Plasma-Derived Exosomes from NAFLD Patients Modulate the Cannabinoid Receptors’ Expression in Cultured HepaRG Cells
Abstract
1. Introduction
2. Results
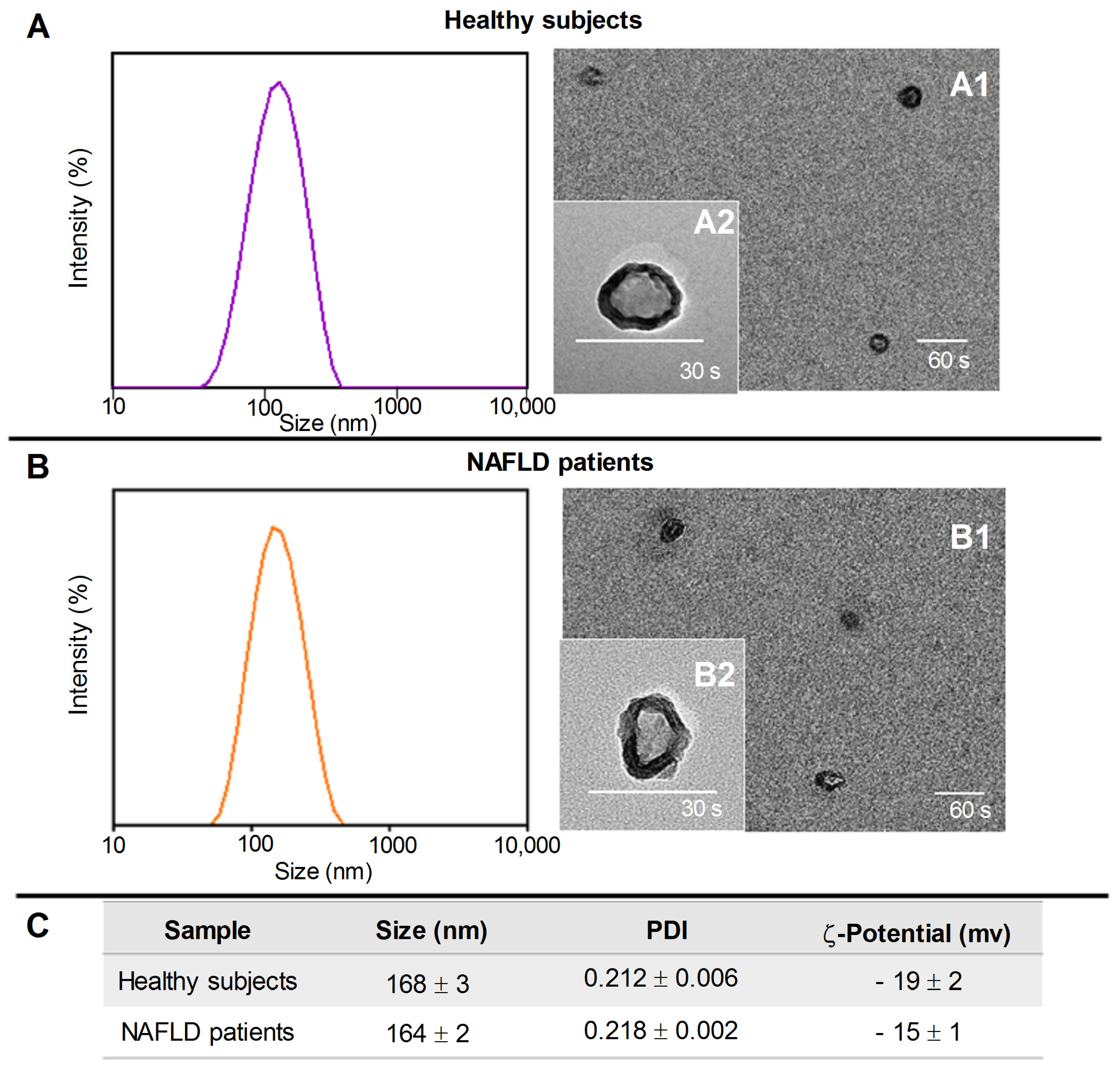
2.1. Exosomes’ Isolation and Characterization
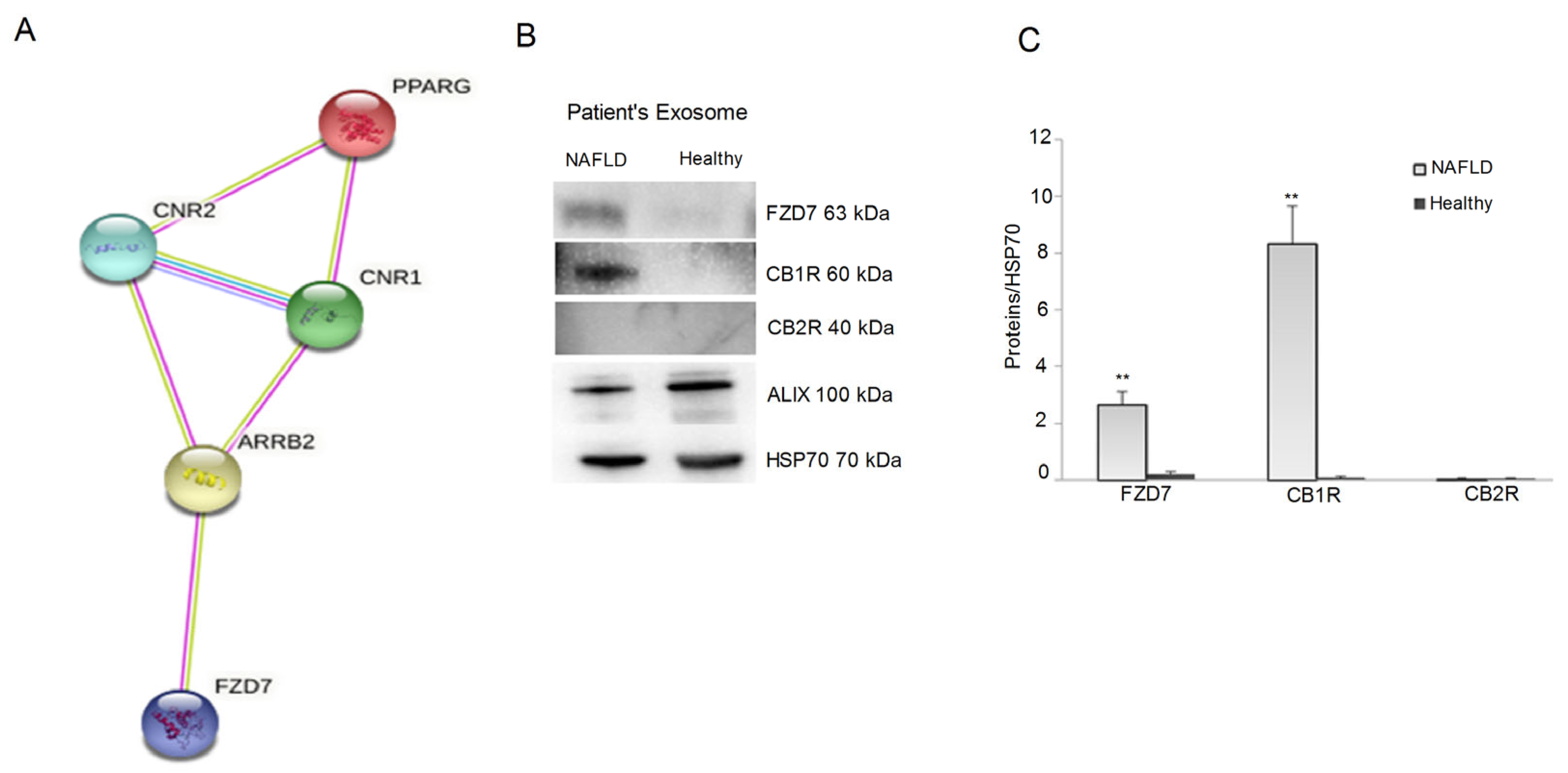
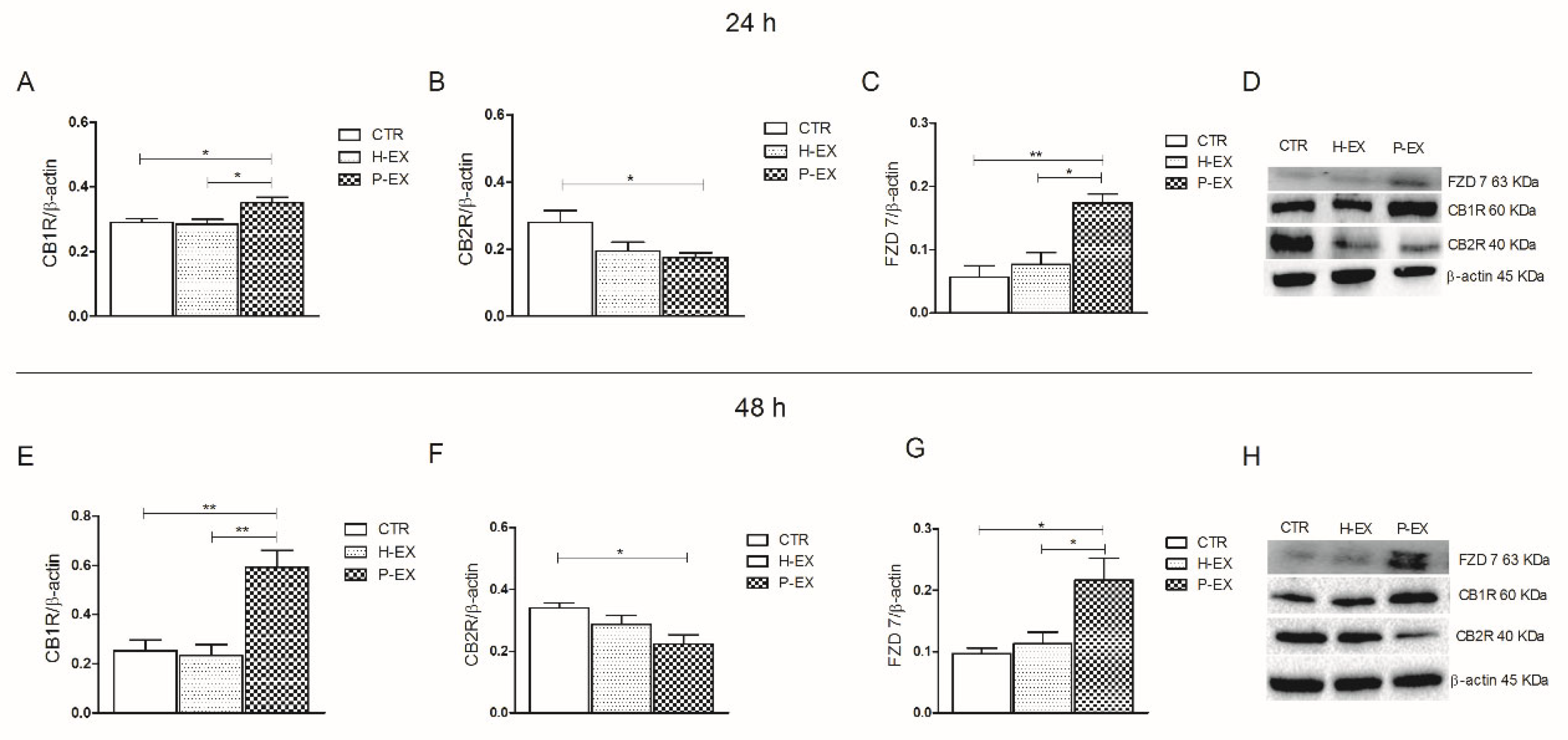
2.2. Protein Expression of CB1, CB2 Receptors, FZD 7, and IL-1β
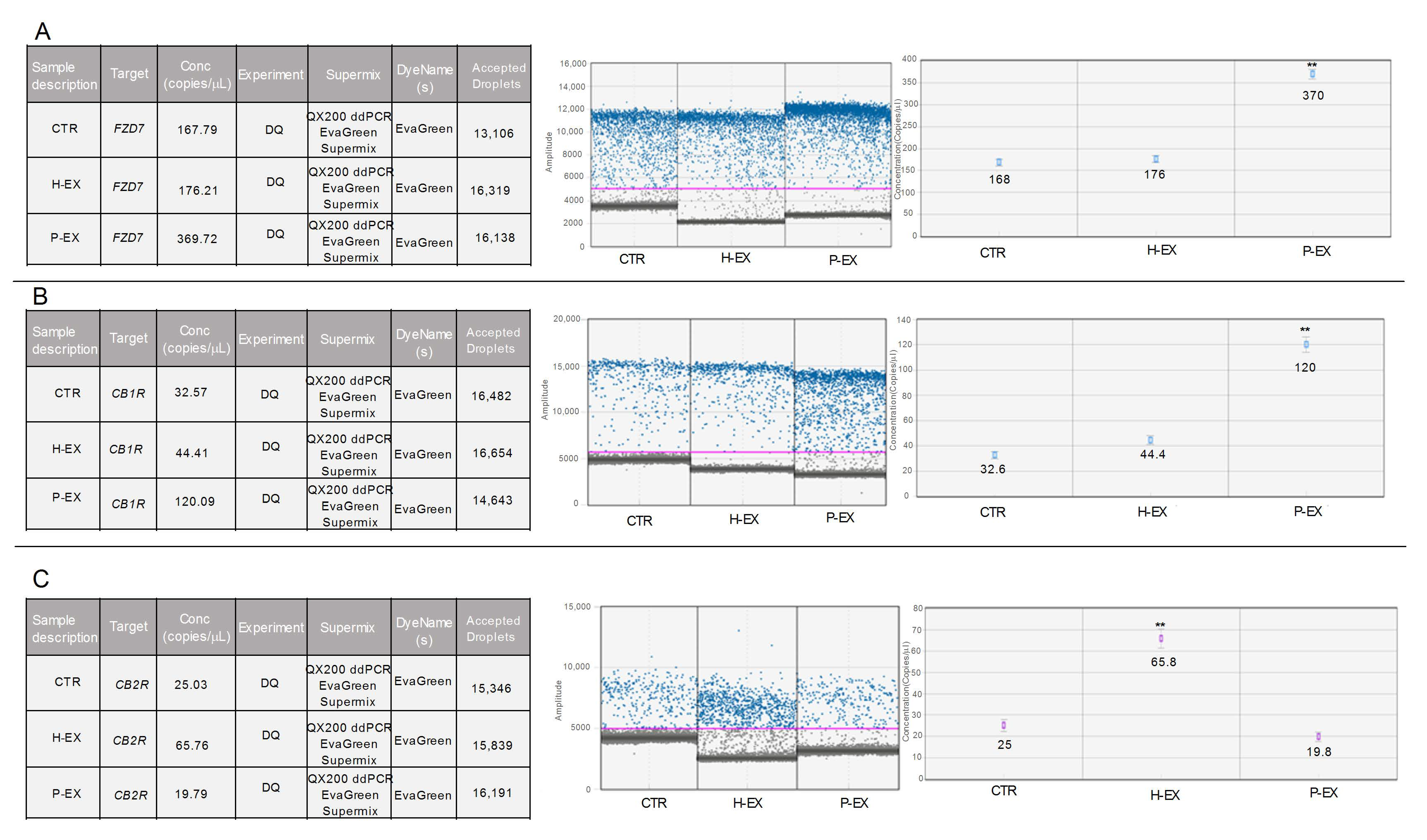
2.3. Gene Expression, CB1, CB2 Receptor, and FZD 7
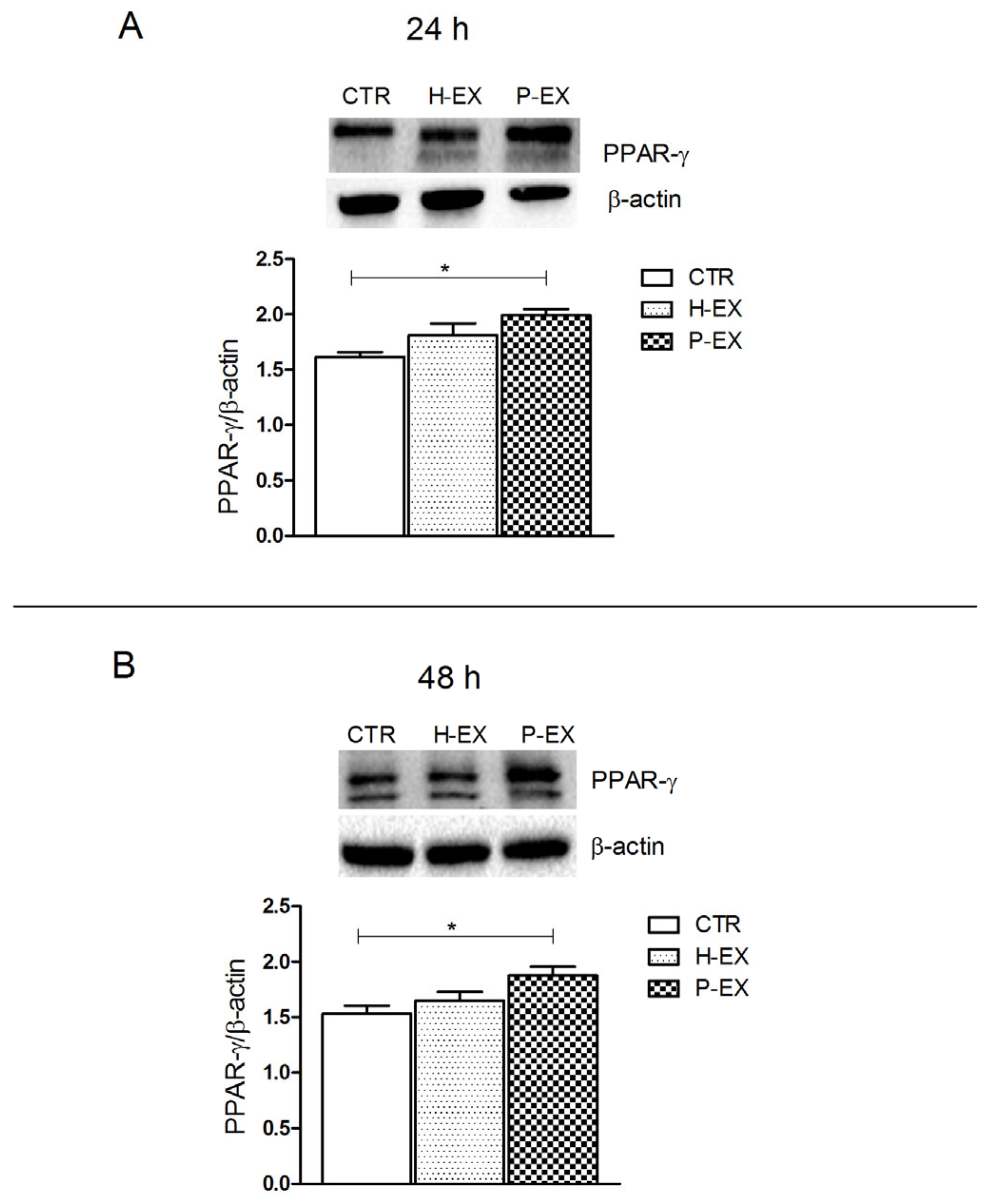
2.4. PPAR-γ Expression
2.5. Fatty Acid Analysis
3. Discussion
4. Materials and Methods
4.1. Exosome Isolation
4.2. Dynamic Light Scattering (DLS) and ζ-Potential Analysis
4.3. Transmission Electronic Microscopy (TEM) Analysis
4.4. Cell cultures and Exosomes Administration
4.5. Western Blotting Analysis
4.6. Immunofluorescence Assay
4.7. RNA Extraction and cDNA Synthesis
4.8. ddPCR Analysis
4.9. IL-1β Assay
4.10. Lipidomic Analysis
4.11. Bioinformatic and Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scavo, M.P.; Depalo, N.; Tutino, V.; De Nunzio, V.; Ingrosso, C.; Rizzi, F.; Notarnicola, M.; Curri, M.L.; Giannelli, G. Exosomes for Diagnosis and Therapy in Gastrointestinal Cancers. Int. J. Mol. Sci. 2020, 21, 367. [Google Scholar] [CrossRef]
- Sasaki, R.; Kanda, T.; Yokosuka, O.; Kato, N.; Matsuoka, S.; Moriyama, M. Exosomes and Hepatocellular Carcinoma: From Bench to Bedside. Int. J. Mol. Sci. 2019, 20, 1406. [Google Scholar] [CrossRef]
- Srinivas, A.N.; Suresh, D.; Santhekadur, P.K.; Suvarna, D.; Kumar, D.P. Extracellular Vesicles as Inflammatory Drivers in NAFLD. Front. Immunol. 2021, 11, 627424. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Cheng, X.; Pan, X.; Li, J. Emerging role of exosomes in liver physiology and pathology. Hepatol. Res. 2016, 47, 194–203. [Google Scholar] [CrossRef]
- Garcia-Martinez, I.; Alen, R.; Rada, P.; Valverde, A.M. Insights Into Extracellular Vesicles as Biomarker of NAFLD Pathogenesis. Front. Med. 2020, 7, 395. [Google Scholar] [CrossRef] [PubMed]
