Absence of Scaffold Protein Tks4 Disrupts Several Signaling Pathways in Colon Cancer Cells
Abstract
1. Introduction
2. Results
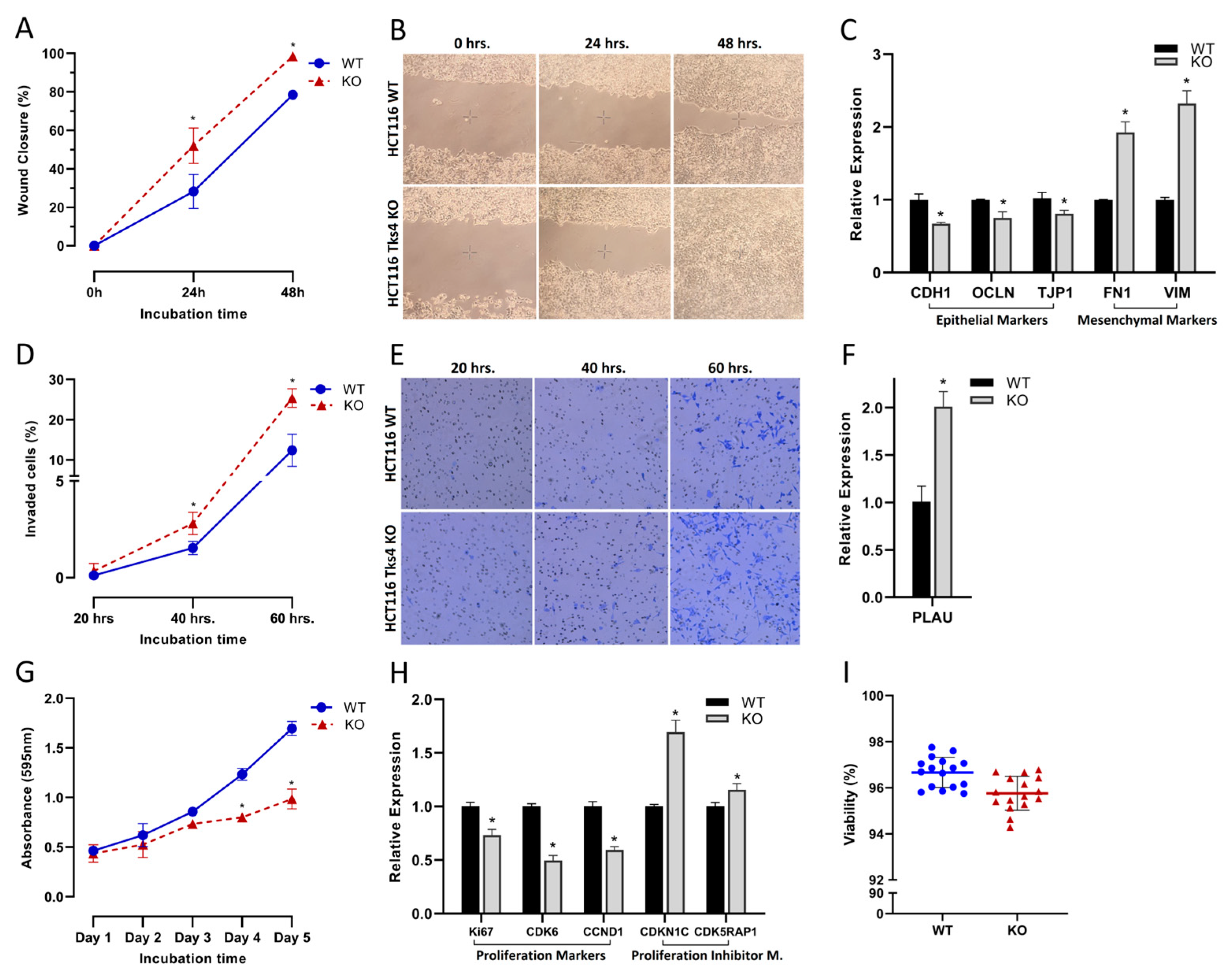
2.1. Changes in Cell Motility and Proliferation
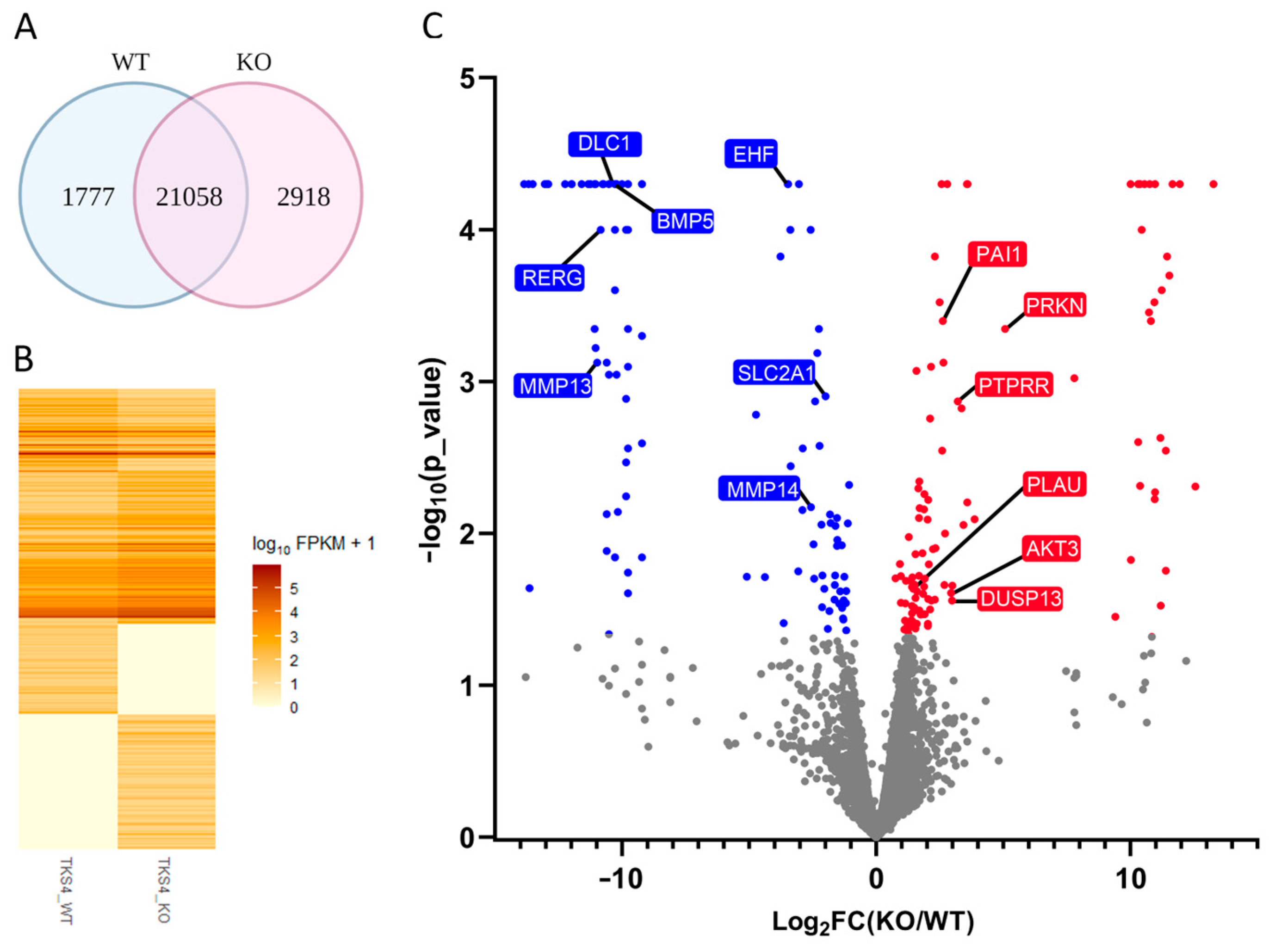
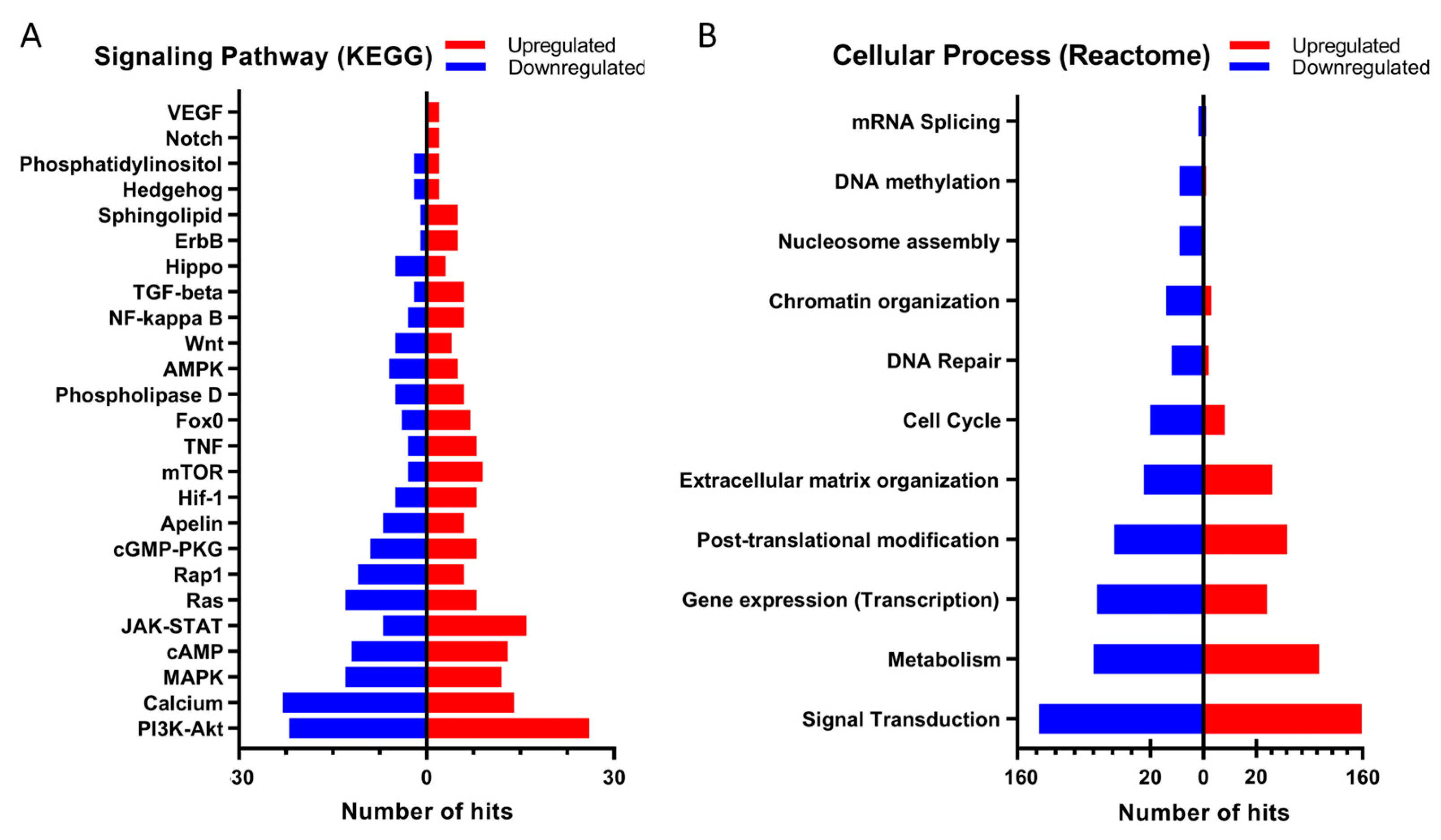
2.2. Changes in mRNA Levels
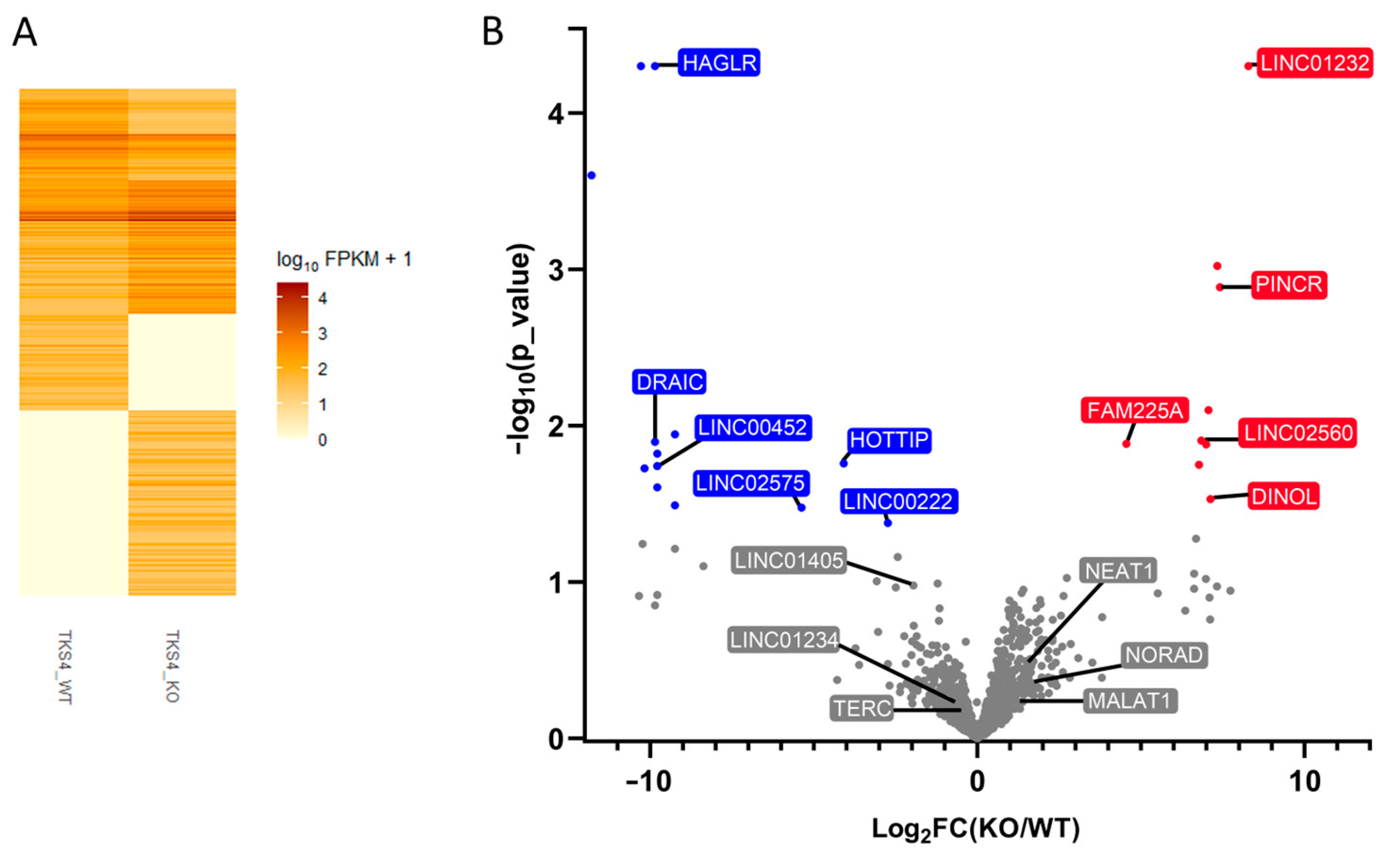
2.3. Changes in lncRNA Levels
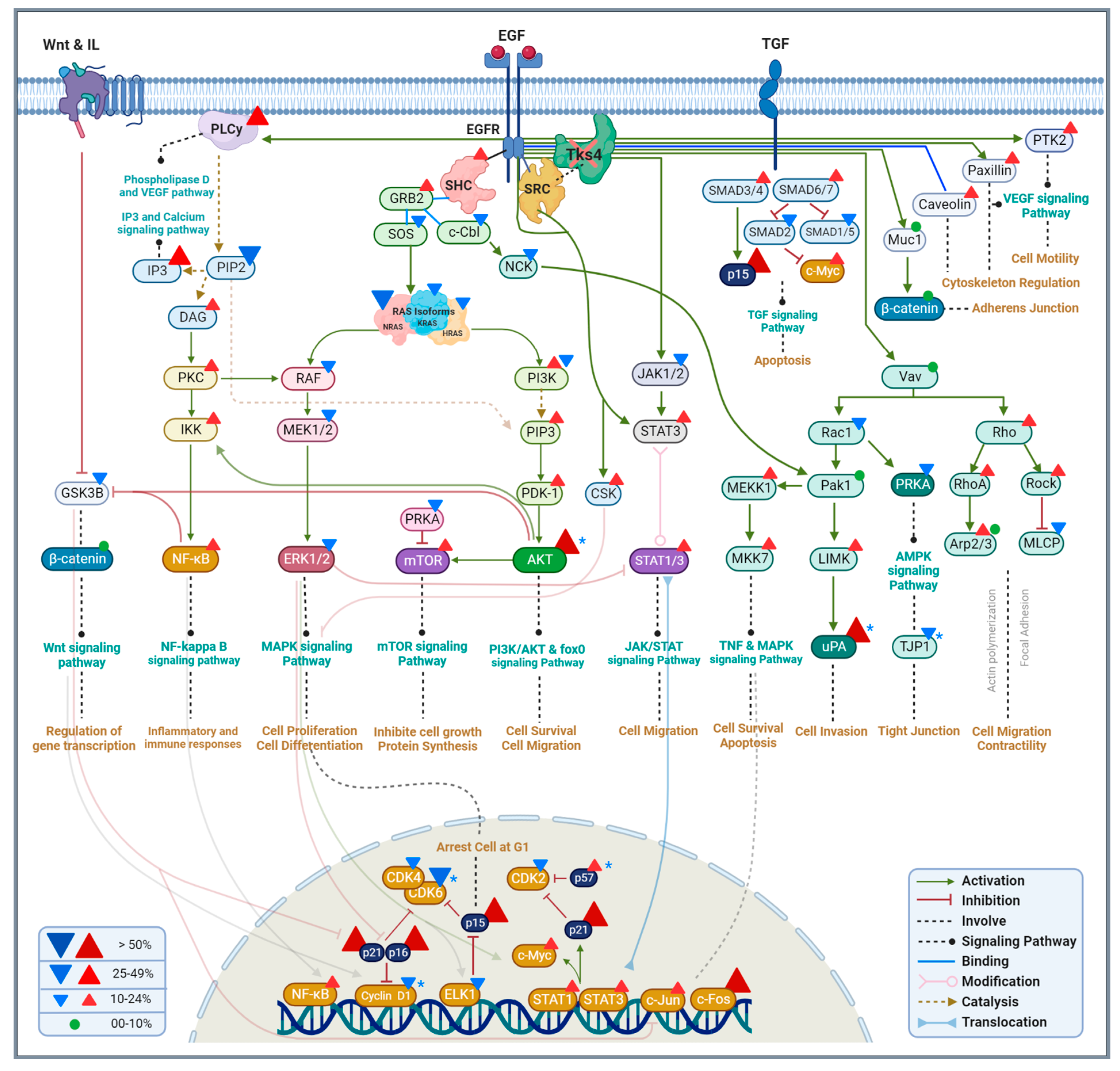
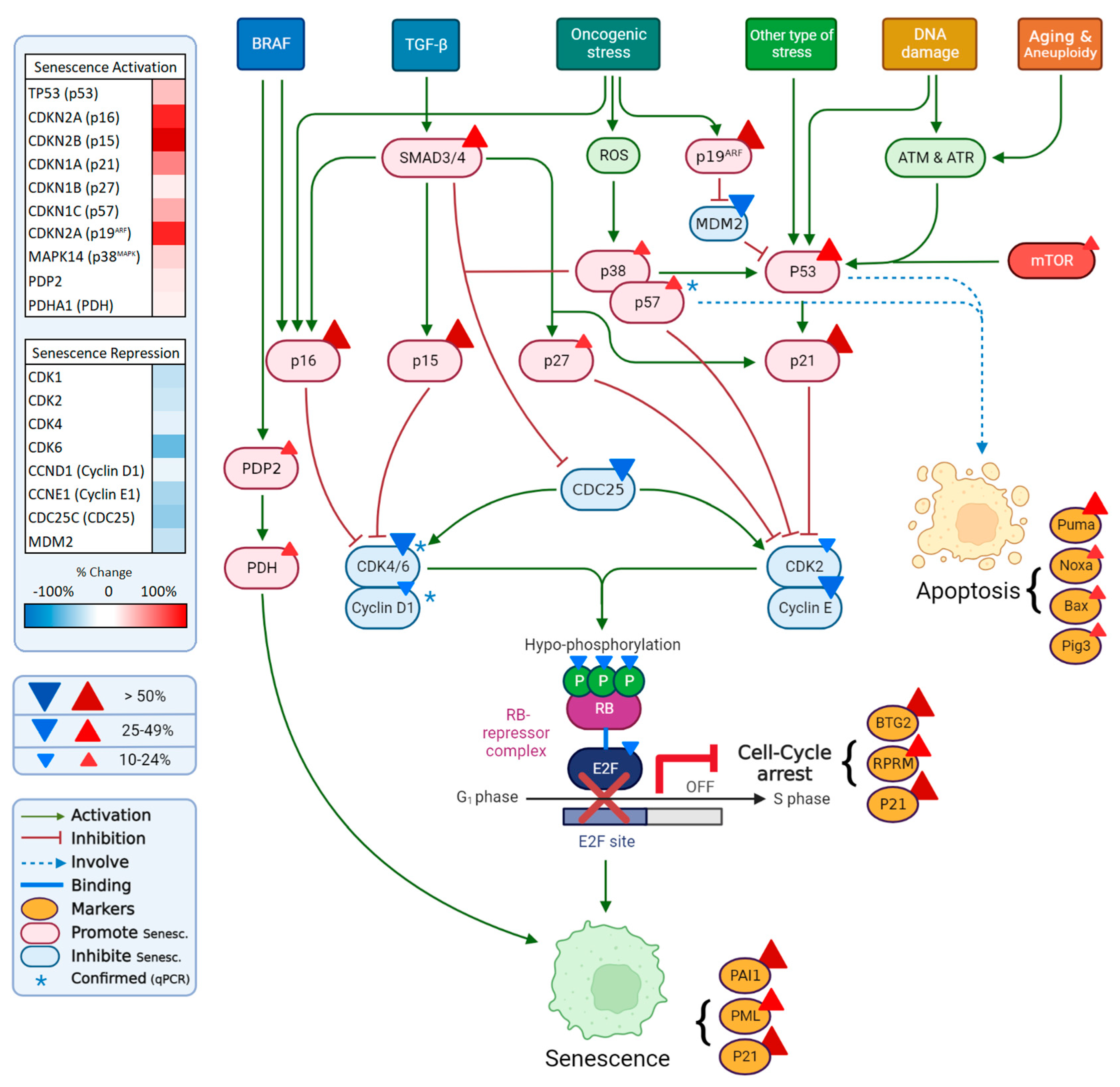
3. Discussion
3.1. The PI3K/AKT Pathway
3.2. The MAPK/ERK Pathway
3.3. Epithelial Mesenchymal Transition
3.4. Podosome Formation
3.5. Proliferation, Senescence and Apoptosis
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Proliferation Assay
4.3. Cell Migration Assay
4.4. Transwell Invasion Assay
4.5. Viability Assay
4.6. Total RNA Extraction and Real-Time Quantitative PCR
4.7. RNA Sequencing
4.8. RNA Sequencing Data Analysis
4.9. Protein Extraction and Western Blot Analysis
4.10. Immunocytochemistry and Fluorescence Microscopy
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cipak, L. Protein Kinases: Function, Substrates, and Implication in Diseases. Int. J. Mol. Sci. 2022, 23, 3560. [Google Scholar] [CrossRef]
- Buday, L.; Tompa, P. Functional classification of scaffold proteins and related molecules. FEBS J. 2010, 277, 4348–4355. [Google Scholar] [CrossRef]
- Buschman, M.D.; Bromann, P.A.; Cejudo-Martin, P.; Wen, F.; Pass, I.; Courtneidge, S.A. The novel adaptor protein Tks4 (SH3PXD2B) is required for functional podosome formation. Mol. Biol. Cell 2009, 20, 1302–1311. [Google Scholar] [CrossRef]
- Kudlik, G.; Takács, T.; Radnai, L.; Kurilla, A.; Szeder, B.; Koprivanacz, K.; Merő, B.L.; Buday, L.; Vas, V. Advances in Understanding TKS4 and TKS5: Molecular Scaffolds Regulating Cellular Processes from Podosome and Invadopodium Formation to Differentiation and Tissue Homeostasis. Int. J. Mol. Sci. 2020, 21, 8117. [Google Scholar] [CrossRef]
- Merő, B.; Koprivanacz, K.; Cserkaszky, A.; Radnai, L.; Vas, V.; Kudlik, G.; Gógl, G.; Sok, P.; Póti, Á.L.; Szeder, B.; et al. Characterization of the Intramolecular Interactions and Regulatory Mechanisms of the Scaffold Protein Tks4. Int. J. Mol. Sci. 2021, 22, 8103. [Google Scholar] [CrossRef]
- Kurochkina, N.; Guha, U. SH3 domains: Modules of protein-protein interactions. Biophys. Rev. 2013, 5, 29–39. [Google Scholar] [CrossRef]
- Bromann, P.A.; Korkaya, H.; Courtneidge, S.A. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene 2004, 23, 7957–7968. [Google Scholar] [CrossRef]
- Dülk, M.; Szeder, B.; Glatz, G.; Merő, B.L.; Koprivanacz, K.; Kudlik, G.; Vas, V.; Sipeki, S.; Cserkaszky, A.; Radnai, L.; et al. EGF Regulates the Interaction of Tks4 with Src through Its SH2 and SH3 Domains. Biochemistry 2018, 57, 4186–4196. [Google Scholar] [CrossRef]
- Bögel, G.; Gujdár, A.; Geiszt, M.; Lányi, Á.; Fekete, A.; Sipeki, S.; Downward, J.; Buday, L. Frank-ter Haar syndrome protein Tks4 regulates epidermal growth factor-dependent cell migration. J. Biol. Chem. 2012, 287, 31321–31329. [Google Scholar] [CrossRef]
- Iqbal, Z.; Cejudo-Martin, P.; de Brouwer, A.; van der Zwaag, B.; Ruiz-Lozano, P.; Scimia, M.C.; Lindsey, J.D.; Weinreb, R.; Albrecht, B.; Megarbane, A.; et al. Disruption of the podosome adaptor protein TKS4 (SH3PXD2B) causes the skeletal dysplasia, eye, and cardiac abnormalities of Frank-Ter Haar Syndrome. Am. J. Hum. Genet. 2010, 86, 254–261. [Google Scholar] [CrossRef]
- Durand, B.; Stoetzel, C.; Schaefer, E.; Calmels, N.; Scheidecker, S.; Kempf, N.; De Melo, C.; Guilbert, A.-S.; Timbolschi, D.; Donato, L.; et al. A severe case of Frank-ter Haar syndrome and literature review: Further delineation of the phenotypical spectrum. Eur. J. Med. Genet. 2020, 63, 103857. [Google Scholar] [CrossRef]
