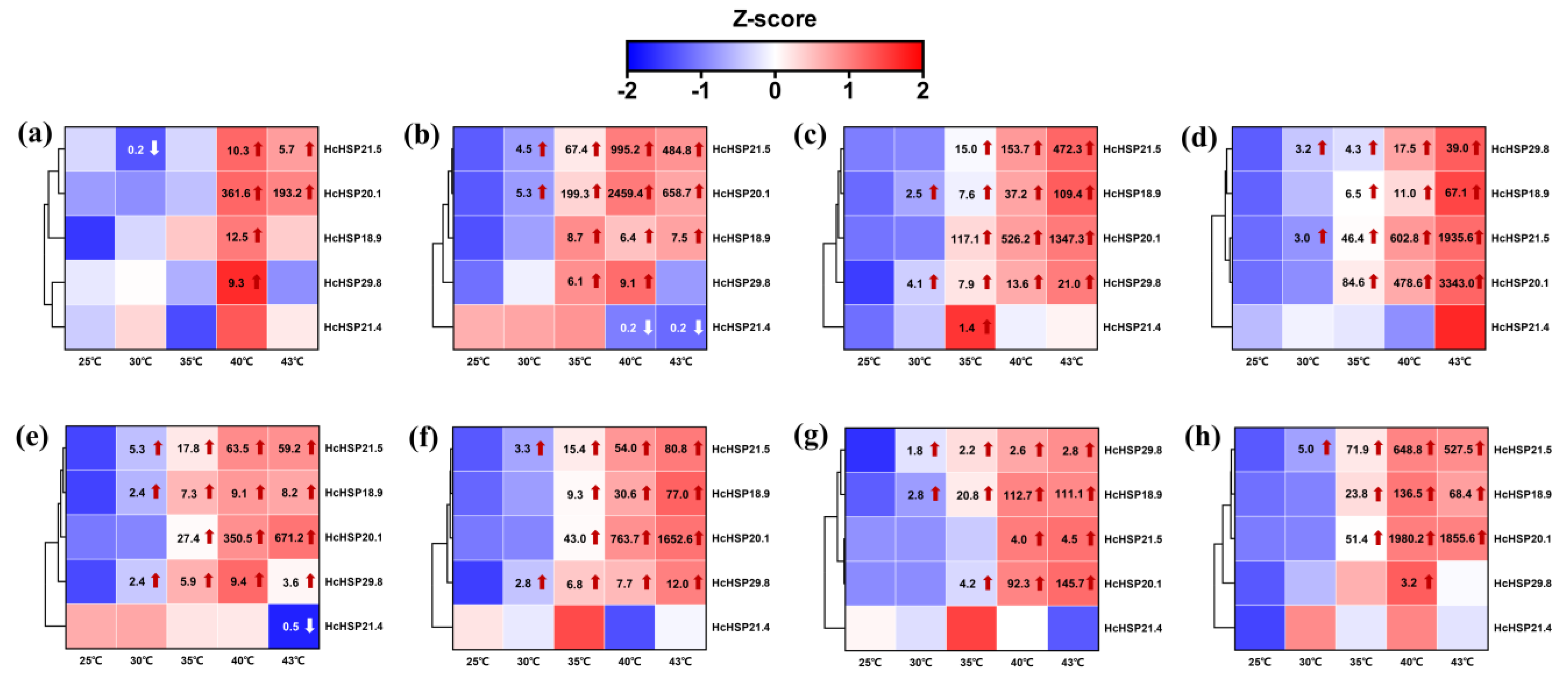
2.5.1. Heat-Induced Expression Profiles of HcHSPs in Different Developmental Stages
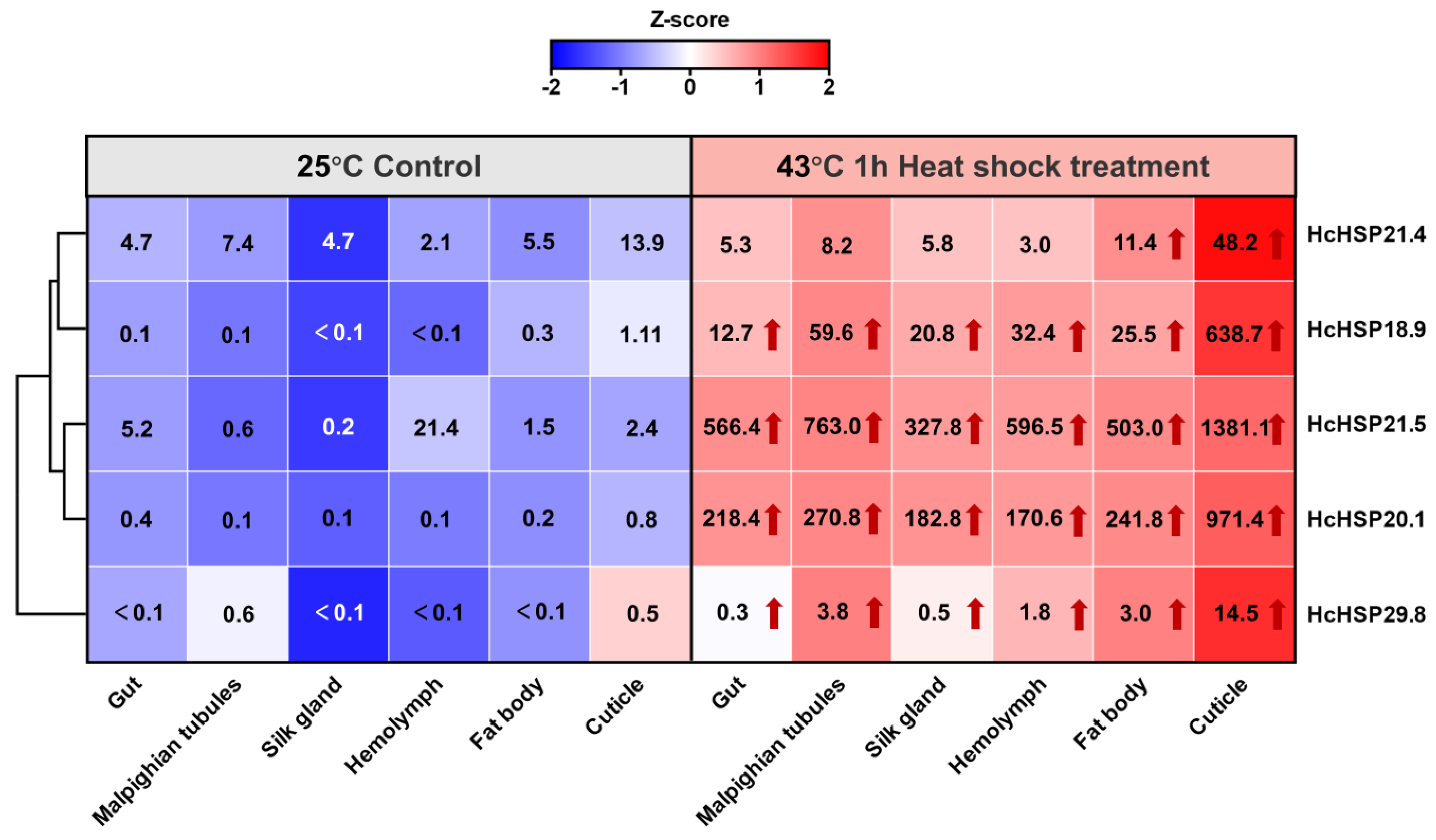
2.5.2. Heat-Induced Expression Profiles of HcHSPs in Various Tissues
3. Discussion
4. Materials and Methods
4.1. Insects
4.2. Sample Preparation
4.3. RNA Extraction and cDNA Synthesis
4.4. HcHSPs Sequence Cloning
4.5. Bioinformatics and Phylogenetic Analyses
4.6. RT-qPCR
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lindquist, S.; Craig, E.A. The Heat-Shock Proteins. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef]
- Ritossa, F. A New Puffing Pattern Induced by Temperature Shock and DNP in Drosophila. Experientia 1962, 18, 571–573. [Google Scholar] [CrossRef]
- Zagoriti, Z.; Georgitsi, M.; Giannakopoulou, O.; Ntellos, F.; Tzartos, S.J.; Patrinos, G.P.; Poulas, K. Genetics of Myasthenia Gravis: A Case-Control Association Study in the Hellenic Population. J. Immunol. Res. 2012, 2012, e484919. [Google Scholar] [CrossRef] [PubMed]
- Parsell, D.A.; Lindquist, S. The Function of Heat-Shock Proteins in Stress Tolerance: Degradation and Reactivation of Damaged Proteins. Annu. Rev. Genet. 1993, 27, 437–496. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.E.; Zhang, Y.L. Identification of Multiple Small Heat-Shock Protein Genes in Plutella Xylostella (L.) and Their Expression Profiles in Response to Abiotic Stresses. Cell Stress Chaperones 2015, 20, 23–35. [Google Scholar] [CrossRef]
- Miano, F.N.; Jiang, T.; Zhang, J.; Zhang, W.-N.; Peng, Y.; Xiao, H.-J. Identification and Up-Regulation of Three Small Heat Shock Proteins in Summer and Winter Diapause in Response to Temperature Stress in Pieris Melete. Int. J. Biol. Macromol. 2022, 209, 1144–1154. [Google Scholar] [CrossRef]
- Feder, M.E.; Hofmann, G.E. Heat-Shock Proteins, Molecular Chaperones, and the Stress Response: Evolutionary and Ecological Physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef]
- King, A.M.; MacRae, T.H. Insect Heat Shock Proteins During Stress and Diapause. Annu. Rev. Entomol. 2015, 60, 59–75. [Google Scholar] [CrossRef]
- Haslbeck, M. SHsps and Their Role in the Chaperone Network. Cell. Mol. Life Sci. 2002, 59, 1649–1657. [Google Scholar] [CrossRef]
- Franck, E.; Madsen, O.; van Rheede, T.; Ricard, G.; Huynen, M.A.; de Jong, W.W. Evolutionary Diversity of Vertebrate Small Heat Shock Proteins. J. Mol. Evol. 2004, 59, 792–805. [Google Scholar] [CrossRef]
