Approaching the Dimerization Mechanism of Small Molecule Inhibitors Targeting PD-L1 with Molecular Simulation
Abstract
1. Introduction
2. Results and Discussion
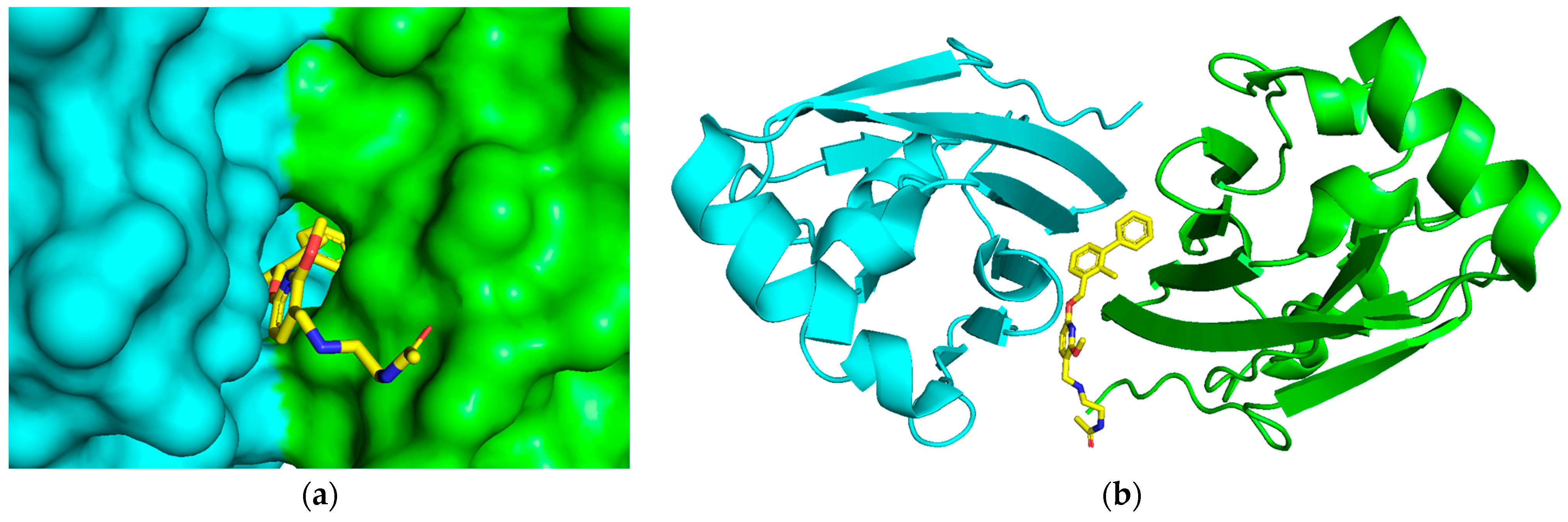
2.1. Acquisition of the Initial Structure by Molecular Docking
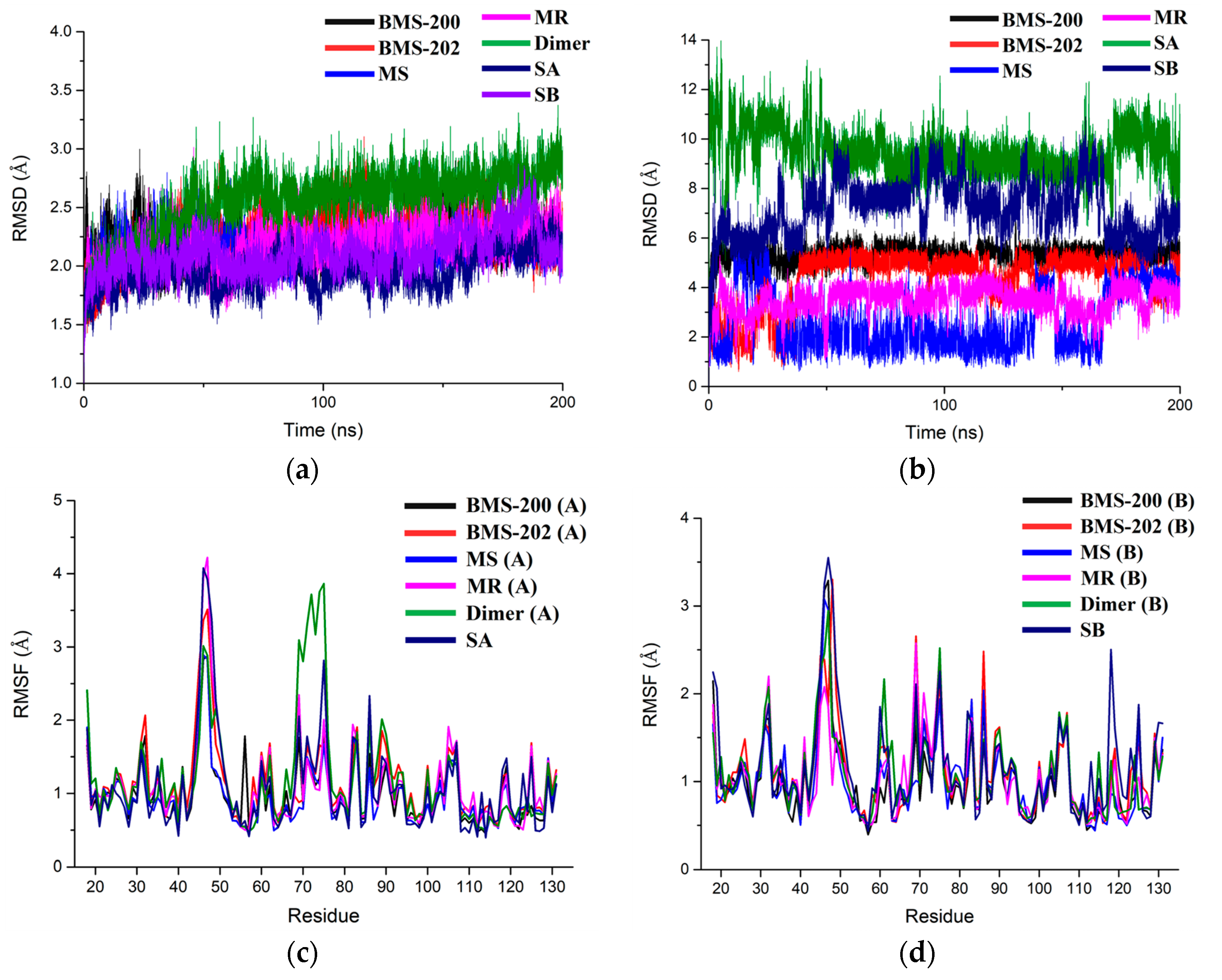
2.2. Stability Evaluation of the Molecular Dynamics Simulation Results
2.3. Comparative Analysis of Binding Free Energy
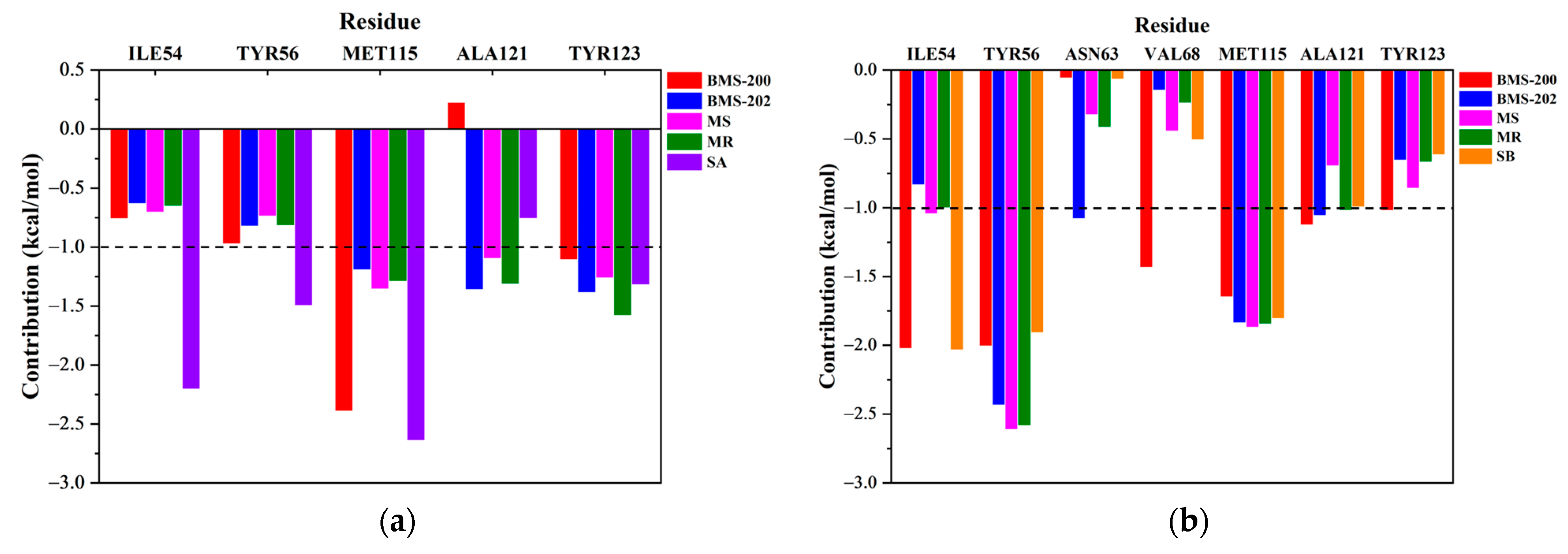
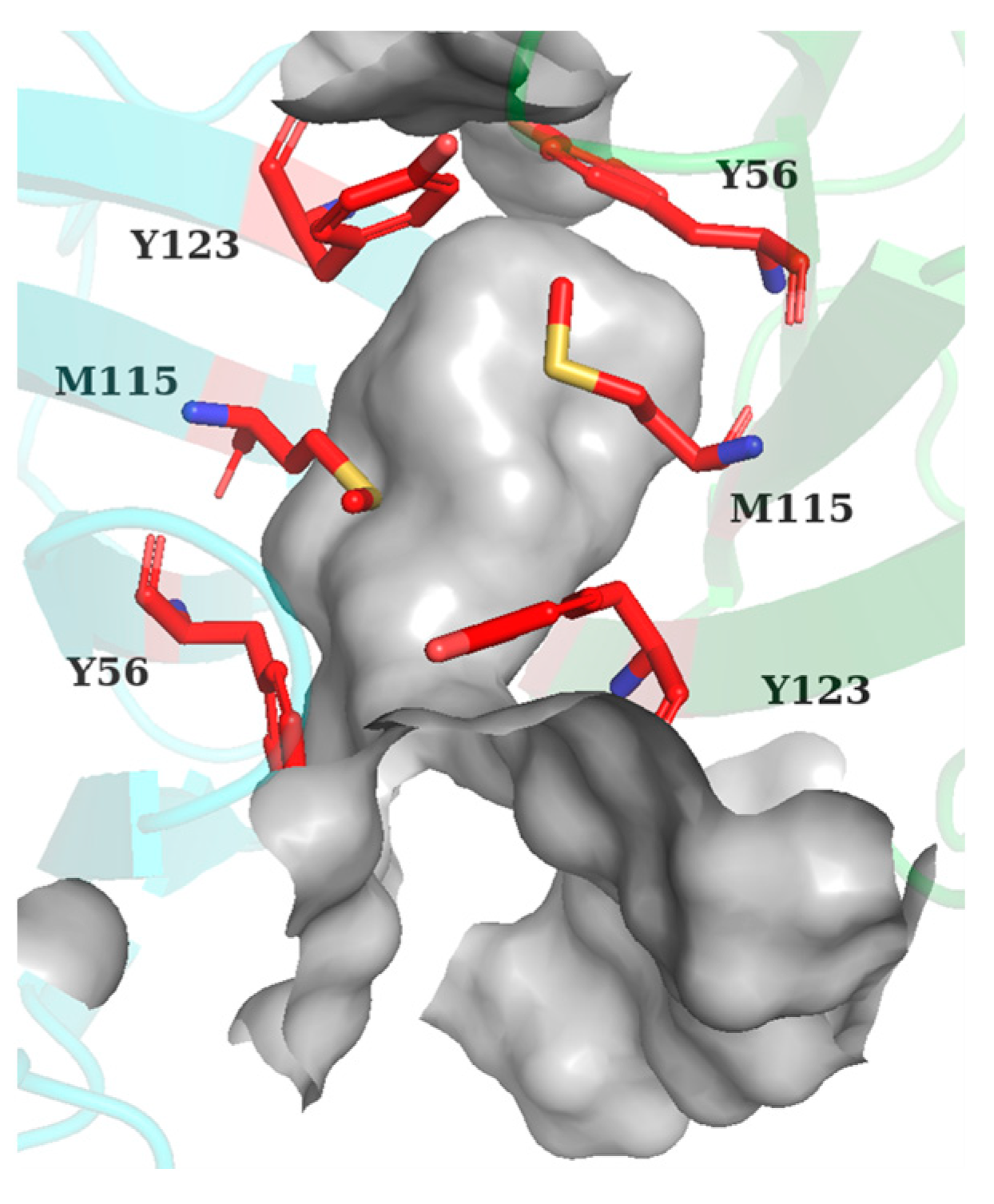
2.4. Key Residue Recognition
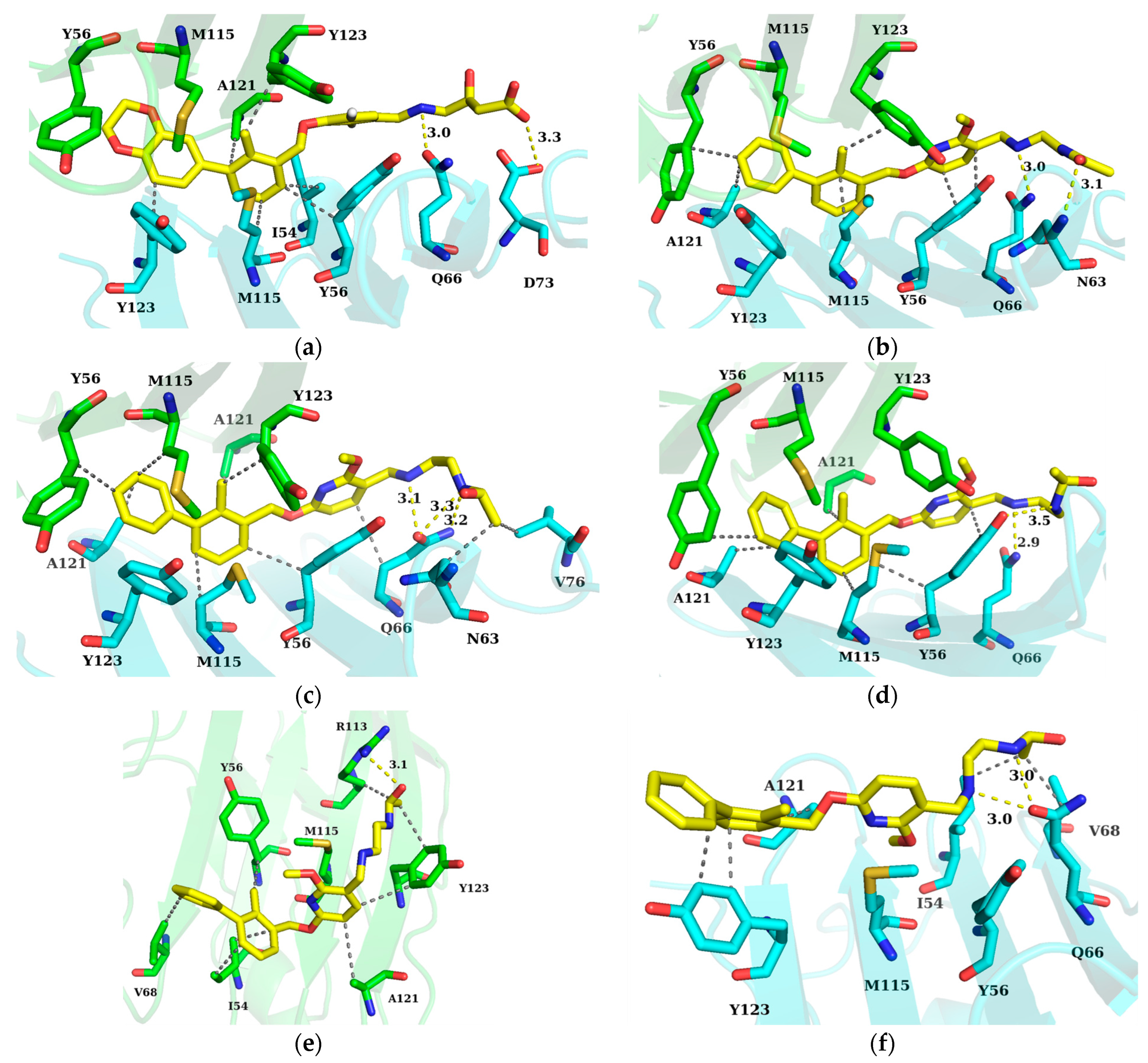
2.5. Hydrogen Bond Analysis
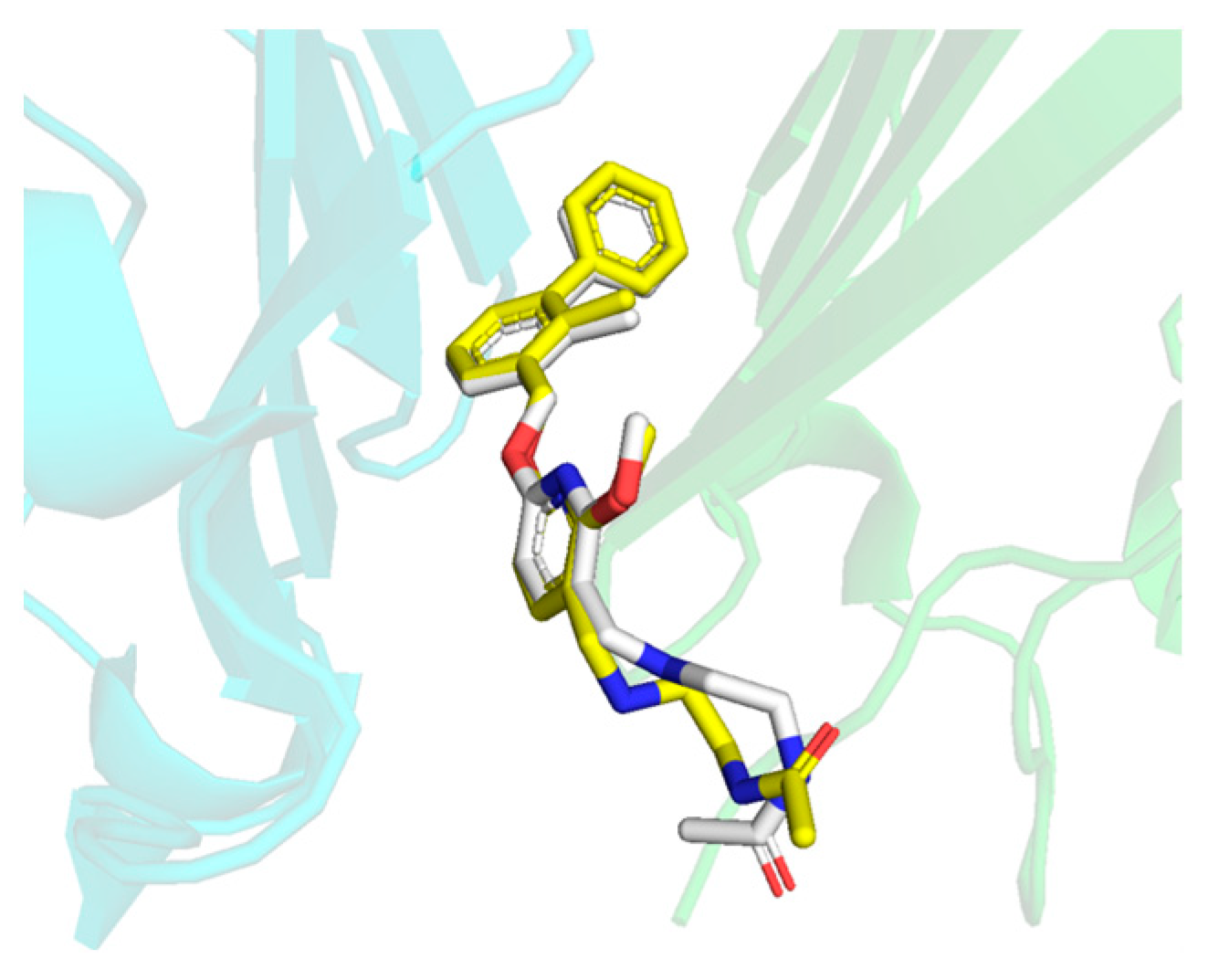
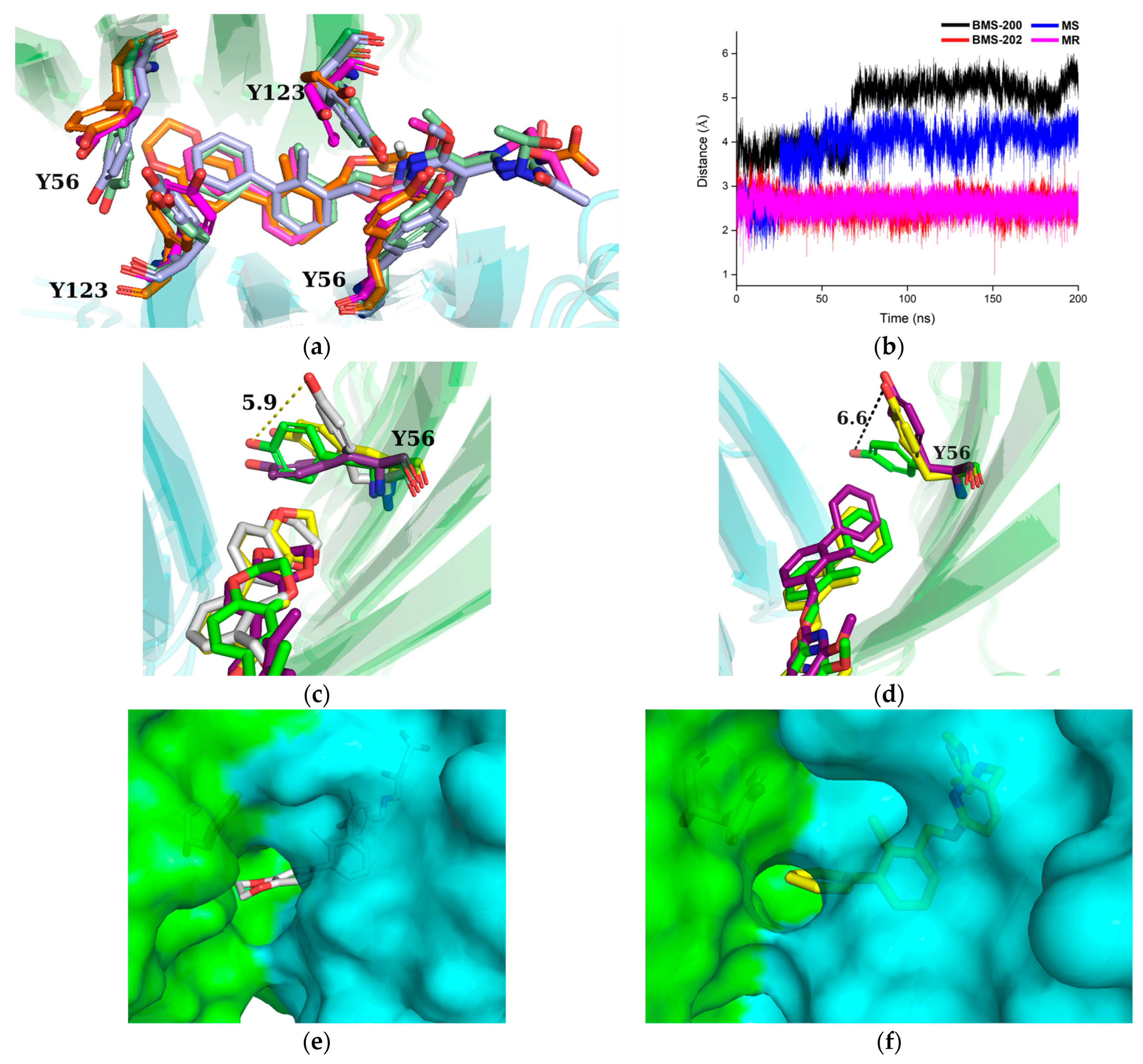
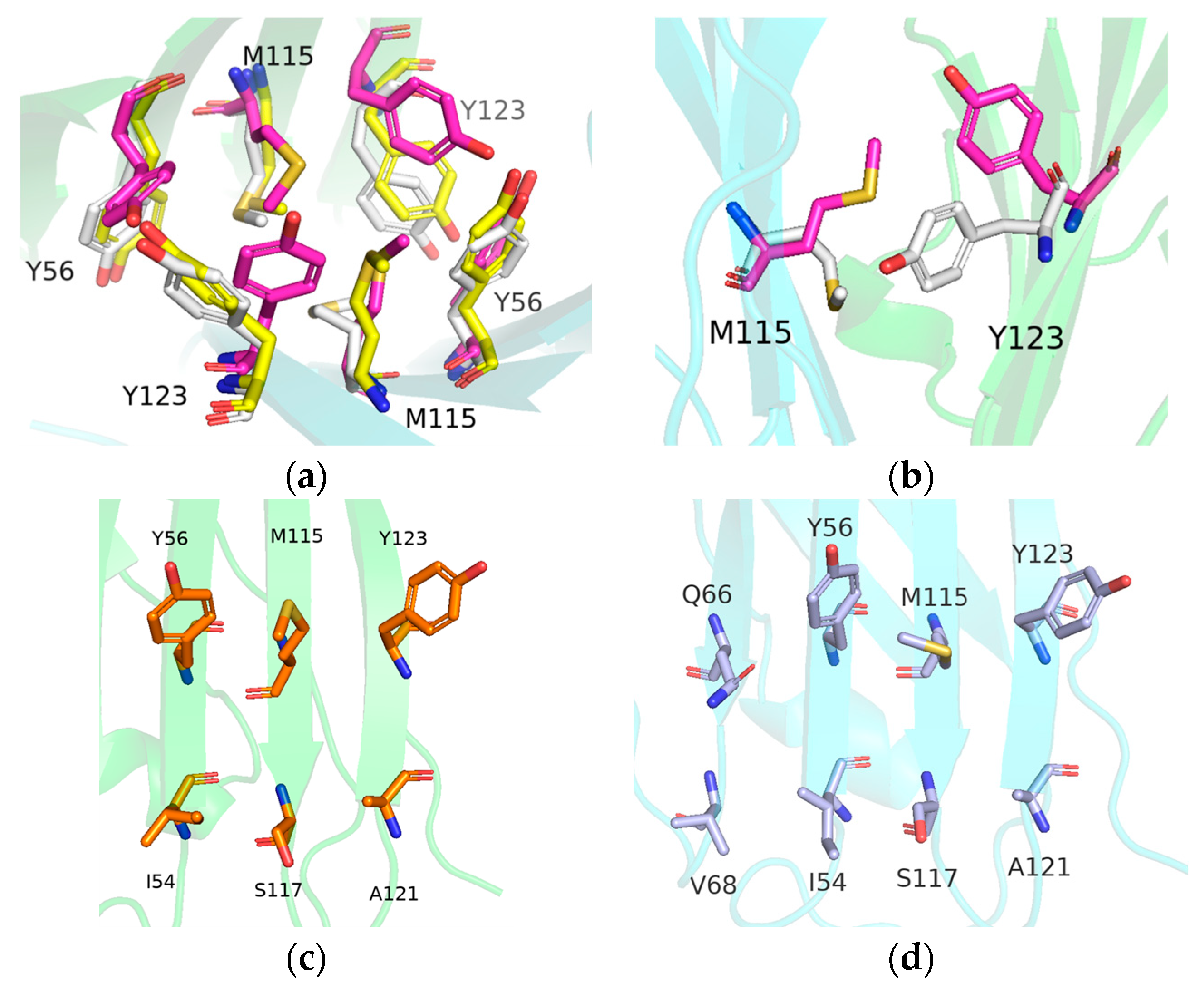
2.6. Binding Mode Analysis and Residue Rearrangement Investigation
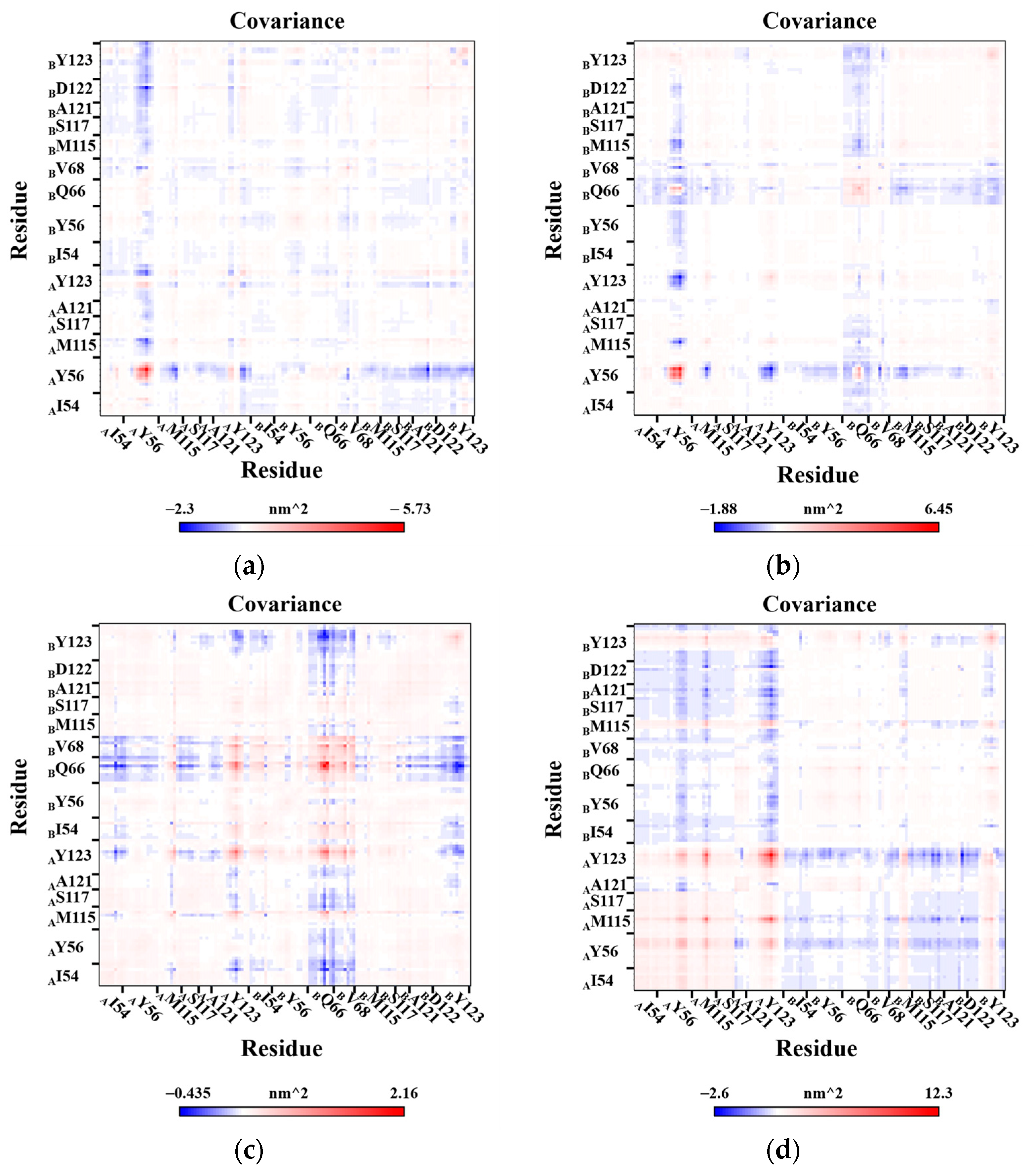
2.7. Correlation Analysis of Residues Motion
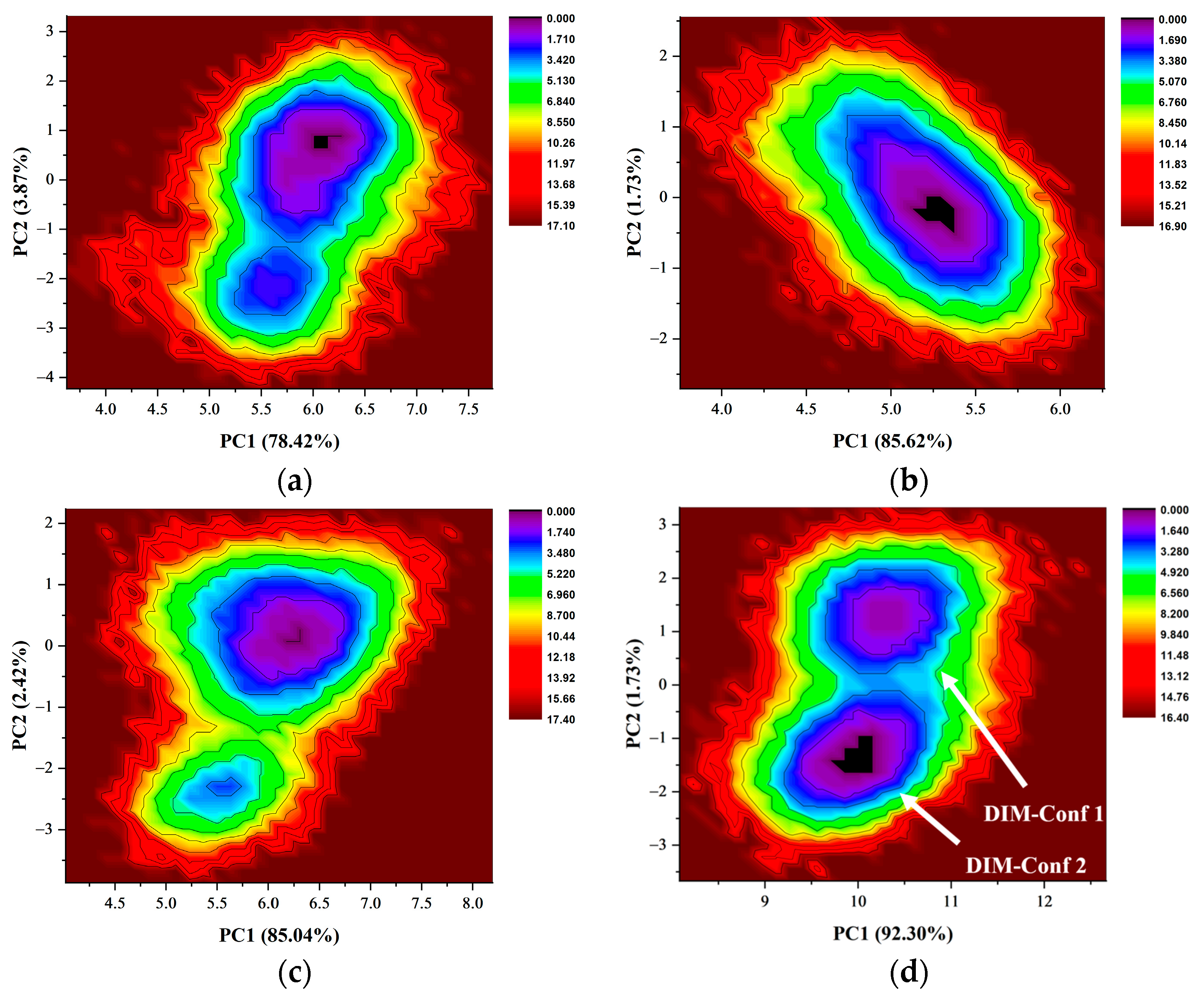
2.8. Free Energy Landscapes and New Insight into PD-L1 Dimerization
3. Materials and Methods
3.1. Molecular Modeling
3.2. Molecular Docking
3.3. Molecular Dynamics Simulation
3.4. Binding Free Energy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharpe, A.H.; Wherry, E.J.; Ahmed, R.; Freeman, G.J. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat. Immunol. 2007, 8, 239–245. [Google Scholar] [CrossRef]
- Fife, B.T.; Pauken, K.E. The role of the PD-1 pathway in autoimmunity and peripheral tolerance. Ann. N. Y. Acad. Sci. 2011, 1217, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Mezzadra, R.; Schumacher, T.N. Regulation and Function of the PD-L1 Checkpoint. Immunity 2018, 48, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.L.; Kuchroo, J.R.; Sage, P.T.; Liang, D.; Francisco, L.M.; Buck, J.; Thaker, Y.R.; Zhang, Q.; McArdel, S.L.; Juneja, V.R.; et al. PD-1 restraint of regulatory T cell suppressive activity is critical for immune tolerance. J. Exp. Med. 2021, 218, e20182232. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Taube, J.M.; Klein, A.; Brahmer, J.R.; Xu, H.; Pan, X.; Kim, J.H.; Chen, L.; Pardoll, D.M.; Topalian, S.L.; Anders, R.A. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin. Cancer Res. 2014, 20, 5064–5074. [Google Scholar] [CrossRef]
- Kamphorst, A.O.; Wieland, A.; Nasti, T.; Yang, S.; Zhang, R.; Barber, D.L.; Konieczny, B.T.; Daugherty, C.Z.; Koenig, L.; Yu, K.; et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 2017, 355, 1423–1427. [Google Scholar] [CrossRef]
- Brahmer, J.R.; Tykodi, S.S.; Chow, L.Q.; Hwu, W.J.; Topalian, S.L.; Hwu, P.; Drake, C.G.; Camacho, L.H.; Kauh, J.; Odunsi, K.; et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 2012, 366, 2455–2465. [Google Scholar] [CrossRef]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Postow, M.A.; Callahan, M.K.; Wolchok, J.D. Immune Checkpoint Blockade in Cancer Therapy. J. Clin. Oncol. 2015, 33, 1974–1982. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hu, L. Immunomodulators targeting the PD-1/PD-L1 protein-protein interaction: From antibodies to small molecules. Med. Res. Rev. 2019, 39, 265–301. [Google Scholar] [CrossRef]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Alsaab, H.O.; Sau, S.; Alzhrani, R.; Tatiparti, K.; Bhise, K.; Kashaw, S.K.; Iyer, A.K. PD-1 and PD-L1 Checkpoint Signaling Inhibition for Cancer Immunotherapy: Mechanism, Combinations, and Clinical Outcome. Front. Pharmacol. 2017, 8, 561. [Google Scholar] [CrossRef]