- Devhare, P.B.; Ray, R.B. Extracellular vesicles: Novel mediator for cell to cell communications in liver pathogenesis. Mol. Asp. Med. 2018, 60, 115–122. [Google Scholar] [CrossRef]
- Newman, L.A.; Sorich, M.J.; Rowland, A. Role of Extracellular Vesicles in the Pathophysiology, Diagnosis and Tracking of Non-Alcoholic Fatty Liver Disease. J. Clin. Med. 2020, 9, 2032. [Google Scholar] [CrossRef]
- Malhi, H.; Barreyro, F.J.; Isomoto, H.; Bronk, S.F.; Gores, G.J. Free fatty acids sensitise hepatocytes to TRAIL mediated cytotoxicity. Gut 2007, 56, 1124–1131. [Google Scholar] [CrossRef]
- Saeed, A.; Dullaart, R.P.F.; Schreuder, T.C.M.A.; Blokzijl, H.; Faber, K.N. Disturbed Vitamin A Metabolism in Non-Alcoholic Fatty Liver Disease (NAFLD). Nutrients 2017, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, F.M.; Disciglio, V.; Franco, I.; Sorino, P.; Bonfiglio, C.; Bianco, A.; Campanella, A.; Lippolis, T.; Pesole, P.L.; Polignano, M.; et al. A Low Glycemic Index Mediterranean Diet Combined with Aerobic Physical Activity Rearranges the Gut Microbiota Signature in NAFLD Patients. Nutrients 2022, 14, 1773. [Google Scholar] [CrossRef] [PubMed]
- Notarnicola, M.; Osella, A.; Caruso, M.; Pesole, P.; Lippolis, A.; Tutino, V.; Bonfiglio, C.; De Nunzio, V.; Scavo, M.; Mirizzi, A.; et al. Nonalcoholic Fatty Liver Disease: Focus on New Biomarkers and Lifestyle Interventions. Int. J. Mol. Sci. 2021, 22, 3899. [Google Scholar] [CrossRef] [PubMed]
- Notarnicola, M.; Caruso, M.G.; Tutino, V.; Bonfiglio, C.; Cozzolongo, R.; Giannuzzi, V.; De Nunzio, V.; De Leonardis, G.; Abbrescia, D.I.; Franco, I.; et al. Significant decrease of saturation index in erythrocytes membrane from subjects with non-alcoholic fatty liver disease (NAFLD). Lipids Health Dis. 2017, 16, 160. [Google Scholar] [CrossRef] [PubMed]
- Tutino, V.; De Nunzio, V.; Caruso, M.G.; Bonfiglio, C.; Franco, I.; Mirizzi, A.; De Leonardis, G.; Cozzolongo, R.; Giannuzzi, V.; Giannelli, G.; et al. Aerobic Physical Activity and a Low Glycemic Diet Reduce the AA/EPA Ratio in Red Blood Cell Membranes of Patients with NAFLD. Nutrients 2018, 10, 1299. [Google Scholar] [CrossRef] [PubMed]