- Poulain, P.; Gelly, J.C.; Flatters, D. Detection and Architecture of Small Heat Shock Protein Monomers. PLoS ONE 2010, 5, e9990. [Google Scholar] [CrossRef]
- Basha, E.; O’Neill, H.; Vierling, E. Small Heat Shock Proteins and α-Crystallins: Dynamic Proteins with Flexible Functions. Trends Biochem. Sci. 2012, 37, 106–117. [Google Scholar] [CrossRef]
- Basha, E.; Friedrich, K.L.; Vierling, E. The N-Terminal Arm of Small Heat Shock Proteins Is Important for Both Chaperone Activity and Substrate Specificity. J. Biol. Chem. 2006, 281, 39943–39952. [Google Scholar] [CrossRef]
- Horwitz, J. Alpha-Crystallin Can Function as a Molecular Chaperone. Proc. Natl. Acad. Sci. USA 1992, 89, 10449–10453. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.W.; Song, H.X.; Chang, Y.W.; Yang, F.; Xie, H.F.; Gong, W.R.; Du, Y.Z. Identification, Expression Analysis and Functional Verification of Two Genes Encoding Small Heat Shock Proteins in the Western Flower Thrips, Frankliniella Occidentalis (Pergande). Int. J. Biol. Macromol. 2022, 211, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.H.; Wang, C.Z.; Kang, L. Cloning and Expression of Five Heat Shock Protein Genes in Relation to Cold Hardening and Development in the Leafminer, Liriomyza Sativa. J. Insect Physiol. 2009, 55, 279–285. [Google Scholar] [CrossRef]
- Sonoda, S.; Ashfaq, M.; Tsumuki, H. Cloning and Nucleonde Sequencing of Three Heat Shock Protein Genes (Hsp90, Hsc70, and Hsp19.5) from the Diamondback Moth, Plutella Xylostella (L.) and Their Expression in Relation to Developmental Stage and Temperature. Arch. Insect Biochem. Physiol. 2006, 62, 80–90. [Google Scholar] [CrossRef]
- Takahashi, K.H.; Rako, L.; Takano-Shimizu, T.; Hoffmann, A.A.; Lee, S.F. Effects of Small Hsp genes on Developmental Stability and Microenvironmental Canalization. BMC Evol. Biol. 2010, 10, 284. [Google Scholar] [CrossRef]
- Li, H.; Hao, D.J.; Xu, T.; Dai, L.L. The Effects of Heat Stress on Herbivorous Insects: An Overview and Future Direction. J. Nanjing For. Univ. Sci. Ed. 2022, 46, 215–224. [Google Scholar] [CrossRef]