- Gong, J.; Chehrazi-Raffle, A.; Reddi, S.; Salgia, R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: A comprehensive review of registration trials and future considerations. J. Immunother. Cancer 2018, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Zheng, X.; Niu, M.; Zhu, S.; Ge, H.; Wu, K. Combination strategies with PD-1/PD-L1 blockade: Current advances and future directions. Mol. Cancer 2022, 21, 28. [Google Scholar] [CrossRef]
- Zhan, M.M.; Hu, X.Q.; Liu, X.X.; Ruan, B.F.; Xu, J.; Liao, C. From monoclonal antibodies to small molecules: The development of inhibitors targeting the PD-1/PD-L1 pathway. Drug Discov. Today 2016, 21, 1027–1036. [Google Scholar] [CrossRef]
- Khalil, D.N.; Smith, E.L.; Brentjens, R.J.; Wolchok, J.D. The future of cancer treatment: Immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 2016, 13, 273–290. [Google Scholar] [CrossRef]
- Zak, K.M.; Kitel, R.; Przetocka, S.; Golik, P.; Guzik, K.; Musielak, B.; Domling, A.; Dubin, G.; Holak, T.A. Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1. Structure 2015, 23, 2341–2348. [Google Scholar] [CrossRef]
- Chupak, L.S.; Zheng, X. Compounds Useful as Immunomodulators. Patent WO 2015034820 A1, 12 March 2015. [Google Scholar]
- Chupak, L.S.; Ding, M.; Martin, S.W.; Zheng, X.; Hewawasam, P.; Connolly, T.P.; Xu, N.; Yeung, K.; Zhu, J.; Langley, D.R.; et al. Compounds Useful as Immunomodulators. Patent WO 2015160641 A2, 22 October 2015. [Google Scholar]
- Guzik, K.; Zak, K.M.; Grudnik, P.; Magiera, K.; Musielak, B.; Torner, R.; Skalniak, L.; Domling, A.; Dubin, G.; Holak, T.A. Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1. J. Med. Chem. 2017, 60, 5857–5867. [Google Scholar] [CrossRef] [PubMed]
