Genome-Wide Identification, Expression Analysis, and Potential Roles under Abiotic Stress of the YUCCA Gene Family in Mungbean (Vigna radiata L.)
Abstract
1. Introduction
2. Results
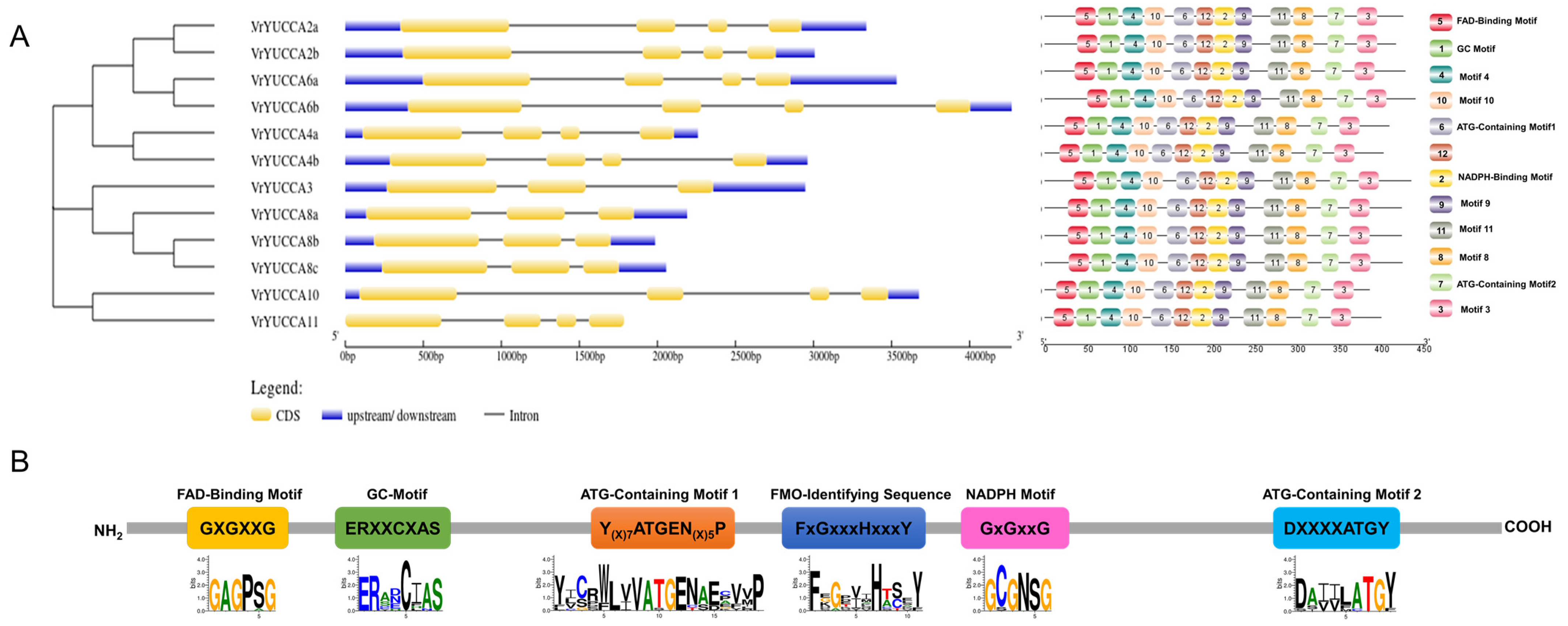
2.1. Identification of the YUCCA Gene Family in Mungbean and Structure Analysis
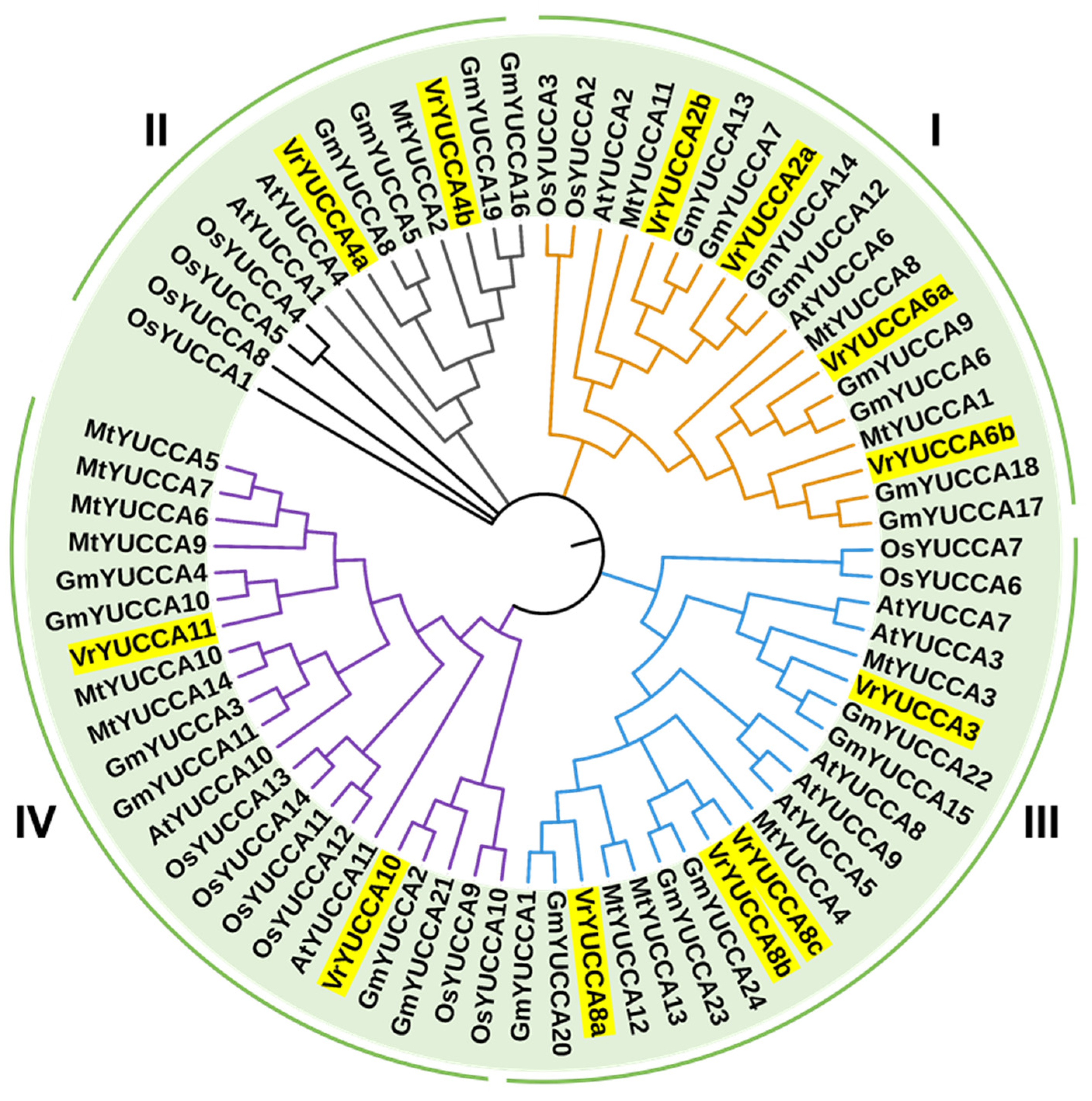
2.2. Phylogenetic Analysis of YUCCAs from Multiple Species
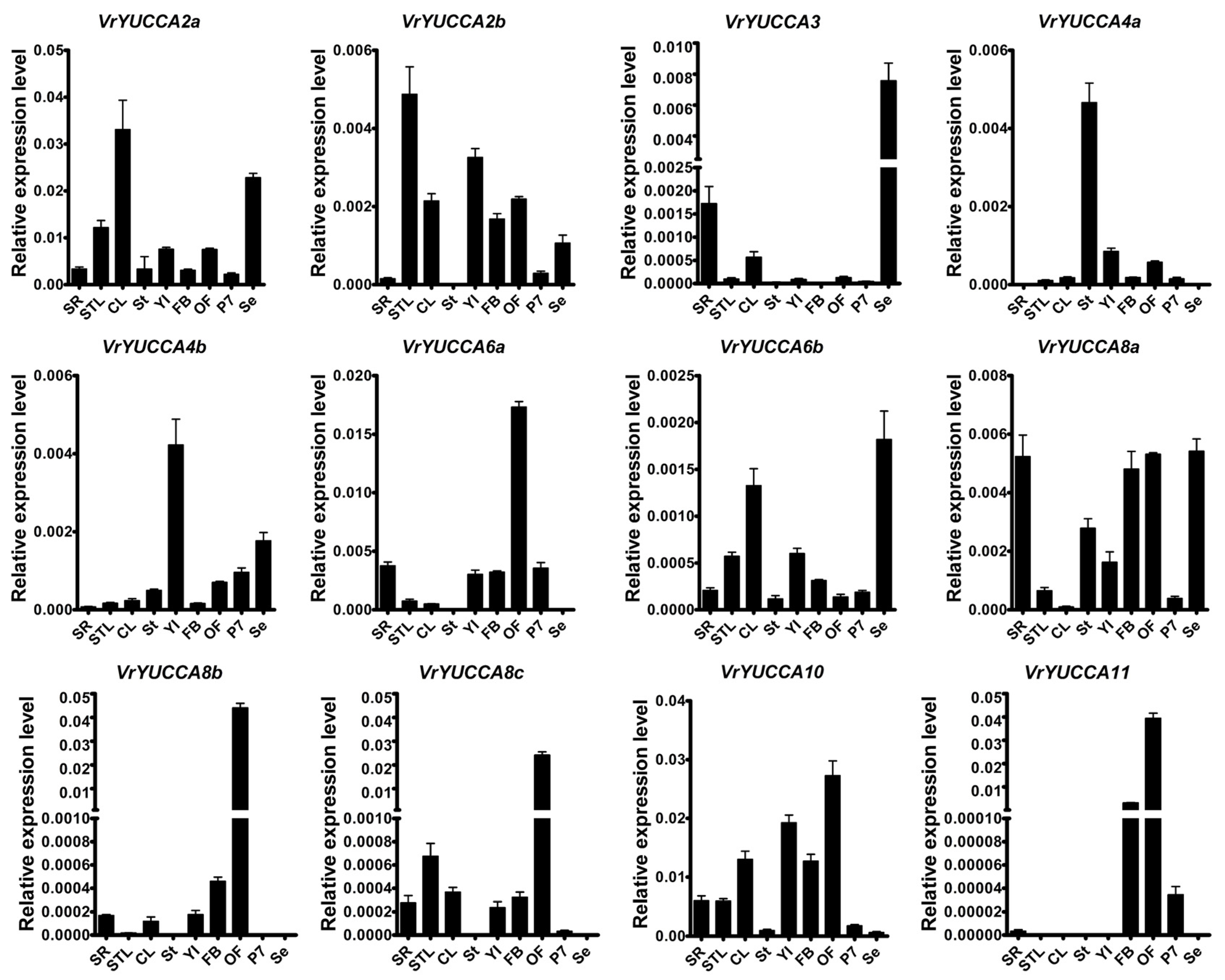
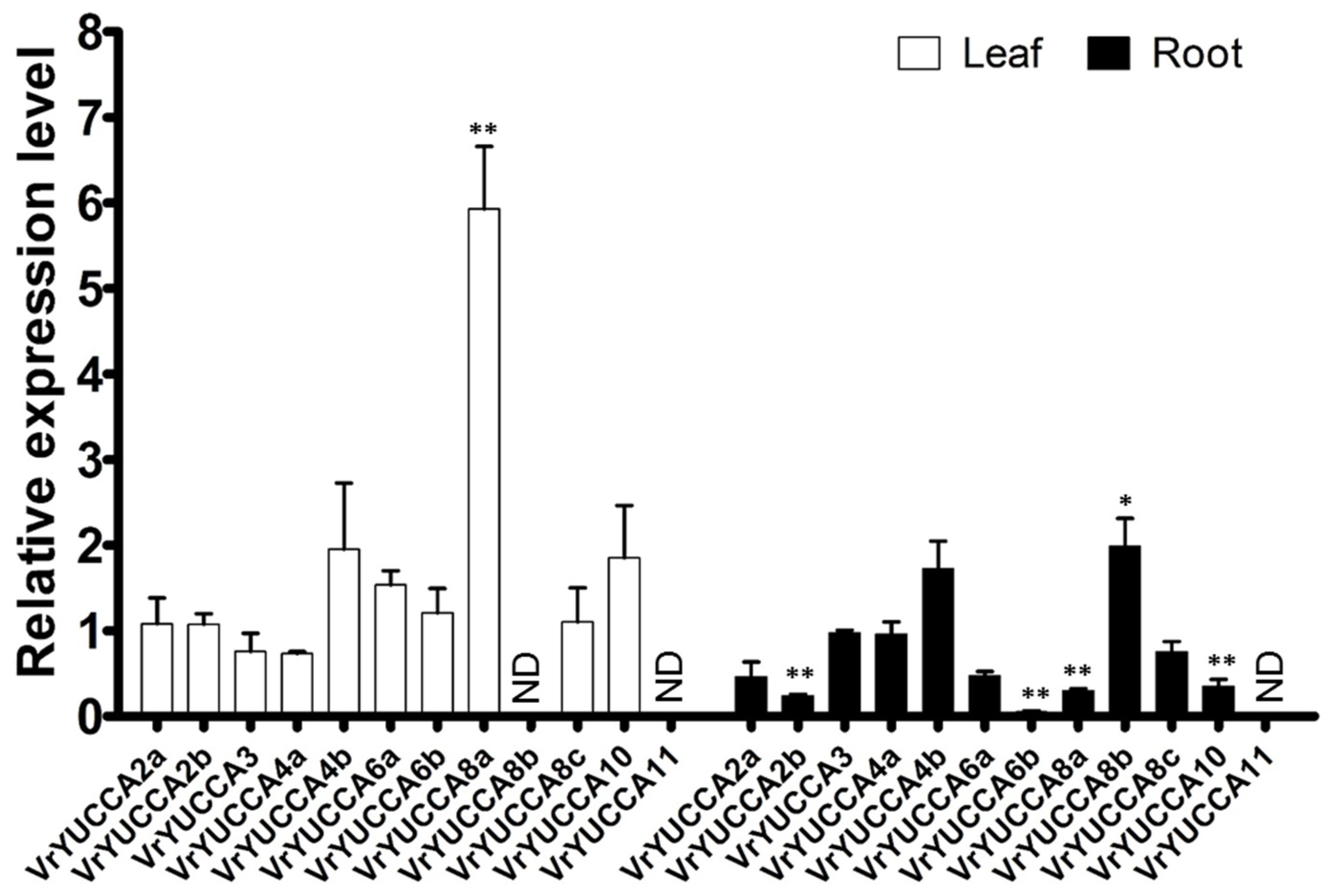
2.3. Expression Pattern of VrYUCCAs in Mungbean
2.4. VrYUCCA Proteins Showed Biological Activity Both In Vitro and In Vivo
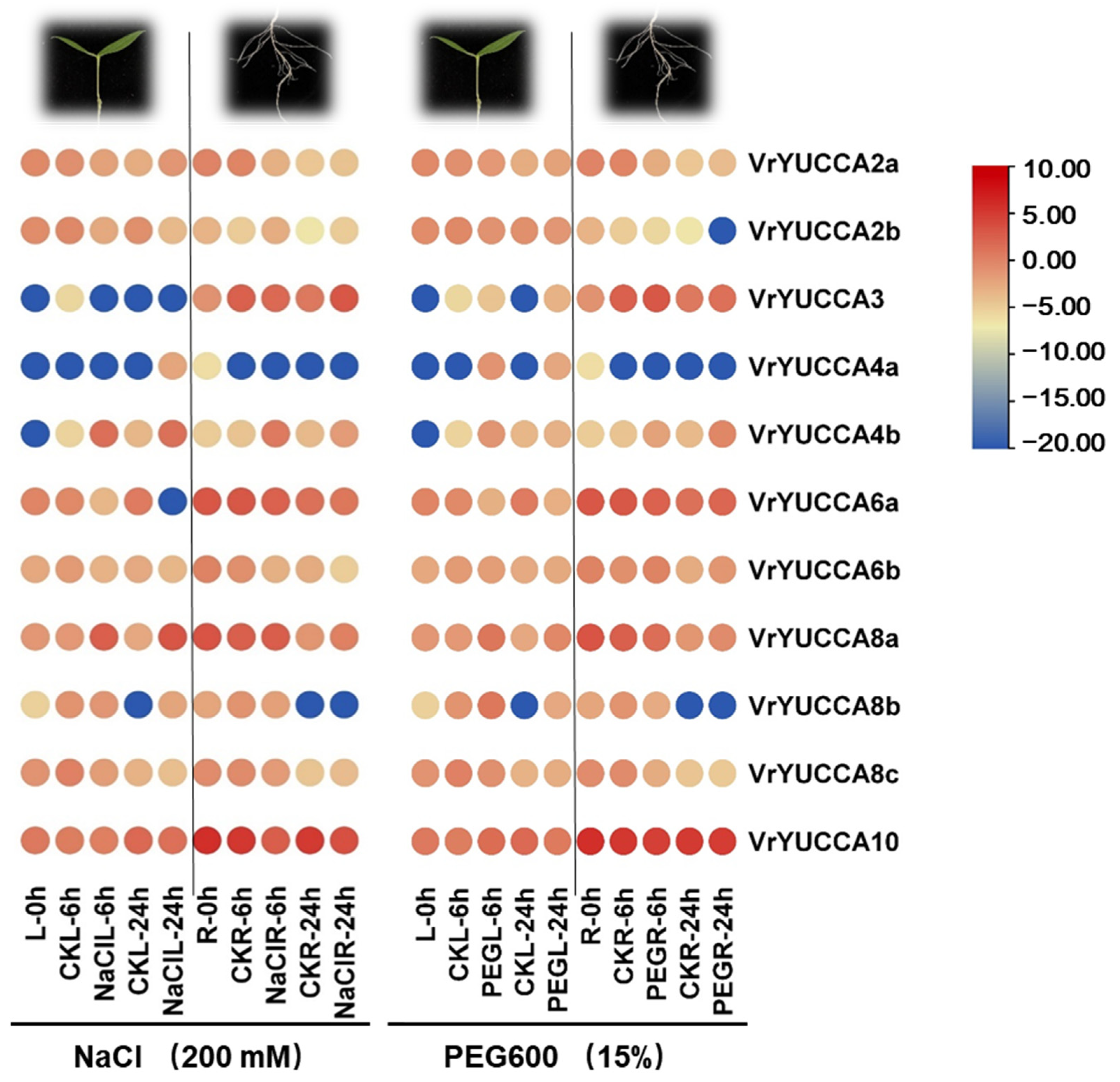
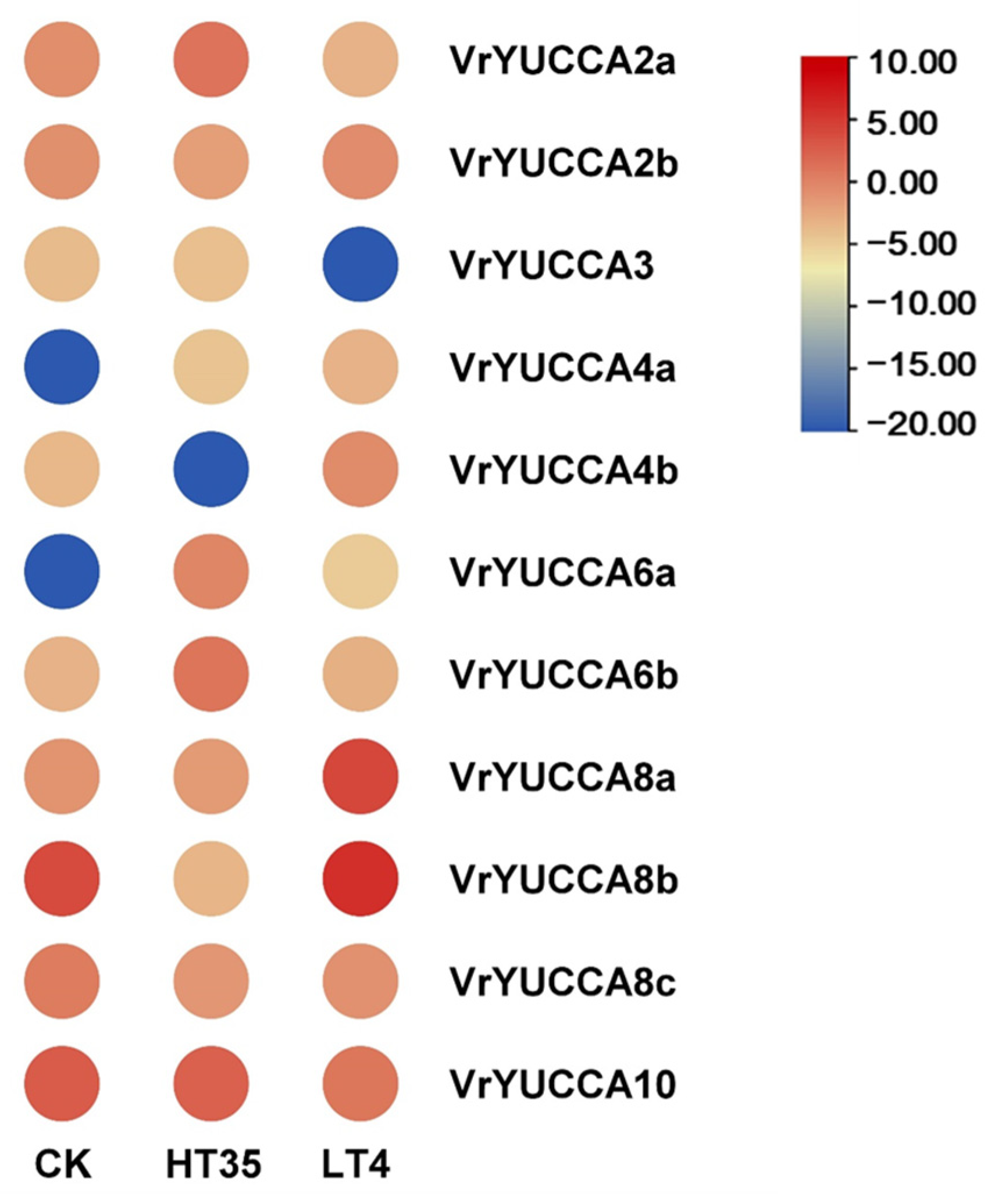
2.5. VrYUCCAs Expression in Response to Abiotic Stress Treatments
3. Discussion
4. Materials and Methods
4.1. Identification, Structure, and Motif Analysis of VrYUCCA Genes
4.2. Phylogenetic Analysis of YUCCA Genes from Multiple Species
4.3. Plant Materials and Treatment
4.4. Expression Analysis Using Real-Time PCR
4.5. Prokaryotic Expression, Purification, and Enzymatic Assays of VrYUCCA
4.6. Arabidopsis Transformation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blakeslee, J.J.; Rossi, T.S.; Kriechbaumer, V. Auxin biosynthesis: Spatial regulation and adaptation to stress. J. Exp. Bot. 2019, 70, 5041–5049. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.S.; Che, G.; Ding, L.; Chen, Z.J.; Liu, X.F.; Wang, H.Y.; Zhao, W.S.; Ning, K.; Zhao, J.Y.; Tesfamichael, K.; et al. Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Sci. Rep. 2016, 6, 20760. [Google Scholar] [CrossRef] [PubMed]