- Dong, C.L.; Zhu, F.; Lu, M.X.; Du, Y.Z. Characterization and Functional Analysis of Cshsp19.0 Encoding a Small Heat Shock Protein in Chilo Suppressalis (Walker). Int. J. Biol. Macromol. 2021, 188, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Garczynski, S.F.; Unruh, T.R.; Guedot, C.; Neven, L.G. Characterization of Three Transcripts Encoding Small Heat Shock Proteins Expressed in the Codling Moth, Cydia Pomonella (Lepidoptera: Tortricidae). Insect Sci. 2011, 18, 473–483. [Google Scholar] [CrossRef]
- Zheng, H.Y.; Qin, P.H.; Yang, K.; Liu, T.X.; Zhang, Y.J.; Chu, D. Genome-Wide Identification and Analysis of the Heat-Shock Protein Gene Superfamily in Bemisia Tabaci and Expression Pattern Analysis under Heat Shock. Insects 2022, 13, 570. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.N.; Zhu, B.J.; Dai, L.S.; Fu, W.W.; Lin, K.Z.; Liu, C.L. Overexpression of Small Heat Shock Protein 21 Protects the Chinese Oak Silkworm Antheraea Pernyi against Thermal Stress. J. Insect Physiol. 2013, 59, 848–854. [Google Scholar] [CrossRef]
- Chu, J.; Jiang, D.L.; Ya, M.W.; Li, Y.J.C.; Wang, J.; Wu, F.A.; Sheng, S. Identifications, Characteristics, and Expression Patterns of Small Heat Shock Protein Genes in a Major Mulberry Pest, Glyphodes Pyloalis (Lepidoptera: Pyralidae). J. Insect Sci. 2020, 20, 2. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Peng, G.F.; Hu, X.X.; Gu, S.S.; Bi, J.X.; Wei, L.T.; Tang, J.; Song, X.W.; Feng, F.; Li, B. Functional Analysis of a Novel Orthologous Small Heat Shock Protein (Shsp) Hsp21.8a and Seven Species-Specific Shsps in Tribolium Castaneum. Genomics 2020, 112, 4474–4485. [Google Scholar] [CrossRef] [PubMed]
- Quan, G.; Duan, J.; Ladd, T.; Krell, P.J. Identification and Expression Analysis of Multiple Small Heat Shock Protein Genes in Spruce Budworm, Choristoneura Fumiferana (L.). Cell Stress Chaperones 2018, 23, 141–154. [Google Scholar] [CrossRef]
- Li, Z.W.; Li, X.; Yu, Q.Y.; Xiang, Z.H.; Kishino, H.; Zhang, Z. The Small Heat Shock Protein (SHSP) Genes in the Silkworm, Bombyx Mori, and Comparative Analysis with Other Insect SHSP Genes. BMC Evol. Biol. 2009, 9, 215. [Google Scholar] [CrossRef]
- Zhao, X.; Geng, Y.; Hu, T.; Xie, C.; Xu, W.; Zuo, Z.; Xue, M.; Hao, D. Ecological Strategies of Hyphantria Cunea (Lepidoptera: Arctiidae) Response to Different Larval Densities. Front. Ecol. Evol. 2023, 11, 1177029. [Google Scholar] [CrossRef]