- Ratukondla, B.; Prakash, S.; Reddy, S.; Puthuran, G.V.; Kannan, N.B.; Pillai, M.R. A Rare Case Report of Frank Ter Haar Syndrome in a Sibling Pair Presenting with Congenital Glaucoma. J. Glaucoma 2020, 29, 236–238. [Google Scholar] [CrossRef]
- Mao, M.; Thedens, D.R.; Chang, B.; Harris, B.S.; Zheng, Q.Y.; Johnson, K.R.; Donahue, L.R.; Anderson, M.G. The podosomal-adaptor protein SH3PXD2B is essential for normal postnatal development. Mamm. Genome 2009, 20, 462–475. [Google Scholar] [CrossRef]
- Vas, V.; Háhner, T.; Kudlik, G.; Ernszt, D.; Kvell, K.; Kuti, D.; Kovács, K.J.; Tóvári, J.; Trexler, M.; Merő, B.L.; et al. Analysis of Tks4 Knockout Mice Suggests a Role for Tks4 in Adipose Tissue Homeostasis in the Context of Beigeing. Cells 2019, 8, 831. [Google Scholar] [CrossRef]
- Hynes, N.E.; Lane, H.A. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat. Rev. Cancer 2005, 5, 341–354. [Google Scholar] [CrossRef]
- Hallberg, B.; Rayter, S.I.; Downward, J. Interaction of Ras and Raf in intact mammalian cells upon extracellular stimulation. J. Biol. Chem. 1994, 269, 3913–3916. [Google Scholar] [CrossRef]
- Vivanco, I.; Sawyers, C.L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer 2002, 2, 489–501. [Google Scholar] [CrossRef]
- Patterson, R.L.; van Rossum, D.B.; Nikolaidis, N.; Gill, D.L.; Snyder, S.H. Phospholipase C-gamma: Diverse roles in receptor-mediated calcium signaling. Trends Biochem. Sci. 2005, 30, 688–697. [Google Scholar] [CrossRef]
- Tomar, A.; Schlaepfer, D.D. A PAK-activated linker for EGFR and FAK. Dev. Cell 2010, 18, 170–172. [Google Scholar] [CrossRef]
- Dülk, M.; Kudlik, G.; Fekete, A.; Ernszt, D.; Kvell, K.; Pongrácz, J.E.; Merő, B.L.; Szeder, B.; Radnai, L.; Geiszt, M.; et al. The scaffold protein Tks4 is required for the differentiation of mesenchymal stromal cells (MSCs) into adipogenic and osteogenic lineages. Sci. Rep. 2016, 6, 34280. [Google Scholar] [CrossRef]
- Vas, V.; Kovács, T.; Körmendi, S.; Bródy, A.; Kudlik, G.; Szeder, B.; Mező, D.; Kállai, D.; Koprivanacz, K.; Merő, B.L.; et al. Significance of the Tks4 scaffold protein in bone tissue homeostasis. Sci. Rep. 2019, 9, 5781. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, S.; Abdullah, C.; Buschman, M.D.; Diaz, B.; Courtneidge, S.A. The role of Tks adaptor proteins in invadopodia formation, growth and metastasis of melanoma. Oncotarget 2016, 7, 78473–78486. [Google Scholar] [CrossRef] [PubMed]
- Mitre, G.P.; Balbinot, K.M.; Ribeiro, A.L.R.; da Silva Kataoka, M.S.; de Melo Alves Júnior, S.; de Jesus Viana Pinheiro, J. Key proteins of invadopodia are overexpressed in oral squamous cell carcinoma suggesting an important role of MT1-MMP in the tumoral progression. Diagn. Pathol. 2021, 16, 33. [Google Scholar] [CrossRef]
- Kui, X.; Wang, Y.; Zhang, C.; Li, H.; Li, Q.; Ke, Y.; Wang, L. Prognostic value of SH3PXD2B (Tks4) in human hepatocellular carcinoma: A combined multi-omics and experimental study. BMC Med. Genom. 2021, 14, 115. [Google Scholar] [CrossRef] [PubMed]
- Leong, H.S.; Robertson, A.E.; Stoletov, K.; Leith, S.J.; Chin, C.A.; Chien, A.E.; Hague, M.N.; Ablack, A.; Carmine-Simmen, K.; McPherson, V.A.; et al. Invadopodia are required for cancer cell extravasation and are a therapeutic target for metastasis. Cell Rep. 2014, 8, 1558–1570. [Google Scholar] [CrossRef]
- Gianni, D.; Taulet, N.; DerMardirossian, C.; Bokoch, G.M. c-Src-mediated phosphorylation of NoxA1 and Tks4 induces the reactive oxygen species (ROS)-dependent formation of functional invadopodia in human colon cancer cells. Mol. Biol. Cell 2010, 21, 4287–4298. [Google Scholar] [CrossRef]
- Szeder, B.; Tárnoki-Zách, J.; Lakatos, D.; Vas, V.; Kudlik, G.; Merő, B.; Koprivanacz, K.; Bányai, L.; Hámori, L.; Róna, G.; et al. Absence of the Tks4 Scaffold Protein Induces Epithelial-Mesenchymal Transition-Like Changes in Human Colon Cancer Cells. Cells 2019, 8, 1343. [Google Scholar] [CrossRef]
