Impaired Blastocyst Formation in Lnx2-Knockdown Mouse Embryos
Abstract
1. Introduction
2. Results
2.1. Lnx2 Expression in Various Tissues and Early Embryos
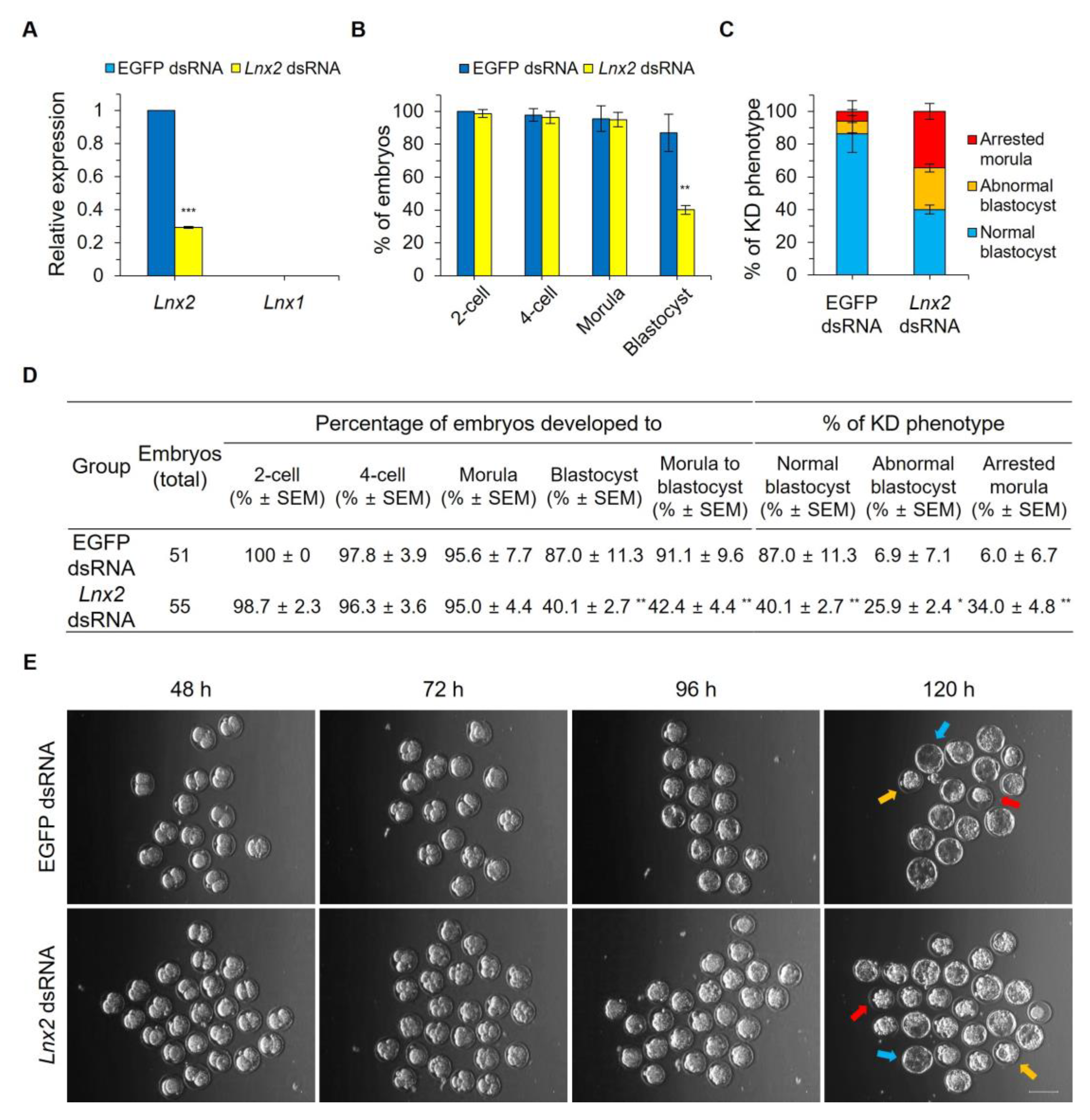
2.2. Lnx2 Knockdown Impairs Blastocyst Formation
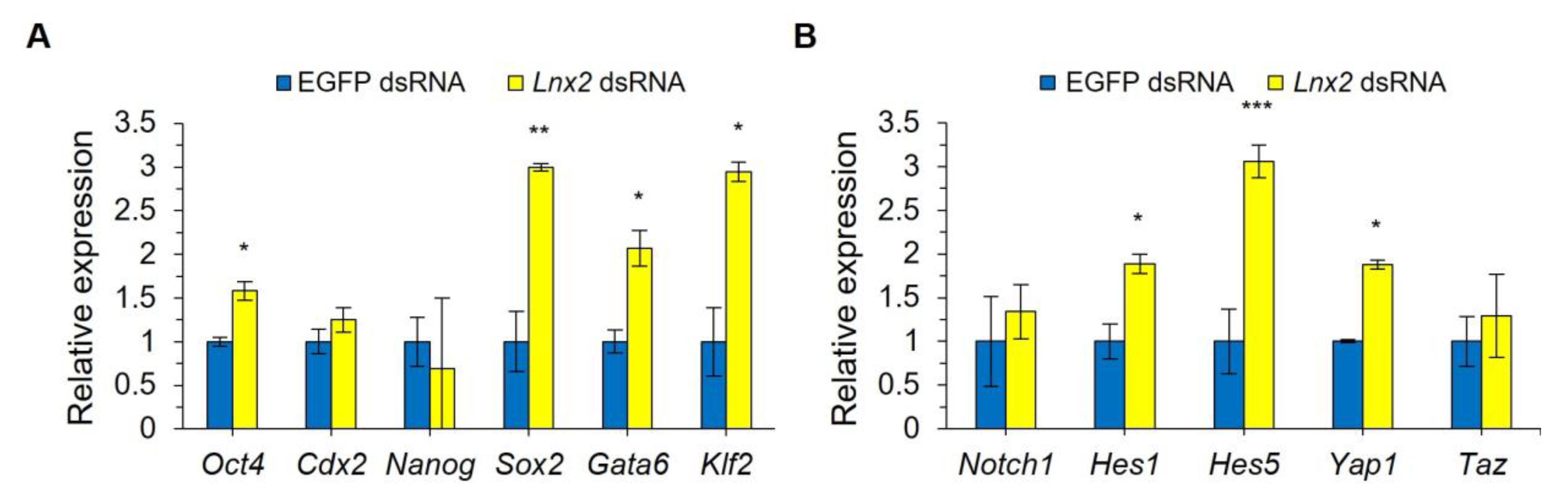
2.3. Expression of ICM-Related Gene in Lnx2 Knockdown Embryos
2.4. Lnx2 Knockdown Alters the Expression Levels of Genes Involved in Cell Signaling Pathways
3. Discussion
4. Materials and Methods
4.1. Mouse Embryo Collection and Culture
4.2. RNA Extraction and RT-PCR
4.3. Real-Time qRT-PCR
4.4. Generation of dsRNA and siRNA
4.5. Microinjection
4.6. Immunofluorescence Staining (IF)
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LNX | Ligand of Numb protein X |
| RING | Really Interesting New Gene |
| PDZ | PSD95, DLGA, ZO-1 |
| ICM | Inner cell mass |
| TE | Trophectoderm |
| EPI | Epiblast |
| PE | Primitive endoderm |
| ZGA | Zygotic gene activation |
| dsRNA | Double-strand RNA |
| siRNA | Small interfering RNA |
| NICD | Notch1 intracellular domain |
| RBPJ | Recombination signal binding protein for immunoglobulin kappa J region |
| CSL | CBF1, Suppressor of Hairless, Lag-1 |
| MAM | Mastermind |
| EGFP | Enhanced green fluorescent protein |
| hCG | Human chorionic gonadotropin |
| KSOM | Potassium-supplemented simplex optimized medium |
| hKSOM | KSOM with HEPES |
References
- Zheng, W.; Liu, K. Maternal control of mouse preimplantation development. Mouse Dev. 2012, 55, 115–139. [Google Scholar]
- Strumpf, D.; Mao, C.-A.; Yamanaka, Y.; Ralston, A.; Chawengsaksophak, K.; Beck, F.; Rossant, J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 2005, 132, 2093–2102. [Google Scholar] [CrossRef]
- Nichols, J.; Zevnik, B.; Anastassiadis, K.; Niwa, H.; Klewe-Nebenius, D.; Chambers, I.; Schöler, H.; Smith, A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998, 95, 379–391. [Google Scholar] [CrossRef]
- Engel-Pizcueta, C.; Pujades, C. Interplay Between Notch and YAP/TAZ Pathways in the Regulation of Cell Fate During Embryo Development. Front. Cell Dev. Biol. 2021, 9, 711531. [Google Scholar] [CrossRef]
- Batista, M.R.; Diniz, P.; Murta, D.; Torres, A.; Lopes-da-Costa, L.; Silva, E. Balanced Notch-Wnt signaling interplay is required for mouse embryo and fetal development. Reproduction 2021, 161, 385–398. [Google Scholar] [CrossRef]