- Wojcikowska, B.; Jaskola, K.; Gasiorek, P.; Meus, M.; Nowak, K.; Gaj, M.D. LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta 2013, 238, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, S.; Friml, J. Auxin: A trigger for change in plant development. Cell 2009, 136, 1005–1016. [Google Scholar] [CrossRef]
- Wang, W.; Xu, B.; Wang, H.; Li, J.Q.; Huang, H.; Xu, L. YUCCA genes are expressed in response to leaf adaxial-abaxial juxtaposition and are required for leaf margin development. Plant Physiol. 2011, 157, 1805–1819. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, M.; Cecchetti, V. Auxin polar transport in stamen formation and development: How many actors? Front. Plant Sci. 2014, 5, 333. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Tian, L.; Yang, J.; Zhao, Y.N.; Zhu, Y.X.; Dai, X.; Zhao, Y.; Yang, Z.N. Auxin production in diploid microsporocytes is necessary and sufficient for early stages of pollen development. PLoS Genet. 2018, 14, e1007397. [Google Scholar] [CrossRef] [PubMed]
- Pandolfini, T.; Molesini, B.; Spena, A. Molecular dissection of the role of auxin in fruit initiation. Trends Plant Sci. 2007, 12, 327–329. [Google Scholar] [CrossRef]
- Ye, X.; Kang, B.G.; Osburn, L.D.; Li, Y.; Zong-Ming, C. Identification of the flavin-dependent monooxygenase-encoding YUCCA gene family in Populus trichocarpa and their expression in vegetative tissues and in response to hormone and environmental stresses. Plant Cell Tiss. Org. 2009, 97, 271–283. [Google Scholar] [CrossRef]
- Lee, M.; Jung, J.H.; Han, D.Y.; Seo, P.J.; Park, W.J.; Park, C.M. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 2012, 235, 923–938. [Google Scholar] [CrossRef]