- Skalniak, L.; Zak, K.M.; Guzik, K.; Magiera, K.; Musielak, B.; Pachota, M.; Szelazek, B.; Kocik, J.; Grudnik, P.; Tomala, M.; et al. Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells. Oncotarget 2017, 8, 72167–72181. [Google Scholar] [CrossRef] [PubMed]
- Muszak, D.; Surmiak, E.; Plewka, J.; Magiera-Mularz, K.; Kocik-Krol, J.; Musielak, B.; Sala, D.; Kitel, R.; Stec, M.; Weglarczyk, K.; et al. Terphenyl-Based Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction. J. Med. Chem. 2021, 64, 11614–11636. [Google Scholar] [CrossRef] [PubMed]
- Zak, K.M.; Grudnik, P.; Guzik, K.; Zieba, B.J.; Musielak, B.; Domling, A.; Dubin, G.; Holak, T.A. Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1). Oncotarget 2016, 7, 30323–30335. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, P.; Gao, F.; Cheng, H.; Qi, J.; Gao, G.F. A dimeric structure of PD-L1: Functional units or evolutionary relics? Protein Cell 2010, 1, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, Y.; Guo, Y.; Pan, Z.; Zhong, S.; Jin, X.; Zhuang, W.; Chen, S.; Gao, J.; Huang, W.; et al. Discovery of phenyl-linked symmetric small molecules as inhibitors of the programmed cell death-1/programmed cell death-ligand 1 interaction. Eur. J. Med. Chem. 2021, 223, 113637. [Google Scholar] [CrossRef] [PubMed]
- Kawashita, S.; Aoyagi, K.; Yamanaka, H.; Hantani, R.; Naruoka, S.; Tanimoto, A.; Hori, Y.; Toyonaga, Y.; Fukushima, K.; Miyazaki, S.; et al. Symmetry-based ligand design and evaluation of small molecule inhibitors of programmed cell death-1/programmed death-ligand 1 interaction. Bioorg. Med. Chem. Lett. 2019, 29, 2464–2467. [Google Scholar] [CrossRef]