- Luna, A.C.D.L.; Forti, F.L. Modulation of SCD1 activity in hepatocyte cell lines: Evaluation of genomic stability and proliferation. Mol. Cell. Biochem. 2021, 476, 3393–3405. [Google Scholar] [CrossRef]
- Scavo, M.P.; Depalo, N.; Rizzi, F.; Carrieri, L.; Serino, G.; Franco, I.; Bonfiglio, C.; Pesole, P.L.; Cozzolongo, R.; Gianuzzi, V.; et al. Exosomal FZD-7 Expression Is Modulated by Different Lifestyle Interventions in Patients with NAFLD. Nutrients 2022, 14, 1133. [Google Scholar] [CrossRef]
- Wang, C.; Liu, J.; Yan, Y.; Tan, Y. Role of Exosomes in Chronic Liver Disease Development and Their Potential Clinical Applications. J. Immunol. Res. 2022, 2022, 1695802. [Google Scholar] [CrossRef]
- Jiao, Y.; Xu, P.; Shi, H.; Chen, D.; Shi, H. Advances on liver cell-derived exosomes in liver diseases. J. Cell. Mol. Med. 2020, 25, 15–26. [Google Scholar] [CrossRef]
- Caraceni, P.; Domenicali, M.; Bernardi, M. The Endocannabinoid System and Liver Diseases. J. Neuroendocr. 2008, 20 (Suppl. 1), 47–52. [Google Scholar] [CrossRef]
- Julien, B.; Grenard, P.; Teixeira-Clerc, F.; Van Nhieu, J.T.; Li, L.; Karsak, M.; Zimmer, A.; Mallat, A.; Lotersztajn, S. Antifibrogenic role of the cannabinoid receptor CB2 in the liver. Gastroenterology 2005, 128, 742–755. [Google Scholar] [CrossRef]
- Sigel, E.; Baur, R.; Rácz, I.; Marazzi, J.; Smart, T.G.; Zimmer, A.; Gertsch, J. The major central endocannabinoid directly acts at GABA A receptors. Proc. Natl. Acad. Sci. USA 2011, 108, 18150–18155. [Google Scholar] [CrossRef]
- Gabrielli, M.; Battista, N.; Riganti, L.; Prada, I.; Antonucci, F.; Cantone, L.; Matteoli, M.; Maccarrone, M.; Verderio, C. Active endocannabinoids are secreted on extracellular membrane vesicles. EMBO Rep. 2015, 16, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Ji, Q.; Li, Q. The role and mechanism of β-arrestins