- Cao, X.; Yang, H.L.; Shang, C.Q.; Ma, S.; Liu, L.; Cheng, J.L. The roles of auxin biosynthesis YUCCA gene family in plants. Int. J. Mol. Sci. 2019, 20, 6343. [Google Scholar] [CrossRef] [PubMed]
- Thodberg, S.; Neilson, E.J.H. The “Green” FMOs: Diversity, functionality and application of plant flavoproteins. Catalysts 2020, 10, 329. [Google Scholar] [CrossRef]
- Di, D.W.; Zhang, C.; Luo, P.; An, C.W.; Guo, G.Q. The biosynthesis of auxin: How many paths truly lead to IAA? Plant Growth Regul. 2016, 78, 275–285. [Google Scholar] [CrossRef]
- Yang, Y.L.; Xu, T.; Wang, H.G.; Feng, D.S. Genome-wide identification and expression analysis of the TaYUCCA gene family in wheat. Mol. Biol. Rep. 2021, 48, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Krueger, S.K.; Williams, D.E. Mammalian flavin-containing monooxygenases: Structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol. Ther. 2005, 106, 357–387. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kamiya, N.; Morinaka, Y.; Matsuoka, M.; Sazuka, T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 2007, 143, 1362–1371. [Google Scholar] [CrossRef] [PubMed]
- Li, W.L.; Zhao, X.Y.; Zhang, X.S. Genome-wide analysis and expression patterns of the YUCCA genes in maize. J Genet. Genom. 2015, 42, 707–710. [Google Scholar] [CrossRef]
- Wang, Y.G.; Liu, H.H.; Wang, S.P.; Li, H.J. Genome-wide identification and expression analysis of the YUCCA gene family in soybean (Glycine max L.). Plant Growth Regul. 2017, 81, 265–275. [Google Scholar] [CrossRef]