- Zhao, X.; Geng, Y.; Hu, T.; Li, W.; Liang, Y.; Hao, D. Comparing the Performance of Hyphantria Cunea (Lepidoptera: Arctiidae) on Artificial and Natural Diets: Feasibility of Mass-Rearing on Artificial Diets. J. Econ. Entomol. 2023, 116, 181–191. [Google Scholar] [CrossRef]
- Edosa, T.T.; Jo, Y.H.; Keshavarz, M.; Anh, Y.S.; Noh, M.Y.; Han, Y.S. Current Status of the Management of Fall Webworm, Hyphantria Cunea: Towards the Integrated Pest Management Development. J. Appl. Entomol. 2019, 143, 1–10. [Google Scholar] [CrossRef]
- Sullivan, G.T.; Karaca, I.; Ozman-Sullivan, S.K.; Kara, K. Tachinid (Diptera: Tachinidae) Parasitoids of Overwintered Hyphantria Cunea (Drury) (Lepidoptera: Arctiidae) Pupae in Hazelnut Plantations in Samsun Province, Turkey. J. Entomol. Res. Soc. 2012, 14, 21–30. [Google Scholar] [CrossRef]
- Zhao, L.Q.; Wang, X.M.; Liu, Z.; Torson, A.S. Energy Consumption and Cold Hardiness of Diapausing Fall Webworm Pupae. Insects 2022, 13, 853. [Google Scholar] [CrossRef]
- Li, Z.; Yin, H.; Li, Y.; Wang, Y.; Yu, W.; Feng, B.; Zhang, S. Hyphantria Cunea (Drury) Showed a Stronger Oviposition Preference for Native Plants after Invading the Subtropical Region of China. Agronomy 2023, 13, 1360. [Google Scholar] [CrossRef]
- Rehnberg, B.G. Temperature Profiles inside Webs of the Fall Webworm, Hyphantria Cunea (Lepidoptera: Arctiidae): Influence of Weather, Compass Orientation, and Time of Day. J. Therm. Biol. 2006, 31, 274–279. [Google Scholar] [CrossRef]
- Li, H.; Tao, R.; Qiao, H.; Zhao, X.D.; Li, S.Y.; Dai, L.L.; Hao, D.J. Functional Analysis of Small Heat Shock Proteins Providing Evidence of Temperature Tolerance in Hyphantria Cunea. J. Appl. Entomol. 2022, 146, 130–143. [Google Scholar] [CrossRef]
- Haslbeck, M.; Kastenmüller, A.; Buchner, J.; Weinkauf, S.; Braun, N. Structural Dynamics of Archaeal Small Heat Shock Proteins. J. Mol. Biol. 2008, 378, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Y.; Pan, L.; Wang, Q.; Han, Y.; Niu, H.; Shan, D.; Hoffmann, A.; Fang, J. Induced Expression of Small Heat Shock Proteins Is Associated with Thermotolerance in Female Laodelphax Striatellus Planthoppers. Cell Stress Chaperones 2019, 24, 115–123. [Google Scholar] [CrossRef]
- Yang, W.J.; Xu, K.K.; Cao, Y.; Meng, Y.L.; Liu, Y.; Li, C. Identification and Expression Analysis of Four Small Heat Shock Protein Genes in Cigarette Beetle, Lasioderma Serricorne (Fabricius). Insects 2019, 10, 139. [Google Scholar] [CrossRef]