- Basu, S.; Yang, J.; Xu, B.; Magiera-Mularz, K.; Skalniak, L.; Musielak, B.; Kholodovych, V.; Holak, T.A.; Hu, L. Design, Synthesis, Evaluation, and Structural Studies of C2-Symmetric Small Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction. J. Med. Chem. 2019, 62, 7250–7263. [Google Scholar] [CrossRef]
- Ahmed, M.; Barakat, K. The Too Many Faces of PD-L1: A Comprehensive Conformational Analysis Study. Biochemistry 2017, 56, 5428–5439. [Google Scholar] [CrossRef]
- Lim, H.; Chun, J.; Jin, X.; Kim, J.; Yoon, J.; No, K.T. Investigation of protein-protein interactions and hot spot region between PD-1 and PD-L1 by fragment molecular orbital method. Sci. Rep. 2019, 9, 16727. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, X.; Yao, H.; Lin, K. The Characteristics of PD-L1 Inhibitors, from Peptides to Small Molecules. Molecules 2019, 24, 1940. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Jin, Y.; Wang, B.; Liu, B. Molecular Mechanism of Small-Molecule Inhibitors in Blocking the PD-1/PD-L1 Pathway through PD-L1 Dimerization. Int. J. Mol. Sci. 2021, 22, 4766. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; An, X.; Bai, Q.; Bing, Z.; Zhou, S.; Liu, H.; Yao, X. Computational Insight into the Small Molecule Intervening PD-L1 Dimerization and the Potential Structure-Activity Relationship. Front. Chem. 2019, 7, 764. [Google Scholar] [CrossRef] [PubMed]
- Cramer, C.J.; Truhlar, D.G. An SCF Solvation Model for the Hydrophobic Effect and Absolute Free Energies of Aqueous Solvation. Science 1992, 256, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Kollman, P.A. Application of RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J. Am. Chem. Soc. 2002, 115, 9620–9631. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Agarwal, R.; Smith, J.C. Speed vs accuracy: Effect on ligand pose accuracy of varying box size and exhaustiveness in AutoDock Vina. Mol. Inform. 2022, 2200188. [Google Scholar] [CrossRef]
- Pham, T.N.H.; Nguyen, T.H.; Tam, N.M.; Vu, T.Y.; Pham, N.T.; Huy, N.T.; Mai, B.K.; Tung, N.T.; Pham, M.Q.; Vu, V.V.; et al. Improving ligand-ranking of AutoDock Vina by changing the empirical parameters. J. Comput. Chem. 2022, 43, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Tanchuk, V.Y.; Tanin, V.O.; Vovk, A.I.; Poda, G. A New, Improved Hybrid Scoring Function for Molecular Docking and Scoring Based on AutoDock and AutoDock Vina. Chem. Biol. Drug Des. 2016, 87, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Rester, U. Dock around the Clock—Current Status of Small Molecule Docking and Scoring. QSAR Comb. Sci. 2006, 25, 605–615. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Sprenger, K.G.; Jaeger, V.W.; Pfaendtner, J. The general AMBER force field (GAFF) can accurately predict thermodynamic and transport properties of many ionic liquids. J. Phys. Chem. B 2015, 119, 5882–5895. [Google Scholar] [CrossRef] [PubMed]
- Massova, I.; Kollman, P.A. Combined molecular mechanical and continuum solvent approach (MM-PBSAGBSA) to predict ligand binding. Perspect. Drug Discov. Des. 2000, 18, 113–135. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Open Source Drug Discovery Consortium; Lynn, A. g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Sun, H.; Li, Y.; Tian, S.; Xu, L.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys. Chem. Chem. Phys. 2014, 16, 16719–16729. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, B.; Gerber, P.; Schulz-Gasch, T.; Stahl, M. Validation and use of the MM-PBSA approach for drug discovery. J. Med. Chem. 2005, 48, 4040–4048. [Google Scholar] [CrossRef] [PubMed]