- Zeisberg, M.; Neilson, E.G. Biomarkers for epithelial-mesenchymal transitions. J. Clin. Invest. 2009, 119, 1429–1437. [Google Scholar] [CrossRef]
- Mahmood, N.; Mihalcioiu, C.; Rabbani, S.A. Multifaceted Role of the Urokinase-Type Plasminogen Activator (uPA) and Its Receptor (uPAR): Diagnostic, Prognostic, and Therapeutic Applications. Front. Oncol. 2018, 8, 24. [Google Scholar] [CrossRef]
- Andrés-Sánchez, N.; Fisher, D.; Krasinska, L. Physiological functions and roles in cancer of the proliferation marker Ki-67. J. Cell Sci. 2022, 135, 258932. [Google Scholar] [CrossRef]
- Montalto, F.I.; De Amicis, F. Cyclin D1 in Cancer: A Molecular Connection for Cell Cycle Control, Adhesion and Invasion in Tumor and Stroma. Cells 2020, 9, 2648. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, B.; Finn, R.S.; Turner, N.C. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 2016, 13, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, E.; Joseph, B. The hallmarks of CDKN1C (p57, KIP2) in cancer. Biochim. Biophys. Acta 2011, 1816, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Ching, Y.-P.; Pang, A.S.H.; Lam, W.-H.; Qi, R.Z.; Wang, J.H. Identification of a neuronal Cdk5 activator-binding protein as Cdk5 inhibitor. J. Biol. Chem. 2002, 277, 15237–15240. [Google Scholar] [CrossRef] [PubMed]
- Pavón, M.A.; Arroyo-Solera, I.; Céspedes, M.V.; Casanova, I.; León, X.; Mangues, R. uPA/uPAR and SERPINE1 in head and neck cancer: Role in tumor resistance, metastasis, prognosis and therapy. Oncotarget 2016, 7, 57351–57366. [Google Scholar] [CrossRef]
- Wilkins-Port, C.E.; Ye, Q.; Mazurkiewicz, J.E.; Higgins, P.J. TGF-beta1 + EGF-initiated invasive potential in transformed human keratinocytes is coupled to a plasmin/MMP-10/MMP-1-dependent collagen remodeling axis: Role for PAI-1. Cancer Res. 2009, 69, 4081–4091. [Google Scholar] [CrossRef]
- Moriya, C.; Jinnin, M.; Yamane, K.; Maruo, K.; Muchemwa, F.C.; Igata, T.; Makino, T.; Fukushima, S.; Ihn, H. Expression of matrix metalloproteinase-13 is controlled by IL-13 via PI3K/Akt3 and PKC-δ in normal human dermal fibroblasts. J. Invest. Dermatol. 2011, 131, 655–661. [Google Scholar] [CrossRef]
- Wang, J.-L.; Wang, Y.; Ren, G.-P. Identification of PTPRR and JAG1 as key genes in castration-resistant prostate cancer by integrated bioinformatics methods. J. Zhejiang Univ. Sci. B 2020, 21, 246–255. [Google Scholar] [CrossRef]
- Park, J.E.; Park, B.C.; Kim, H.-A.; Song, M.; Park, S.G.; Lee, D.H.; Kim, H.-J.; Choi, H.-K.; Kim, J.-T.; Cho, S. Positive regulation of apoptosis signal-regulating kinase 1 by dual-specificity phosphatase 13A. Cell Mol. Life Sci. 2010, 67, 2619–2629. [Google Scholar] [CrossRef]
- Tugores, A.; Le, J.; Sorokina, I.; Snijders, A.J.; Duyao, M.; Reddy, P.S.; Carlee, L.; Ronshaugen, M.; Mushegian, A.; Watanaskul, T.; et al. The epithelium-specific ETS protein EHF/ESE-3 is a context-dependent transcriptional repressor downstream of MAPK signaling cascades. J. Biol. Chem. 2001, 276, 20397–20406. [Google Scholar] [CrossRef]
- Finlin, B.S.; Gau, C.L.; Murphy, G.A.; Shao, H.; Kimel, T.; Seitz, R.S.; Chiu, Y.F.; Botstein, D.; Brown, P.O.; Der, C.J.; et al. RERG is a novel ras-related, estrogen-regulated and growth-inhibitory gene in breast cancer. J. Biol. Chem. 2001, 276, 42259–42267. [Google Scholar] [CrossRef]
- Busuioc, C.; Ciocan-Cartita, C.A.; Braicu, C.; Zanoaga, O.; Raduly, L.; Trif, M.; Muresan, M.-S.; Ionescu, C.; Stefan, C.; Crivii, C.; et al. Epithelial-Mesenchymal Transition Gene Signature Related to Prognostic in Colon Adenocarcinoma. J. Pers. Med. 2021, 11, 476. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Li, Z.; Qin, X.; Zhou, X.; Zhang, H.; Li, S. LINC00114 stimulates growth and glycolysis of esophageal cancer cells by recruiting EZH2 to enhance H3K27me3 of DLC1. Clin. Epigenetics 2022, 14, 51. [Google Scholar] [CrossRef]