- Kriehuber, T.; Rattei, T.; Weinmaier, T.; Bepperling, A.; Haslbeck, M.; Buchner, J. Independent Evolution of the Core Domain and Its Flanking Sequences in Small Heat Shock Proteins. Faseb. J. 2010, 24, 3633–3642. [Google Scholar] [CrossRef] [PubMed]
- Bakthisaran, R.; Tangirala, R.; Rao, C.M. Small Heat Shock Proteins: Role in Cellular Functions and Pathology. Biochim. Biophys. Acta BBA Proteins Proteom. 2015, 1854, 291–319. [Google Scholar] [CrossRef]
- Perlman, D.; Halvorson, H.O. A Putative Signal Peptidase Recognition Site and Sequence in Eukaryotic and Prokaryotic Signal Peptides. J. Mol. Biol. 1983, 167, 391–409. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.X.; Hua, J.; Cui, Y.D.; Du, Y.Z. Five Small Heat Shock Protein Genes from Chilo Suppressalis: Characteristics of Gene, Genomic Organization, Structural Analysis, and Transcription Profiles. Cell Stress Chaperones 2014, 19, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Gu, J.; Huang, L.H.; Zheng, S.C.; Liu, L.; Xu, W.-H.; Feng, Q.-L.; Kang, L. Cloning and Expression Analysis of Six Small Heat Shock Protein Genes in the Common Cutworm, Spodoptera Litura. J. Insect Physiol. 2011, 57, 908–914. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.H.; Wang, H.S.; Kang, L. Different Evolutionary Lineages of Large and Small Heat Shock Proteins in Eukaryotes. Cell Res. 2008, 18, 1074–1076. [Google Scholar] [CrossRef]
- Savard, J.; Tautz, D.; Richards, S.; Weinstock, G.M.; Gibbs, R.A.; Werren, J.H.; Tettelin, H.; Lercher, M.J. Phylogenomic Analysis Reveals Bees and Wasps (Hymenoptera) at the Base of the Radiation of Holometabolous Insects. Genome Res. 2006, 16, 1334–1338. [Google Scholar] [CrossRef] [PubMed]
- Zahiri, R.; Kitching, I.J.; Lafontaine, J.D.; Mutanen, M.; Kaila, L.; Holloway, J.D.; Wahlberg, N. A New Molecular Phylogeny Offers Hope for a Stable Family Level Classification of the Noctuoidea (Lepidoptera). Zool. Scr. 2011, 40, 158–173. [Google Scholar] [CrossRef]
- Luo, J.; Cheng, X.Y.; Yan, X.; Tao, W.Q.; Holland, J.D.; Xu, R.M. Characterization and Polymorphism Analysis of Phosphoglucose Isomerase Gene in the Fall Webwomt (Hyphantria Cunea). Bull. Entomol. Res. 2012, 102, 477–488. [Google Scholar] [CrossRef]
- Jhan, P.K.; Lee, K.Y. Developing Extreme Heat Acclimation in Bemisia Tabaci Mediterranean (Hemiptera: Aleyrodidae). Arch. Insect Biochem. Physiol. 2022, 110, e21890. [Google Scholar] [CrossRef]