- Wang, Y.N.; Yang, W.; Zuo, Y.Y.; Zhu, L.; Hastwell, A.H.; Chen, L.; Tian, Y.P.; Su, C.; Ferguson, B.J.; Li, X. GmYUC2a mediates auxin biosynthesis during root development and nodulation in soybean. J. Exp. Bot. 2019, 70, 3165–3176. [Google Scholar] [CrossRef]
- Chen, M.M.; Li, X.; Cai, Y.M.; Zhang, Y.C.; Gu, J.J.; Yang, L.Y. Identification and expression pattern analysis of YUCCA and ARF gene families during somatic embryogenesis of Lilium spp. Biol. Plant. 2020, 64, 385–394. [Google Scholar] [CrossRef]
- Song, C.H.; Zhang, D.; Zheng, L.W.; Shen, Y.W.; Zuo, X.Y.; Mao, J.P.; Meng, Y.A.; Wu, H.Q.; Zhang, Y.K.; Liu, X.Y.; et al. Genome-wide identification and expression profiling of the YUCCA gene family in Malus domestica. Sci. Rep. 2020, 10, 10866. [Google Scholar] [CrossRef]
- Wang, X.Y.; Chen, B.Z.; Ma, C.K.; Qiao, K.K.; Li, Z.S.; Wang, J.S.; Peng, R.H.; Fan, S.L.; Ma, Q.F. Systematical characterization of YUCCA gene family in five cotton species, and potential functions of YUCCA22 gene in drought resistance of cotton. Ind. Crop. Prod. 2021, 162, 113290. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, J.; Cui, C.; Chai, L.; Zheng, B.; Jiang, J.; Li, H.; Tu, J. Genome-wide identification and expression profiling of the YUCCA gene family in Brassica napus. Oil Crop Sci. 2022, 7, 103–111. [Google Scholar] [CrossRef]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef]
- Cheng, Y.F.; Dai, X.H.; Zhao, Y.D. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef]