- Shav-Tal, Y.; Zipori, D. PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS Lett. 2002, 531, 109–114. [Google Scholar] [CrossRef]
- Barbosa, I.; Haque, N.; Fiorini, F.; Barrandon, C.; Tomasetto, C.; Blanchette, M.; Le Hir, H. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat. Struct. Mol. Biol. 2012, 19, 983–990. [Google Scholar] [CrossRef]
- Levesque, L.; Salazar, N.; Roy, S.W. Distinct Minor Splicing Patterns across Cancers. Genes 2022, 13, 387. [Google Scholar] [CrossRef]
- Lee, M.; Sadowska, A.; Bekere, I.; Ho, D.; Gully, B.S.; Lu, Y.; Iyer, K.S.; Trewhella, J.; Fox, A.H.; Bond, C.S. The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation. Nucleic Acids Res. 2015, 43, 3826–3840. [Google Scholar] [CrossRef]
- Zhou, Y.; Xia, L.; Wang, H.; Oyang, L.; Su, M.; Liu, Q.; Lin, J.; Tan, S.; Tian, Y.; Liao, Q.; et al. Cancer stem cells in progression of colorectal cancer. Oncotarget 2018, 9, 33403–33415. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Snyder, M.; Iraola-Guzmán, S.; Saus, E.; Gabaldón, T. Discovery and Validation of Clinically Relevant Long Non-Coding RNAs in Colorectal Cancer. Cancers 2022, 14, 3866. [Google Scholar] [CrossRef]
- Goenka, A.; Tiek, D.M.; Song, X.; Iglesia, R.P.; Lu, M.; Hu, B.; Cheng, S.-Y. The Role of Non-Coding RNAs in Glioma. Biomedicines 2022, 10, 2031. [Google Scholar] [CrossRef]
- Liu, H.; Wang, D.; Kan, S.; Hao, M.; Chang, L.; Lu, P.; Liu, Y.; Jin, Y.; Liu, W. The role of lncRNAs and in oral cancer. Front. Cell Dev. Biol. 2022, 10, 826650. [Google Scholar] [CrossRef]
- Han, X.; Li, B. The emerging role of noncoding RNAs in the Hedgehog signaling pathway in cancer. Biomed. Pharmacother. 2022, 154, 113581. [Google Scholar] [CrossRef]
- Aprile, M.; Costa, V.; Cimmino, A.; Calin, G.A. Emerging role of oncogenic long non-coding RNA as cancer biomarkers. Int. J. Cancer 2022, 152, 822–834. [Google Scholar] [CrossRef]
- Chaudhary, R.; Gryder, B.; Woods, W.S.; Subramanian, M.; Jones, M.F.; Li, X.L.; Jenkins, L.M.; Shabalina, S.A.; Mo, M.; Dasso, M.; et al. Prosurvival long noncoding RNA regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3. Elife 2017, 6, 23244. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Garcia, J.T.; Hung, T.; Flynn, R.A.; Shen, Y.; Qu, K.; Payumo, A.Y.; Peres-da-Silva, A.; Broz, D.K.; Baum, R.; et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 2016, 48, 1370–1376. [Google Scholar] [CrossRef]
- Mello, S.S.; Sinow, C.; Raj, N.; Mazur, P.K.; Bieging-Rolett, K.; Broz, D.K.; Imam, J.F.C.; Vogel, H.; Wood, L.D.; Sage, J.; et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017, 31, 1095–1108. [Google Scholar] [CrossRef]
- Xiao, B.; Liu, L.; Chen, Z.; Li, A.; Wang, P.; Xiang, C.; Zeng, Y.; Li, H.; Xiao, T. Identification of epithelial-mesenchymal transition-related prognostic lncRNAs biomarkers associated with melanoma microenvironment. Front. Cell Dev. Biol. 2021, 9, 679133. [Google Scholar] [CrossRef]
- Meng, L.-D.; Shi, G.-D.; Ge, W.-L.; Huang, X.-M.; Chen, Q.; Yuan, H.; Wu, P.-F.; Lu, Y.-C.; Shen, P.; Zhang, Y.-H.; et al. Linc01232 promotes the metastasis of pancreatic cancer by suppressing the ubiquitin-mediated degradation of HNRNPA2B1 and activating the A-Raf-induced MAPK/ERK signaling pathway. Cancer Lett. 2020, 494, 107–120. [Google Scholar] [CrossRef]
- Li, Q.; Lei, C.; Lu, C.; Wang, J.; Gao, M.; Gao, W. LINC01232 exerts oncogenic activities in pancreatic adenocarcinoma via regulation of TM9SF2. Cell Death Dis. 2019, 10, 698. [Google Scholar] [CrossRef]
- Shi, L.; Magee, P.; Fassan, M.; Sahoo, S.; Leong, H.S.; Lee, D.; Sellers, R.; Brullé-Soumaré, L.; Cairo, S.; Monteverde, T.; et al. A KRAS-responsive long non-coding RNA controls microRNA processing. Nat. Commun. 2021, 12, 2038. [Google Scholar] [CrossRef]