- Hirate, Y.; Hirahara, S.; Inoue, K.-i.; Suzuki, A.; Alarcon, V.B.; Akimoto, K.; Hirai, T.; Hara, T.; Adachi, M.; Chida, K. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr. Biol. 2013, 23, 1181–1194. [Google Scholar] [CrossRef] [PubMed]
- Rice, D.S.; Northcutt, G.M.; Kurschner, C. The Lnx family proteins function as molecular scaffolds for Numb family proteins. Mol. Cell Neurosci. 2001, 18, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Back to the future with ubiquitin. Cell 2004, 116, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Nourry, C.; Grant, S.G.; Borg, J.-P. PDZ domain proteins: Plug and play! Sci. STKE 2003, 2003, re7–re7. [Google Scholar] [CrossRef]
- Lv, B.; Liu, C.; Chen, Y.; Qi, L.; Wang, L.; Ji, Y.; Xue, Z. Light-induced injury in mouse embryos revealed by single-cell RNA sequencing. Biol. Res. 2019, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; McGill, M.A.; Dermer, M.; Dho, S.E.; Wolting, C.D.; McGlade, C.J. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 2002, 21, 93–102. [Google Scholar] [CrossRef]
- McGill, M.A.; McGlade, C.J. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 2003, 278, 23196–23203. [Google Scholar] [CrossRef]
- Zhou, J.; Fujiwara, T.; Ye, S.; Li, X.; Zhao, H. Ubiquitin E3 ligase LNX2 is critical for osteoclastogenesis in vitro by regulating M-CSF/RANKL signaling and Notch2. Calcif. Tissue Int. 2015, 96, 465–475. [Google Scholar] [CrossRef]
- Won, M.; Ro, H.; Dawid, I.B. Lnx2 ubiquitin ligase is essential for exocrine cell differentiation in the early zebrafish pancreas. Proc. Natl. Acad. Sci. USA 2015, 112, 12426–12431. [Google Scholar] [CrossRef]
- Li, L.; Fridley, B.L.; Kalari, K.; Niu, N.; Jenkins, G.; Batzler, A.; Abo, R.P.; Schaid, D.; Wang, L. Discovery of genetic biomarkers contributing to variation in drug response of cytidine analogues using human lymphoblastoid cell lines. BMC Genom. 2014, 15, 93. [Google Scholar] [CrossRef]
- Wang, H.; Kane, A.W.; Lee, C.; Ahn, S. Gli3 repressor controls cell fates and cell adhesion for proper establishment of neurogenic niche. Cell Rep. 2014, 8, 1093–1104. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.; Hreinsson, J.; Strand, M.L.; Hovatta, O.; Soder, O.; Philipson, L.; Pettersson, R.F.; Sollerbrant, K. Coxsackievirus and adenovirus receptor (CAR) is expressed in male germ cells and forms a complex with the differentiation factor JAM-C in mouse testis. Exp. Cell Res. 2006, 312, 817–830. [Google Scholar] [CrossRef]
- Mittal, V. Improving the efficiency of RNA interference in mammals. Nat. Rev. Genet. 2004, 5, 355–365. [Google Scholar] [CrossRef]