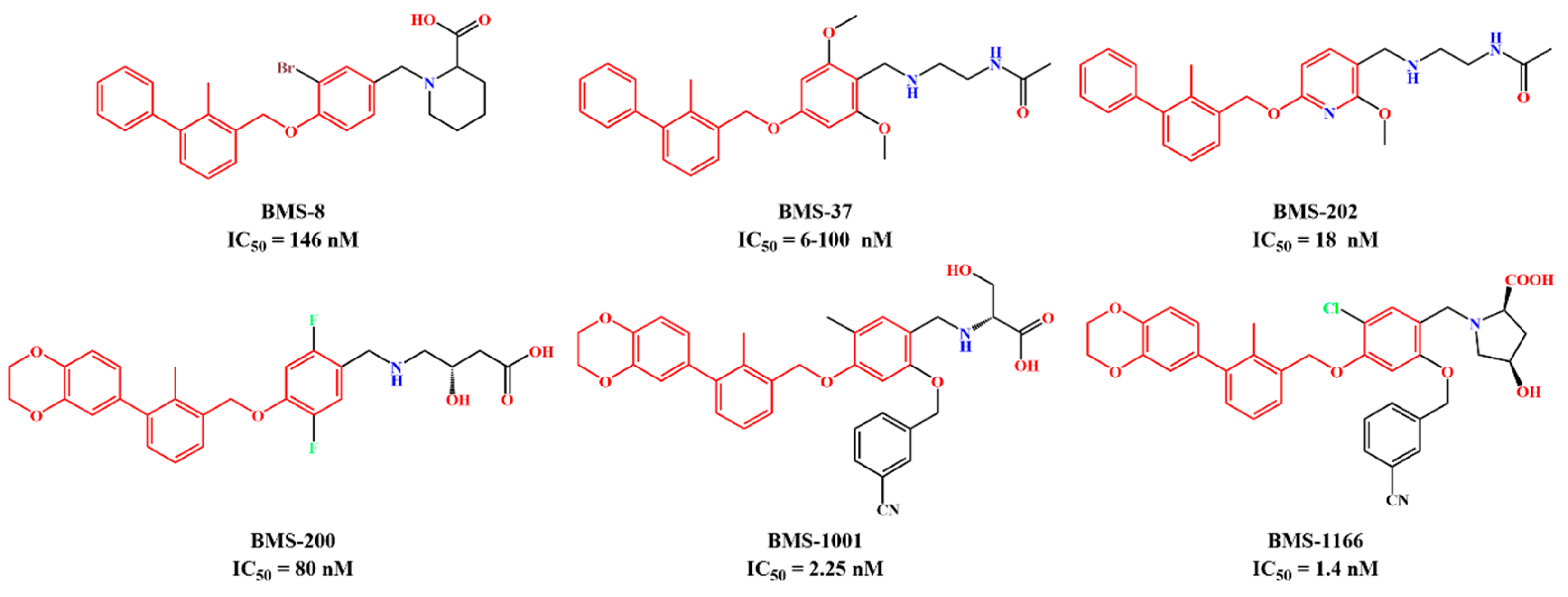
| System | Receptor | Inhibitor | Structure of Inhibitor |
|---|---|---|---|
| BMS-200 | PD-L1 Dimer | BMS-200 | 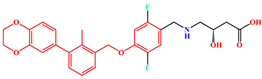 |
| BMS-202 | PD-L1 Dimer | BMS-202 | 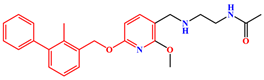 |
| MR | PD-L1 Dimer | R-202 | 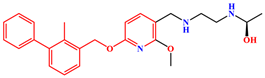 |
| MS | PD-L1 Dimer | S-202 |  |
| SA | APD-L1 | ||
| SB | BPD-L1 | ||
| Dimer | PD-L1 Dimer | / | / |
| Contribution a | BMS-200 | BMS-202 | MS | MR | Dimer i | SA | SB |
|---|---|---|---|---|---|---|---|
| ΔEvdw b | −66.45 ± 0.15 | −64.58 ± 0.13 | −62.57 ± 0.17 | −63.30 ± 0.12 | −50.76 ± 0.29 | −34.17 ± 0.17 | −32.32 ± 0.16 |
| ΔEele c | −10.93 ± 0.23 | −7.82 ± 0.09 | −4.28 ± 0.09 | −4.18 ± 0.09 | −168.16 ± 1.03 | −4.84 ± 0.17 | −2.31 ± 0.09 |
| ΔEpol,sol d | 40.83 ± 0.29 | 34.47 ± 0.16 | 26.38 ± 0.14 | 28.38 ± 0.12 | 193.76 ± 1.32 | 14.68 ± 0.28 | 12.46 ± 0.18 |
| ΔEnonpl,sol e | −5.77 ± 0.01 | −5.23 ± 0.01 | −5.22 ± 0.01 | −5.02 ± 0.01 | −7.23 ± 0.03 | −3.62 ± 0.02 | −3.53 ± 0.01 |
| ΔGpol,total f | 29.91 ± 0.52 | 26.65 ± 0.26 | 22.10 ± 0.24 | 24.20 ± 0.21 | 25.60 ± 2.35 | 9.83 ± 0.45 | 10.14 ± 0.27 |
| ΔGnonpl,total g | −72.22 ± 0.16 | −69.82 ± 0.14 | −67.80 ± 0.19 | −68.32 ± 0.13 | −57.99 ± 0.32 | −37.80 ± 0.19 | −35.84 ± 0.17 |
| ΔG h | −42.32 ± 0.16 | −43.17 ± 0.13 | −45.70 ± 0.17 | −44.13 ± 0.13 | −32.38 ± 0.53 | −27.96 ± 0.16 | −25.71 ± 0.17 |
| System | Donor | Receptor | Occupancy (%) a |