- Yang, M.-H.; Zhao, L.; Wang, L.; Ou-Yang, W.; Hu, S.-S.; Li, W.-L.; Ai, M.-L.; Wang, Y.-Q.; Han, Y.; Li, T.-T.; et al. Nuclear lncRNA HOXD-AS1 suppresses colorectal carcinoma growth and metastasis via inhibiting HOXD3-induced integrin β3 transcriptional activating and MAPK/AKT signalling. Mol. Cancer 2019, 18, 31. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Lu, J.; Zhao, Y. Long non-coding RNA LINC00222 regulates GSK3β activity and promotes cell apoptosis in lung adenocarcinoma. Biomed. Pharmacother. 2018, 106, 755–762. [Google Scholar] [CrossRef]
- Tang, Y.; Ji, F. lncRNA HOTTIP facilitates osteosarcoma cell migration, invasion and epithelial-mesenchymal transition by forming a positive feedback loop with c-Myc. Oncol. Lett. 2019, 18, 1649–1656. [Google Scholar] [CrossRef]
- Danielsen, S.A.; Eide, P.W.; Nesbakken, A.; Guren, T.; Leithe, E.; Lothe, R.A. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim. Biophys. Acta 2015, 1855, 104–121. [Google Scholar] [CrossRef]
- Pavón, M.A.; Arroyo-Solera, I.; Téllez-Gabriel, M.; León, X.; Virós, D.; López, M.; Gallardo, A.; Céspedes, M.V.; Casanova, I.; López-Pousa, A.; et al. Enhanced cell migration and apoptosis resistance may underlie the association between high SERPINE1 expression and poor outcome in head and neck carcinoma patients. Oncotarget 2015, 6, 29016–29033. [Google Scholar] [CrossRef]
- Buikhuisen, J.Y.; Gomez Barila, P.M.; Torang, A.; Dekker, D.; de Jong, J.H.; Cameron, K.; Vitale, S.; Stassi, G.; van Hooff, S.R.; Castro, M.A.A.; et al. AKT3 Expression in Mesenchymal Colorectal Cancer Cells Drives Growth and Is Associated with Epithelial-Mesenchymal Transition. Cancers 2021, 13, 801. [Google Scholar] [CrossRef]
- Sadeghalvad, M.; Mansouri, K.; Mohammadi-Motlagh, H.-R.; Noorbakhsh, F.; Mostafaie, A.; Alipour, S.; Rezaei, N. Long non-coding RNA HOTAIR induces the PI3K/AKT/mTOR signaling pathway in breast cancer cells. Rev. Assoc. Med. Bras. 2022, 68, 456–462. [Google Scholar] [CrossRef]
- Wan, Y.; Yao, Z.; Chen, W.; Li, D. The lncRNA NORAD/miR-520a-3p Facilitates Malignancy in Non-Small Cell Lung Cancer via PI3k/Akt/mTOR Signaling Pathway. Onco Targets Ther. 2020, 13, 1533–1544. [Google Scholar] [CrossRef]
- Peng, W.; Wang, Z.; Fan, H. LncRNA NEAT1 Impacts Cell Proliferation and Apoptosis of Colorectal Cancer via Regulation of Akt Signaling. Pathol. Oncol. Res. 2017, 23, 651–656. [Google Scholar] [CrossRef]
- Wang, R.; Lu, X.; Yu, R. lncRNA MALAT1 Promotes EMT Process and Cisplatin Resistance of Oral Squamous Cell Carcinoma via PI3K/AKT/m-TOR Signal Pathway. Onco Targets Ther. 2020, 13, 4049–4061. [Google Scholar] [CrossRef]
- Dai, Q.; Zhang, T.; Li, C. LncRNA MALAT1 Regulates the Cell Proliferation and Cisplatin Resistance in Gastric Cancer via PI3K/AKT Pathway. Cancer Manag. Res. 2020, 12, 1929–1939. [Google Scholar] [CrossRef]
- Oster, B.; Thorsen, K.; Lamy, P.; Wojdacz, T.K.; Hansen, L.L.; Birkenkamp-Demtröder, K.; Sørensen, K.D.; Laurberg, S.; Orntoft, T.F.; Andersen, C.L. Identification and validation of highly frequent CpG island hypermethylation in colorectal adenomas and carcinomas. Int. J. Cancer 2011, 129, 2855–2866. [Google Scholar] [CrossRef]
- Luo, X.; Huang, R.; Sun, H.; Liu, Y.; Bi, H.; Li, J.; Yu, H.; Sun, J.; Lin, S.; Cui, B.; et al. Methylation of a panel of genes in peripheral blood leukocytes is associated with colorectal cancer. Sci. Rep. 2016, 6, 29922. [Google Scholar] [CrossRef]
- Li, X.; Zhao, X.; Yang, B.; Li, Y.; Liu, T.; Pang, L.; Fan, Z.; Ma, W.; Liu, Z.; Li, Z. Long non-coding RNA HOXD-AS1 promotes tumor progression and predicts poor prognosis in colorectal cancer. Int. J. Oncol. 2018, 53, 21–32. [Google Scholar] [CrossRef]
- Yiwei, T.; Hua, H.; Hui, G.; Mao, M.; Xiang, L. HOTAIR Interacting with MAPK1 Regulates Ovarian Cancer skov3 Cell Proliferation, Migration, and Invasion. Med. Sci. Monit. 2015, 21, 1856–1863. [Google Scholar] [CrossRef]