in cancer invasion and metastasis (Review). Int. J. Mol. Med. 2017, 41, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Iannotti, F.; Vitale, R. The Endocannabinoid System and PPARs: Focus on Their Signalling Crosstalk, Action and Transcriptional Regulation. Cells 2021, 10, 586. [Google Scholar] [CrossRef] [PubMed]
- Azar, S.; Udi, S.; Drori, A.; Hadar, R.; Nemirovski, A.; Vemuri, K.V.; Miller, M.; Sherill-Rofe, D.; Arad, Y.; Gur-Wahnon, D.; et al. Reversal of diet-induced hepatic steatosis by peripheral CB1 receptor blockade in mice is p53/miRNA-22/SIRT1/PPARα dependent. Mol. Metab. 2020, 42, 101087. [Google Scholar] [CrossRef]
- Iannotti, F.A.; Di Marzo, V.; Petrosino, S. Endocannabinoids and endocannabinoid-related mediators: Targets, metabolism and role in neurological disorders. Prog. Lipid Res. 2016, 62, 107–128. [Google Scholar] [CrossRef]
- Di Marzo, V. New approaches and challenges to targeting the endocannabinoid system. Nat. Rev. Drug Discov. 2018, 17, 623–639. [Google Scholar] [CrossRef]
- Fajas, L.; Auboeuf, D.; Raspé, E.; Schoonjans, K.; Lefebvre, A.-M.; Saladin, R.; Najib, J.; Laville, M.; Fruchart, J.-C.; Deeb, S.; et al. The Organization, Promoter Analysis, and Expression of the Human PPARγ Gene. J. Biol. Chem. 1997, 272, 18779–18789. [Google Scholar] [CrossRef]
- Memon, R.A.; Tecott, L.H.; Nonogaki, K.; Beigneux, A.; Moser, A.H.; Grunfeld, C.; Feingold, K.R. Up-Regulation of Peroxisome Proliferator-Activated Receptors (PPAR-α) and PPAR-γ Messenger Ribonucleic Acid Expression in the Liver in Murine Obesity: Troglitazone Induces Expression of PPAR-γ-Responsive Adipose Tissue-Specific Genes in the Liver of Obese Diabetic Mice. Endocrinology 2000, 141, 4021–4031. [Google Scholar] [CrossRef]
- Morán-Salvador, E.; López-Parra, M.; García-Alonso, V.; Titos, E.; Martínez-Clemente, M.; González-Périz, A.; López-Vicario, C.; Barak, Y.; Arroyo, V.; Clària, J. Role for PPARγ in obesity-induced hepatic steatosis as determined by hepatocyte- and macrophage-specific conditional knockouts. FASEB J. 2011, 25, 2538–2550. [Google Scholar] [CrossRef]