- Vicentini, F.T.M.d.C.; Borgheti-Cardoso, L.N.; Depieri, L.V.; de Macedo Mano, D.; Abelha, T.F.; Petrilli, R.; Bentley, M.V.L.B. Delivery systems and local administration routes for therapeutic siRNA. Pharm. Res. 2013, 30, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, J.; Jeong, J.; Hong, S.H.; Kim, D.; Choi, S.; Choi, I.; Oh, J.S.; Cho, C. Identification of a novel embryo-prevalent gene, Gm11545, involved in preimplantation embryogenesis in mice. FASEB J. 2019, 33, 11326–11337. [Google Scholar] [CrossRef] [PubMed]
- Young, P.W. LNX1/LNX2 proteins: Functions in neuronal signalling and beyond. Neuronal Signal. 2018, 2, NS20170191. [Google Scholar] [CrossRef]
- Chazaud, C.; Yamanaka, Y.; Pawson, T.; Rossant, J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell 2006, 10, 615–624. [Google Scholar] [CrossRef]
- Masui, S.; Nakatake, Y.; Toyooka, Y.; Shimosato, D.; Yagi, R.; Takahashi, K.; Okochi, H.; Okuda, A.; Matoba, R.; Sharov, A.A. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 2007, 9, 625–635. [Google Scholar] [CrossRef]
- Mitsui, K.; Tokuzawa, Y.; Itoh, H.; Segawa, K.; Murakami, M.; Takahashi, K.; Maruyama, M.; Maeda, M.; Yamanaka, S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003, 113, 631–642. [Google Scholar] [CrossRef]
- Boroviak, T.; Loos, R.; Bertone, P.; Smith, A.; Nichols, J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 2014, 16, 513–525. [Google Scholar] [CrossRef]
- Schrode, N.; Saiz, N.; Di Talia, S.; Hadjantonakis, A.-K. GATA6 levels modulate primitive endoderm cell fate choice and timing in the mouse blastocyst. Dev. Cell 2014, 29, 454–467. [Google Scholar] [CrossRef]
- Hall, J.; Guo, G.; Wray, J.; Eyres, I.; Nichols, J.; Grotewold, L.; Morfopoulou, S.; Humphreys, P.; Mansfield, W.; Walker, R. Oct4 and LIF/Stat3 additively induce Krüppel factors to sustain embryonic stem cell self-renewal. Cell Stem. Cell 2009, 5, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Chan, Y.-S.; Loh, Y.-H.; Cai, J.; Tong, G.-Q.; Lim, C.-A.; Robson, P.; Zhong, S.; Ng, H.-H. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 2008, 10, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Cave, J.W. Selective repression of Notch pathway target gene transcription. Dev. Biol. 2011, 360, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Ro, H.; Dawid, I.B. Modulation of Tcf3 repressor complex composition regulates cdx4 expression in zebrafish. EMBO J. 2011, 30, 2894–2907. [Google Scholar] [CrossRef]