- Geng, Q.; Li, Z.; Li, X.; Wu, Y.; Chen, N. LncRNA NORAD, sponging miR-363-3p, promotes invasion and EMT by upregulating PEAK1 and activating the ERK signaling pathway in NSCLC cells. J. Bioenerg. Biomembr. 2021, 53, 321–332. [Google Scholar] [CrossRef]
- Liu, F.; Tai, Y.; Ma, J. LncRNA NEAT1/let-7a-5p axis regulates the cisplatin resistance in nasopharyngeal carcinoma by targeting Rsf-1 and modulating the Ras-MAPK pathway. Cancer Biol. Ther. 2018, 19, 534–542. [Google Scholar] [CrossRef]
- Zhang, F.; Wu, L.; Qian, J.; Qu, B.; Xia, S.; La, T.; Wu, Y.; Ma, J.; Zeng, J.; Guo, Q.; et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J. Autoimmun. 2016, 75, 96–104. [Google Scholar] [CrossRef]
- Gao, R.; Zhang, R.; Zhang, C.; Zhao, L.; Zhang, Y. Long noncoding RNA CCAT1 promotes cell proliferation and metastasis in human medulloblastoma via MAPK pathway. Tumori 2018, 104, 43–50. [Google Scholar] [CrossRef]
- Xie, S.-J.; Diao, L.-T.; Cai, N.; Zhang, L.-T.; Xiang, S.; Jia, C.-C.; Qiu, D.-B.; Liu, C.; Sun, Y.-J.; Lei, H.; et al. mascRNA and its parent lncRNA MALAT1 promote proliferation and metastasis of hepatocellular carcinoma cells by activating ERK/MAPK signaling pathway. Cell Death Discov. 2021, 7, 110. [Google Scholar] [CrossRef]
- Nabeshima, K.; Inoue, T.; Shimao, Y.; Sameshima, T. Matrix metalloproteinases in tumor invasion: Role for cell migration. Pathol. Int. 2002, 52, 255–264. [Google Scholar] [CrossRef]
- Liu, D.; Liu, J.; Li, Y.; Liu, H.; Hassan, H.M.; He, W.; Li, M.; Zhou, Y.; Fu, X.; Zhan, J.; et al. Upregulated bone morphogenetic protein 5 enhances proliferation and epithelial-mesenchymal transition process in benign prostatic hyperplasia via BMP/Smad signaling pathway. Prostate 2021, 81, 1435–1449. [Google Scholar] [CrossRef]
- Mae, J.; Nagaya, K.; Okamatsu-Ogura, Y.; Tsubota, A.; Matsuoka, S.; Nio-Kobayashi, J.; Kimura, K. Adipocytes and Stromal Cells Regulate Brown Adipogenesis Through Secretory Factors During the Postnatal White-to-Brown Conversion of Adipose Tissue in Syrian Hamsters. Front. Cell Dev. Biol. 2021, 9, 698692. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y. Long non-coding RNA NORAD contributes to the proliferation, invasion and EMT progression of prostate cancer via the miR-30a-5p/RAB11A/WNT/β-catenin pathway. Cancer Cell Int. 2020, 20, 571. [Google Scholar] [CrossRef]
- Huang, C.; Liu, J.; He, L.; Wang, F.; Xiong, B.; Li, Y.; Yang, X. The long noncoding RNA noncoding RNA activated by DNA damage (NORAD)-microRNA-496-Interleukin-33 axis affects carcinoma-associated fibroblasts-mediated gastric cancer development. Bioengineered 2021, 12, 11738–11755. [Google Scholar] [CrossRef]
- Yu, S.-Y.; Peng, H.; Zhu, Q.; Wu, Y.-X.; Wu, F.; Han, C.-R.; Yan, B.; Li, Q.; Xiang, H.-G. Silencing the long noncoding RNA NORAD inhibits gastric cancer cell proliferation and invasion by the RhoA/ROCK1 pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3760–3770. [Google Scholar]
- Chen, Y.; Li, J.; Xiao, J.-K.; Xiao, L.; Xu, B.-W.; Li, C. The lncRNA NEAT1 promotes the epithelial-mesenchymal transition and metastasis of osteosarcoma cells by sponging miR-483 to upregulate STAT3 expression. Cancer Cell. Int. 2021, 21, 90. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, Y.; Hu, Y.; Zhong, J.; Jiang, C.; Zhang, H. LncRNA CCAT1 sponges miR-218-5p to promote EMT, cellular migration and invasion of retinoblastoma by targeting MTF2. Cell Signal. 2021, 86, 110088. [Google Scholar] [CrossRef]
- Jarroux, J.; Foretek, D.; Bertrand, C.; Gabriel, M.; Szachnowski, U.; Saci, Z.; Guo, S.; Londoño-Vallejo, A.; Pinskaya, M.; Morillon, A. HOTAIR lncRNA promotes epithelial-mesenchymal transition by redistributing LSD1 at regulatory chromatin regions. EMBO Rep. 2021, 22, e50193. [Google Scholar] [CrossRef]