- Challa, K.R.; Aggarwal, P.; Nath, U. Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 2016, 28, 2117–2130. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.Y.; Miao, J.H.; Yumoto, E.; Yokota, T.; Asahina, M.; Watahiki, M. YUCCA9-mediated auxin biosynthesis and polar auxin transport synergistically regulate regeneration of root systems following root cutting. Plant Cell Physiol. 2017, 58, 1710–1723. [Google Scholar] [CrossRef]
- Cha, J.-Y.; Kim, W.-Y.; Kang, S.B.; Kim, J.I.; Baek, D.; Jung, I.J.; Kim, M.R.; Li, N.; Kim, H.-J.; Nakajima, M.; et al. A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nat. Commun. 2015, 6, 8041. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.Q.; Qi, L.L.; Li, Y.N.; Chu, J.F.; Li, C.Y. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012, 8, e1002594. [Google Scholar] [CrossRef] [PubMed]
- Perez-Alonso, M.M.; Sanchez-Parra, B.; Ortiz-Garcia, P.; Santamaria, M.E.; Diaz, I.; Pollmann, S. Jasmonic acid-dependent MYC transcription factors bind to a tandem G-box motif in the YUCCA8 and YUCCA9 promoters to regulate biotic stress responses. Int. J. Mol. Sci. 2021, 22, 9768. [Google Scholar] [CrossRef]
- Zhang, T.; Li, R.N.; Xing, J.L.; Yan, L.; Wang, R.C.; Zhao, Y.D. The YUCCA-Auxin-WOX11 module controls crown root development in rice. Front. Plant Sci. 2018, 9, 523. [Google Scholar] [CrossRef]
- Fujino, K.; Matsuda, Y.; Ozawa, K.; Nishimura, T.; Koshiba, T.; Fraaije, M.W.; Sekiguchi, H. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genom. 2008, 279, 499–507. [Google Scholar] [CrossRef]
- Woo, Y.M.; Park, H.J.; Su’udi, M.; Yang, J.I.; Park, J.J.; Back, K.; Park, Y.M.; An, G. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 2007, 65, 125–136. [Google Scholar] [CrossRef]