- Chang, H.-Y.; Chen, J.-R.; Chen, Y.-H.; Xiao, Q.; Chen, Y.-L.; Yang, S.-C. The Preliminary Results for Evaluating Cocoa Butter’s Hepatoprotective Effects against Lipid Accumulation and Inflammation in Adult Male Rats Chronically Fed Ethanol. Bioengineering 2022, 9, 526. [Google Scholar] [CrossRef]
- Sundararajan, V.; Sarkar, F.H.; Ramasamy, T.S. Correction to: The versatile role of exosomes in cancer progression: Diagnostic and therapeutic implications. Cell. Oncol. 2018, 41, 463. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ding, X.; Nan, L.; Wang, Y.; Wang, J.; Yan, Z.; Zhang, W.; Sun, J.; Zhu, W.; NI, B.; et al. Investigation of the roles of exosomes in colorectal cancer liver metastasis. Oncol. Rep. 2015, 33, 2445–2453. [Google Scholar] [CrossRef]
- Batkai, S.; Osei-Hyiaman, D.; Pan, H.; El-Assal, O.; Rajesh, M.; Mukhopadhyay, P.; Hong, F.; Harvey-White, J.; Jafri, A.; Haskó, G.; et al. Cannabinoid-2 receptor mediates protection against hepatic ischemia/reperfusion injury. FASEB J. 2007, 21, 1788–1800. [Google Scholar] [CrossRef] [PubMed]
- Buckley, N.E.; Hansson, S.; Harta, G.; Mezey, É. Expression of the CB1 and CB2 receptor messenger RNAs during embryonic development in the rat. Neuroscience 1997, 82, 1131–1149. [Google Scholar] [CrossRef] [PubMed]
- Mallat, A.; Lotersztajn, S. Endocannabinoids as novel mediators of liver diseases. J. Endocrinol. Investig. 2006, 29, 58–65. [Google Scholar]
- Scavo, M.P.; Rizzi, F.; DePalo, N.; Fanizza, E.; Ingrosso, C.; Curri, M.L.; Giannelli, G. A Possible Role of FZD10 Delivering Exosomes Derived from Colon Cancers Cell Lines in Inducing Activation of Epithelial–Mesenchymal Transition in Normal Colon Epithelial Cell Line. Int. J. Mol. Sci. 2020, 21, 6705. [Google Scholar] [CrossRef]
- Knopfová, L.; Šmarda, J. The use of Cox-2 and PPARγ signaling in anti-cancer therapies. Exp. Ther. Med. 2010, 1, 257–264. [Google Scholar] [CrossRef]