- Queitsch, C.; Sangster, T.A.; Lindquist, S. Hsp90 as a Capacitor of Phenotypic Variation. Nature 2002, 417, 618–624. [Google Scholar] [CrossRef]
- Concha, C.; Edman, R.M.; Belikoff, E.J.; Schiemann, A.H.; Carey, B.; Scott, M.J. Organization and Expression of the Australian Sheep Blowfly (Lucilia Cuprina) Hsp23, Hsp24, Hsp70 and Hsp83 Genes. Insect Mol. Biol. 2012, 21, 169–180. [Google Scholar] [CrossRef]
- Feliciello, I.; Akrap, I.; Ugarković, Đ. Satellite DNA Modulates Gene Expression in the Beetle Tribolium Castaneum after Heat Stress. PLOS Genet. 2015, 11, e1005466. [Google Scholar] [CrossRef]
- Liu, W.W.; Yang, P.; Chen, X.M.; Xu, D.L.; Hu, Y.H. Cloning and Expression Analysis of Four Heat Shock Protein Genes in Ericerus Pela (Homoptera: Coccidae). J. Insect Sci. 2014, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Regier, J.C.; Mazur, G.D.; Kafatos, F.C. The Silkmoth Chorion: Morphological and Biochemical Characterization of Four Surface Regions. Dev. Biol. 1980, 76, 286–304. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.L.; Meng, J.Y.; Zhou, L.; Yao, M.S.; Zhang, C.Y. Identification of Five Small Heat Shock Protein Genes in Spodoptera Frugiperda and Expression Analysis in Response to Different Environmental Stressors. Cell Stress Chaperones 2021, 26, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Xiong, W.; Hu, X.; Gu, S.; Zhang, S.; Gao, S.; Song, X.; Bi, J.; Li, B. Characterization and Function Alanalysis of Hsp21.8b: An Orthologous Small Heat Shock Protein Gene in Tribolium Castaneum. J. Appl. Entomol. 2018, 142, 654–666. [Google Scholar] [CrossRef]
- Chang, Y.-W.; Zhang, X.-X.; Lu, M.-X.; Du, Y.-Z.; Zhu-Salzman, K. Molecular Cloning and Characterization of Small Heat Shock Protein Genes in the Invasive Leaf Miner Fly, Liriomyza Trifolii. Genes 2019, 10, 775. [Google Scholar] [CrossRef]
- Tao, R.; Li, H.; Sun, Y.H.; Yu, X.H.; Zhu, H.; Hao, D.J. Indentification and Screening of Internal Reference Genes of Hyphantria Cunea (Lepidoptera: Arctiidae). Sci. Silvae Sin. 2019, 55, 111–120. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods Companion Methods Enzymol. 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | GenBank Accession Number | ORF (bp) | Protein Length (aa) | Molecular Weight (kDa) | Isoelectric Point (IP) | Instability Index (II) | Signal Peptide (AA) | Subcellular Location |