- Song, S.; Chen, Y.; Liu, L.; Benjamin, S.Y.H.; Mao, C.; Gan, Y.; Yu, H. OsFTIP7 determines auxin-mediated anther dehiscence in rice. Nat. Plants 2018, 4, 495–504. [Google Scholar] [CrossRef]
- Lambrides, C.J.; Godwin, I.D. Mungbean. In Pulses, Sugar and Tuber Crops. Genome Mapping & Molecular Breeding in Plants; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 3, pp. 69–90. [Google Scholar]
- Chen, J.; Somta, P.; Chen, X.; Cui, X.; Yuan, X.; Srinives, P. Gene mapping of a mutant mungbean (Vigna radiata L.) using new molecular markers suggests a gene encoding a YUC4-like protein regulates the chasmogamous flower trait. Front. Plant Sci. 2016, 7, 830. [Google Scholar] [CrossRef] [PubMed]
- Exposito-Rodriguez, M.; Borges, A.A.; Borges-Perez, A.; Perez, J.A. Gene structure and spatiotemporal expression profile of tomato genes encoding YUCCA-like flavin monooxygenases: The ToFZY gene family. Plant Physiol. Biochem. 2011, 49, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Tobena-Santamaria, R.; Bliek, M.; Ljung, K.; Sandberg, G.; Mol, J.N.M.; Souer, E.; Koes, R. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 2002, 16, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.M.; Wang, J.; Zhang, T.Y.; Hu, X.Y.; Liu, R.; Gao, T.E.; Zhao, S.J.; Yuan, Y.L.; Zheng, J.Y.; Wang, Z.R.; et al. Genome-wide identification and analysis on YUCCA gene family in Isatis indigotica Fort. and IiYUCCA6-1 functional exploration. Int. J. Mol. Sci. 2020, 21, 2188. [Google Scholar] [CrossRef]