- Teixeira-Clerc, F.; Julien, B.; Grenard, P.; Tran Van Nhieu, J.; Deveaux, V.; Li, L.; Serriere-Lanneau, V.; Ledent, C.; Mallat, A.; Lotersztajn, S. CB1 cannabinoid receptor antagonism: A new strategy for the treatment of liver fibrosis. Nat. Med. 2006, 12, 671–676. [Google Scholar] [CrossRef]
- Du, H.; Chen, X.; Zhang, J.; Chen, C. Inhibition of COX-2 expression by endocannabinoid 2-arachidonoylglycerol is mediated via PPAR-γ. Br. J. Pharmacol. 2011, 163, 1533–1549. [Google Scholar] [CrossRef]
- Gigante, I.; Tutino, V.; Russo, F.; De Nunzio, V.; Coletta, S.; Armentano, R.; Crovace, A.; Caruso, M.; Orlando, A.; Notarnicola, M. Cannabinoid Receptors Overexpression in a Rat Model of Irritable Bowel Syndrome (IBS) after Treatment with a Ketogenic Diet. Int. J. Mol. Sci. 2021, 22, 2880. [Google Scholar] [CrossRef]
- Chimienti, G.; Orlando, A.; Lezza, A.; D’Attoma, B.; Notarnicola, M.; Gigante, I.; Pesce, V.; Russo, F. The Ketogenic Diet Reduces the Harmful Effects of Stress on Gut Mitochondrial Biogenesis in a Rat Model of Irritable Bowel Syndrome. Int. J. Mol. Sci. 2021, 22, 3498. [Google Scholar] [CrossRef]
- Hernández, A.; Arab, J.P.; Reyes, D.; Lapitz, A.; Moshage, H.; Bañales, J.M.; Arrese, M. Extracellular Vesicles in NAFLD/ALD: From Pathobiology to Therapy. Cells 2020, 9, 817. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, F.; Lai, X.; Liu, H.; Wu, S.; Han, Y.; Shen, Y. Exosomes secreted by palmitic acid-treated hepatocytes promote LX-2 cell activation by transferring miRNA-107. Cell Death Discov. 2021, 7, 174. [Google Scholar] [CrossRef] [PubMed]
- Nakao, Y.; Fukushima, M.; Mauer, A.S.; Liao, C.-Y.; Ferris, A.; Dasgupta, D.; Heppelmann, C.J.; Vanderboom, P.M.; Saraswat, M.; Pandey, A.; et al. A Comparative Proteomic Analysis of Extracellular Vesicles Associated With Lipotoxicity. Front. Cell Dev. Biol. 2021, 9, 735001. [Google Scholar] [CrossRef] [PubMed]