|---|---|---|---|---|---|---|---|---|
| HcHSP18.9 | OP964824 | 501 | 166 | 18.9 | 5.96 | 39.94 | No | Cytoplasmic |
| HcHSP20.1 | OP964825 | 528 | 175 | 20.1 | 6.00 | 49.62 | No | Cytoplasmic |
| HcHSP21.5 | OP964826 | 576 | 191 | 21.5 | 6.38 | 49.80 | No | Cytoplasmic |
| HcHSP21.4 | OP964827 | 564 | 187 | 21.4 | 5.79 | 48.88 | No | Cytoplasmic |
| HcHSP29.8 | OP964828 | 786 | 261 | 29.8 | 5.84 | 46.27 | 15 | Extracellular |
| Application | Primer Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|---|
| PCR | HcHSP18.9 | ATGTCTCTTTTGCCATACTTCT | CTATTTTGGTTCATCAACAGCAACC |
| HcHSP20.1 | ATGTCATTGGTGCCGTATTGG | CTAAGCTTTTTCTTCCTGTGCT | |
| HcHSP21.5 | ATGTCTCTGCTACCATTTGTTTTGG | TTACTTCTTATCTTCAGCGCCG | |
| HcHSP21.4 | ATGGCTGATAGTGGTCTGAAGA | TCAGTGCTTCTGGATAGGGA | |
| HcHSP29.8 | ATGCAGAAATATTTCTTAGTACTCGCA | TTACTCTTCTTGTTCCATTAGTACA | |
| qPCR | HcHSP18.9 | AAGCTGTCTTCGGATGGTGT | GCCTCACGGGTCCTGTATG |
| HcHSP20.1 | CAGTGGCTGGACCAAGAGTC | CCGATCTAAACGCCACCAGA | |
| HcHSP21.5 | GAATCCCGGCTTTCATCCGA | CCTGTCTGCGAAATAGGCAC | |
| HcHSP21.4 | ACATCGTCACAACACAGCGA | GCTTGAGTGACTTGCCGTCT | |
| HcHSP29.8 | GAGGCGAAGACCCATTCTCC | CAGTTGACTCCAAGGCCACA | |
| EF1α | TTATCGTCGCTGCTGGTACA | GAGTGTGAAAGCGAGCAGAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Liu, Y.; Li, H.; Li, Z.; Hao, D. Spatiotemporal Patterns of Five Small Heat Shock Protein Genes in Hyphantria cunea in Response to Thermal Stress. Int. J. Mol. Sci. 2023, 24, 15176. https://doi.org/10.3390/ijms242015176
Zhao S, Liu Y, Li H, Li Z, Hao D. Spatiotemporal Patterns of Five Small Heat Shock Protein Genes in Hyphantria cunea in Response to Thermal Stress. International Journal of Molecular Sciences. 2023; 24(20):15176. https://doi.org/10.3390/ijms242015176
Chicago/Turabian StyleZhao, Shiyue, Yukun Liu, Hui Li, Zichun Li, and Dejun Hao. 2023. "Spatiotemporal Patterns of Five Small Heat Shock Protein Genes in Hyphantria cunea in Response to Thermal Stress" International Journal of Molecular Sciences 24, no. 20: 15176. https://doi.org/10.3390/ijms242015176
APA StyleZhao, S., Liu, Y., Li, H., Li, Z., & Hao, D. (2023). Spatiotemporal Patterns of Five Small Heat Shock Protein Genes in Hyphantria cunea in Response to Thermal Stress. International Journal of Molecular Sciences, 24(20), 15176. https://doi.org/10.3390/ijms242015176