|---|---|---|---|
| BMS-200 | BMS-200@O3 | BAsp73@OD1 | 56.67 ± 1.88 |
| BHis69@NE2 | BMS-200@O2 | 19.71 ± 2.41 | |
| BMS-200@O1 | BAsp73@OD2 | 15.16 ± 2.17 | |
| BMS-202 | BGlu66@NE2 | BMS-202@N1 | 80.55 ± 1.57 |
| BAsn63@N | BMS-202@O2 | 73.75 ± 1.98 | |
| MS | S-202@N2 | BGln66@OE1 | 48.47 ± 2.72 |
| MR | BGln66@NE2 | R-202@N2 | 60.79 ± 1.58 |
| R-202@N1 | BTyr56@OH | 17.15 ± 2.15 | |
| SA | Arg113@NE | S-202@O2 | 12.96 ± 2.74 |
| Arg113@NH2 | S-202@N1 | 11.36 ± 2.16 | |
| Arg113@NH1 | S-202@O2 | 11 ± 2.02 | |
| SB | S-202@N2 | Gln66@OE1 | 27.69 ± 3.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, J.; Wang, B.; Yang, Y.; Liu, B.; Jin, Y. Approaching the Dimerization Mechanism of Small Molecule Inhibitors Targeting PD-L1 with Molecular Simulation. Int. J. Mol. Sci. 2023, 24, 1280. https://doi.org/10.3390/ijms24021280
Liang J, Wang B, Yang Y, Liu B, Jin Y. Approaching the Dimerization Mechanism of Small Molecule Inhibitors Targeting PD-L1 with Molecular Simulation. International Journal of Molecular Sciences. 2023; 24(2):1280. https://doi.org/10.3390/ijms24021280
Chicago/Turabian StyleLiang, Jianhuai, Bingfeng Wang, Yang Yang, Boping Liu, and Yulong Jin. 2023. "Approaching the Dimerization Mechanism of Small Molecule Inhibitors Targeting PD-L1 with Molecular Simulation" International Journal of Molecular Sciences 24, no. 2: 1280. https://doi.org/10.3390/ijms24021280
APA StyleLiang, J., Wang, B., Yang, Y., Liu, B., & Jin, Y. (2023). Approaching the Dimerization Mechanism of Small Molecule Inhibitors Targeting PD-L1 with Molecular Simulation. International Journal of Molecular Sciences, 24(2), 1280. https://doi.org/10.3390/ijms24021280