- Kono, H.; Uesugi, T.; Froh, M.; Rusyn, I.; Bradford, B.U.; Thurman, R.G. ICAM-1 is involved in the mechanism of alcohol-induced liver injury: Studies with knockout mice. Am. J. Physiol. Liver Physiol. 2001, 280, G1289–G1295. [Google Scholar] [CrossRef]
- Tutino, V.; Gigante, I.; Scavo, M.P.; Refolo, M.G.; De Nunzio, V.; Milella, R.A.; Caruso, M.G.; Notarnicola, M. Stearoyl-CoA Desaturase-1 Enzyme Inhibition by Grape Skin Extracts Affects Membrane Fluidity in Human Colon Cancer Cell Lines. Nutrients 2020, 12, 693. [Google Scholar] [CrossRef]
- Scavo, M.P.; Cigliano, A.; DePalo, N.; Fanizza, E.; Bianco, M.G.; Denora, N.; Laquintana, V.; Curri, M.L.; Lorusso, D.; Lotesoriere, C.; et al. Frizzled-10 Extracellular Vesicles Plasma Concentration Is Associated with Tumoral Progression in Patients with Colorectal and Gastric Cancer. J. Oncol. 2019, 2019, 2715968. [Google Scholar] [CrossRef]
- Depalo, N.; Fanizza, E.; Vischio, F.; Denora, N.; Laquintana, V.; Cutrignelli, A.; Striccoli, M.; Giannelli, G.; Agostiano, A.; Curri, M.L.; et al. Imaging modification of colon carcinoma cells exposed to lipid based nanovectors for drug delivery: A scanning electron microscopy investigation. RSC Adv. 2019, 9, 21810–21825. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Notarnicola, M.; Lorusso, D.; Tutino, V.; De Nunzio, V.; De Leonardis, G.; Marangelli, G.; Guerra, V.; Veronese, N.; Caruso, M.G.; Giannelli, G. Differential Tissue Fatty Acids Profiling between Colorectal Cancer Patients with and without Synchronous Metastasis. Int. J. Mol. Sci. 2018, 19, 962. [Google Scholar] [CrossRef]

| CTR | H-EX | P-EX | |
|---|---|---|---|
| IL-1β (pg/mL) | 19.11 ± 2.05 | 29.35 ± 3.452 | 66.83 ± 2.0568 ** |
| A | Exosome Fatty Acids (%) | Healthy Subjects | NAFLD Patients | |
|---|---|---|---|---|
| Stearic acid (C18:0) | 5.49 ± 0.41 | 6.26 ± 0.20 ** | ||
| Oleic acid (C18:1n9) | 26.18 ± 1.02 | 33.12 ± 3.87 ** | ||
| Saturated fatty acids (SFAs) | 30.72 ± 2.54 | 34.53 ± 2.60 | ||
| Monounsaturated fatty acids (MUFAs) | 32.53 ± 0.78 | 39.91 ± 3.17 ** | ||
| Oleic acid/Stearic acid ratio (SCD-1) | 4.78 ± 0.28 | 5.57 ± 0.63 * | ||