- Yi, F.; Pereira, L.; Merrill, B.J. Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cells 2008, 26, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.F.; Johnstone, S.E.; Newman, J.J.; Kagey, M.H.; Young, R.A. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008, 22, 746–755. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Lyu, J.; Wang, F.; Miao, C.; Nan, Z.; Zhang, J.; Xi, Y.; Zhou, Q.; Yang, X.; Ge, W. The histone deacetylase HDAC1 positively regulates Notch signaling during Drosophila wing development. Biol. Open 2018, 7, bio029637. [Google Scholar] [CrossRef]
- Wu, L.M.N.; Wang, J.; Conidi, A.; Zhao, C.; Wang, H.; Ford, Z.; Zhang, L.; Zweier, C.; Ayee, B.G.; Maurel, P. Zeb2 recruits HDAC–NuRD to inhibit Notch and controls Schwann cell differentiation and remyelination. Nat. Neurosci. 2016, 19, 1060–1072. [Google Scholar] [CrossRef]
- Ro, H.; Dawid, I.B. Organizer restriction through modulation of Bozozok stability by the E3 ubiquitin ligase Lnx-like. Nat. Cell Biol. 2009, 11, 1121–1127. [Google Scholar] [CrossRef]
- Chen, J.; Xu, J.; Zhao, W.; Hu, G.; Cheng, H.; Kang, Y.; Xie, Y.; Lu, Y. Characterization of human LNX, a novel ligand of Numb protein X that is downregulated in human gliomas. Int. J. Biochem. Cell Biol. 2005, 37, 2273–2283. [Google Scholar] [CrossRef]
- Nishioka, N.; Yamamoto, S.; Kiyonari, H.; Sato, H.; Sawada, A.; Ota, M.; Nakao, K.; Sasaki, H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 2008, 125, 270–283. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-J.; Kim, J.; Han, G.; Hong, S.-P.; Kim, D.; Cho, C. Impaired Blastocyst Formation in Lnx2-Knockdown Mouse Embryos. Int. J. Mol. Sci. 2023, 24, 1385. https://doi.org/10.3390/ijms24021385
Lee S-J, Kim J, Han G, Hong S-P, Kim D, Cho C. Impaired Blastocyst Formation in Lnx2-Knockdown Mouse Embryos. International Journal of Molecular Sciences. 2023; 24(2):1385. https://doi.org/10.3390/ijms24021385
Chicago/Turabian StyleLee, Seung-Jae, Jaehwan Kim, Gwidong Han, Seung-Pyo Hong, Dayeon Kim, and Chunghee Cho. 2023. "Impaired Blastocyst Formation in Lnx2-Knockdown Mouse Embryos" International Journal of Molecular Sciences 24, no. 2: 1385. https://doi.org/10.3390/ijms24021385
APA StyleLee, S.-J., Kim, J., Han, G., Hong, S.-P., Kim, D., & Cho, C. (2023). Impaired Blastocyst Formation in Lnx2-Knockdown Mouse Embryos. International Journal of Molecular Sciences, 24(2), 1385. https://doi.org/10.3390/ijms24021385