- Lu, W.; Zhang, H.; Niu, Y.; Wu, Y.; Sun, W.; Li, H.; Kong, J.; Ding, K.; Shen, H.-M.; Wu, H.; et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol. Cancer 2017, 16, 118. [Google Scholar] [CrossRef]
- Liu, T.; Wang, H.; Yu, H.; Bi, M.; Yan, Z.; Hong, S.; Li, S. The Long Non-coding RNA HOTTIP Is Highly Expressed in Colorectal Cancer and Enhances Cell Proliferation and Invasion. Mol. Ther. Nucleic Acids 2020, 19, 612–618. [Google Scholar] [CrossRef]
- Gao, L.-F.; Li, W.; Liu, Y.-G.; Zhang, C.; Gao, W.-N.; Wang, L. Inhibition of MIR4435-2HG on Invasion, Migration, and EMT of Gastric Carcinoma Cells by Mediating MiR-138-5p/Sox4 Axis. Front. Oncol. 2021, 11, 661288. [Google Scholar] [CrossRef]
- Lv, W.; Jia, Y.; Wang, J.; Duan, Y.; Wang, X.; Liu, T.; Hao, S.; Liu, L. Long non-coding RNA SNHG10 upregulates BIN1 to suppress the tumorigenesis and epithelial-mesenchymal transition of epithelial ovarian cancer via sponging miR-200a-3p. Cell Death Discov. 2022, 8, 60. [Google Scholar] [CrossRef]
- Gimona, M.; Buccione, R.; Courtneidge, S.A.; Linder, S. Assembly and biological role of podosomes and invadopodia. Curr. Opin. Cell Biol. 2008, 20, 235–241. [Google Scholar] [CrossRef]
- Stölting, M.; Wiesner, C.; van Vliet, V.; Butt, E.; Pavenstädt, H.; Linder, S.; Kremerskothen, J. Lasp-1 regulates podosome function. PLoS ONE 2012, 7, e35340. [Google Scholar] [CrossRef]
- Buccione, R.; Orth, J.D.; McNiven, M.A. Foot and mouth: Podosomes, invadopodia and circular dorsal ruffles. Nat. Rev. Mol. Cell Biol. 2004, 5, 647–657. [Google Scholar] [CrossRef]
- Bo, H.; Gong, Z.; Zhang, W.; Li, X.; Zeng, Y.; Liao, Q.; Chen, P.; Shi, L.; Lian, Y.; Jing, Y.; et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget 2015, 6, 20404–20418. [Google Scholar] [CrossRef]
- Ma, Z.; Peng, P.; Zhou, J.; Hui, B.; Ji, H.; Wang, J.; Wang, K. Long Non-Coding RNA SH3PXD2A-AS1 Promotes Cell Progression Partly Through Epigenetic Silencing P57 and KLF2 in Colorectal Cancer. Cell Physiol. Biochem. 2018, 46, 2197–2214. [Google Scholar] [CrossRef]
- Chou, J.; Wang, B.; Zheng, T.; Li, X.; Zheng, L.; Hu, J.; Zhang, Y.; Xing, Y.; Xi, T. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem. Biophys. Res. Commun. 2016, 472, 262–269. [Google Scholar] [CrossRef]
- Peng, X.; Li, X.; Yang, S.; Huang, M.; Wei, S.; Ma, Y.; Li, Y.; Wu, B.; Jin, H.; Li, B.; et al. LINC00511 drives invasive behavior in hepatocellular carcinoma by regulating exosome secretion and invadopodia formation. J. Exp. Clin. Cancer Res. 2021, 40, 183. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Ming, T.; Tang, S.; Ren, S.; Yang, H.; Liu, M.; Tao, Q.; Xu, H. Wnt signaling in colorectal cancer: Pathogenic role and therapeutic target. Mol. Cancer 2022, 21, 144. [Google Scholar] [CrossRef] [PubMed]
- Lowe, S.W.; Schmitt, E.M.; Smith, S.W.; Osborne, B.A.; Jacks, T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 1993, 362, 847–849. [Google Scholar] [CrossRef]
- Ghosh, A.; Stewart, D.; Matlashewski, G. Regulation of human p53 activity and cell localization by alternative splicing. Mol. Cell Biol. 2004, 24, 7987–7997. [Google Scholar] [CrossRef] [PubMed]
- Michael, D.; Oren, M. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 2003, 13, 49–58. [Google Scholar] [CrossRef]
- Jiang, Z.; Zeng, Q.; Li, D.; Ding, L.; Lu, W.; Bian, M.; Wu, J. Long non-coding RNA MALAT1 promotes high glucose-induced rat cartilage endplate cell apoptosis via the p38/MAPK signalling pathway. Mol. Med. Rep. 2020, 21, 2220–2226. [Google Scholar] [CrossRef]
- Robson, E.J.D.; Khaled, W.T.; Abell, K.; Watson, C.J. Epithelial-to-mesenchymal transition confers resistance to apoptosis in three murine mammary epithelial cell lines. Differentiation 2006, 74, 254–264. [Google Scholar] [CrossRef]