- Chen, Q.G.; Dai, X.H.; De-Paoli, H.; Cheng, Y.F.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y.D. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef]
- Poulet, A.; Kriechbaumer, V. Bioinformatics analysis of phylogeny and transcription of TAA/YUC auxin biosynthetic genes. Int. J. Mol. Sci. 2017, 18, 1791. [Google Scholar] [CrossRef]
- Pan, L.; Zeng, W.; Niu, L.; Lu, Z.; Liu, H.; Cui, G.; Zhu, Y.; Chu, J.; Li, W.; Fang, W.; et al. PpYUC11, a strong candidate gene for the stony hard phenotype in peach (Prunus persica L. Batsch), participates in IAA biosynthesis during fruit ripening. J. Exp. Bot. 2015, 66, 7031–7044. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef]
- Won, C.; Shen, X.L.; Mashiguchi, K.; Zheng, Z.Y.; Dai, X.H.; Cheng, Y.F.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y.D. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF Arabidopsis and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef]
- Kriechbaumer, V.; Wang, P.W.; Hawes, C.; Abell, B.M. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J. 2012, 70, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Park, C.M. Auxin homeostasis in plant stress adaptation response. Plant Signal. Behav. 2007, 2, 306–307. [Google Scholar] [CrossRef]
- Ogata, Y.; Kimura, N.; Sano, R. Gcorn Plant: A database for retrieving functional and evolutionary traits of plant genes. Plant Physiol. 2019, 180, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.X.; Yan, Q.; Yuan, X.X.; Lin, Y.; Chen, J.B.; Wu, R.R.; Xue, C.C.; Zhu, Y.L.; Chen, X. Two polygalacturonase-inhibiting proteins (VrPGIP) of Vigna radiata confer resistance to bruchids (Callosobruchus spp.). J. Plant Physiol. 