| B | HepaRG membrane Fatty Acids (%) | CTR | H-EX | P-EX |
| Stearic acid (C18:0) | 10.07 ± 1.51 | 10.14 ± 1.70 | 11.07 ± 1.61 | |
| Oleic acid (C18:1n9) | 27.63 ± 1.13 | 27.92 ± 1.53 | 33.28 ± 1.31 *** | |
| Saturated fatty acids (SFAs) | 32.48 ± 2.32 | 34.82 ± 1.09 | 35.93 ± 3.11 | |
| Monounsaturated fatty acids (MUFAs) | 49.63 ± 2.27 | 50.04 ± 2.14 | 56.21 ± 1.85 *** | |
| Oleic acid/Stearic acid ratio (SCD-1) | 2.79 ± 0.41 | 2.80 ± 0.34 | 3.34 ± 0.25 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Nunzio, V.; Carrieri, L.; Scavo, M.P.; Lippolis, T.; Cofano, M.; Caponio, G.R.; Tutino, V.; Rizzi, F.; Depalo, N.; Osella, A.R.; et al. Plasma-Derived Exosomes from NAFLD Patients Modulate the Cannabinoid Receptors’ Expression in Cultured HepaRG Cells. Int. J. Mol. Sci. 2023, 24, 1739. https://doi.org/10.3390/ijms24021739
De Nunzio V, Carrieri L, Scavo MP, Lippolis T, Cofano M, Caponio GR, Tutino V, Rizzi F, Depalo N, Osella AR, et al. Plasma-Derived Exosomes from NAFLD Patients Modulate the Cannabinoid Receptors’ Expression in Cultured HepaRG Cells. International Journal of Molecular Sciences. 2023; 24(2):1739. https://doi.org/10.3390/ijms24021739
Chicago/Turabian StyleDe Nunzio, Valentina, Livianna Carrieri, Maria Principia Scavo, Tamara Lippolis, Miriam Cofano, Giusy Rita Caponio, Valeria Tutino, Federica Rizzi, Nicoletta Depalo, Alberto Ruben Osella, and et al. 2023. "Plasma-Derived Exosomes from NAFLD Patients Modulate the Cannabinoid Receptors’ Expression in Cultured HepaRG Cells" International Journal of Molecular Sciences 24, no. 2: 1739. https://doi.org/10.3390/ijms24021739
APA StyleDe Nunzio, V., Carrieri, L., Scavo, M. P., Lippolis, T., Cofano, M., Caponio, G. R., Tutino, V., Rizzi, F., Depalo, N., Osella, A. R., & Notarnicola, M. (2023). Plasma-Derived Exosomes from NAFLD Patients Modulate the Cannabinoid Receptors’ Expression in Cultured HepaRG Cells. International Journal of Molecular Sciences, 24(2), 1739. https://doi.org/10.3390/ijms24021739