- Franco, D.L.; Mainez, J.; Vega, S.; Sancho, P.; Murillo, M.M.; de Frutos, C.A.; Del Castillo, G.; López-Blau, C.; Fabregat, I.; Nieto, M.A. Snail1 suppresses TGF-beta-induced apoptosis and is sufficient to trigger EMT in hepatocytes. J. Cell Sci. 2010, 123, 3467–3477. [Google Scholar] [CrossRef]
- Siraj, A.K.; Pratheeshkumar, P.; Divya, S.P.; Parvathareddy, S.K.; Bu, R.; Masoodi, T.; Kong, Y.; Thangavel, S.; Al-Sanea, N.; Ashari, L.H.; et al. TGFβ-induced SMAD4-dependent Apoptosis Proceeded by EMT in CRC. Mol. Cancer Ther. 2019, 18, 1312–1322. [Google Scholar] [CrossRef]
- Kortlever, R.M.; Higgins, P.J.; Bernards, R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat. Cell Biol. 2006, 8, 877–884. [Google Scholar] [CrossRef]
- Hsu, K.-S.; Kao, H.-Y. PML: Regulation and multifaceted function beyond tumor suppression. Cell Biosci. 2018, 8, 5. [Google Scholar] [CrossRef]
- Huang, W.; Hickson, L.J.; Eirin, A.; Kirkland, J.L.; Lerman, L.O. Cellular senescence: The good, the bad and the unknown. Nat. Rev. Nephrol. 2022, 18, 611–627. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Panda, A.; Kang, M.-J.; Xu, J.; Selimyan, R.; Yoon, J.-H.; Martindale, J.L.; De, S.; Wood, W.H., 3rd; Becker, K.G.; et al. Senescence-associated lncRNAs: Senescence-associated long noncoding RNAs. Aging Cell 2013, 12, 890–900. [Google Scholar] [CrossRef]
- Bian, W.; Jing, X.; Yang, Z.; Shi, Z.; Chen, R.; Xu, A.; Wang, N.; Jiang, J.; Yang, C.; Zhang, D.; et al. Downregulation of LncRNA NORAD promotes Ox-LDL-induced vascular endothelial cell injury and atherosclerosis. Aging 2020, 12, 6385–6400. [Google Scholar] [CrossRef]
- Ruan, L.; Mendhe, B.; Parker, E.; Kent, A.; Isales, C.M.; Hill, W.D.; McGee-Lawrence, M.; Fulzele, S.; Hamrick, M.W. Long Non-coding RNA MALAT1 Is Depleted with Age in Skeletal Muscle and MALAT1 Silencing Increases Expression of TGF-β1. Front. Physiol. 2021, 12, 742004. [Google Scholar] [CrossRef]
- Haemmig, S.; Yang, D.; Sun, X.; Das, D.; Ghaffari, S.; Molinaro, R.; Chen, L.; Deng, Y.; Freeman, D.; Moullan, N.; et al. Long noncoding RNA integrates a DNA-PK-mediated DNA damage response and vascular senescence. Sci. Transl. Med. 2020, 12, 1868. [Google Scholar] [CrossRef]
- Yuan, D.; Liu, Y.; Li, M.; Zhou, H.; Cao, L.; Zhang, X.; Li, Y. Senescence associated long non-coding RNA 1 regulates cigarette smoke-induced senescence of type II alveolar epithelial cells through sirtuin-1 signaling. J. Int. Med. Res. 2021, 49, 300060520986049. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Goff, L.; Trapnell, C.; Kelley, D. cummeRbund: Analysis, Exploration, Manipulation, and Visualization of Cufflinks High-Throughput Sequencing Data. R Package Version 2.40.0. Bioconductor. 2017. Available online: https://bioconductor.org/packages/release/bioc/html/cummeRbund.html (accessed on 12 September 2022).
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacksi, M.; Schad, E.; Buday, L.; Tantos, A. Absence of Scaffold Protein Tks4 Disrupts Several Signaling Pathways in Colon Cancer Cells. Int. J. Mol. Sci. 2023, 24, 1310. https://doi.org/10.3390/ijms24021310
Jacksi M, Schad E, Buday L, Tantos A. Absence of Scaffold Protein Tks4 Disrupts Several Signaling Pathways in Colon Cancer Cells. International Journal of Molecular Sciences. 2023; 24(2):1310. https://doi.org/10.3390/ijms24021310
Chicago/Turabian StyleJacksi, Mevan, Eva Schad, László Buday, and Agnes Tantos. 2023. "Absence of Scaffold Protein Tks4 Disrupts Several Signaling Pathways in Colon Cancer Cells" International Journal of Molecular Sciences 24, no. 2: 1310. https://doi.org/10.3390/ijms24021310
APA StyleJacksi, M., Schad, E., Buday, L., & Tantos, A. (2023). Absence of Scaffold Protein Tks4 Disrupts Several Signaling Pathways in Colon Cancer Cells. International Journal of Molecular Sciences, 24(2), 1310. https://doi.org/10.3390/ijms24021310