2021, 258–259, 153376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.R.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protocs. 2006, 1, 641–646. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, R.; Chen, J.; Lin, Y.; Jia, Q.; Guo, Y.; Liu, J.; Yan, Q.; Xue, C.; Chen, X.; Yuan, X. Genome-Wide Identification, Expression Analysis, and Potential Roles under Abiotic Stress of the YUCCA Gene Family in Mungbean (Vigna radiata L.). Int. J. Mol. Sci. 2023, 24, 1603. https://doi.org/10.3390/ijms24021603
Wu R, Chen J, Lin Y, Jia Q, Guo Y, Liu J, Yan Q, Xue C, Chen X, Yuan X. Genome-Wide Identification, Expression Analysis, and Potential Roles under Abiotic Stress of the YUCCA Gene Family in Mungbean (Vigna radiata L.). International Journal of Molecular Sciences. 2023; 24(2):1603. https://doi.org/10.3390/ijms24021603
Chicago/Turabian StyleWu, Ranran, Jingbin Chen, Yun Lin, Qiyuan Jia, Yingjian Guo, Jinyang Liu, Qiang Yan, Chenchen Xue, Xin Chen, and Xingxing Yuan. 2023. "Genome-Wide Identification, Expression Analysis, and Potential Roles under Abiotic Stress of the YUCCA Gene Family in Mungbean (Vigna radiata L.)" International Journal of Molecular Sciences 24, no. 2: 1603. https://doi.org/10.3390/ijms24021603
APA StyleWu, R., Chen, J., Lin, Y., Jia, Q., Guo, Y., Liu, J., Yan, Q., Xue, C., Chen, X., & Yuan, X. (2023). Genome-Wide Identification, Expression Analysis, and Potential Roles under Abiotic Stress of the YUCCA Gene Family in Mungbean (Vigna radiata L.). International Journal of Molecular Sciences, 24(2), 1603. https://doi.org/10.3390/ijms24021603




