Functional Relationships between Long Non-Coding RNAs and Estrogen Receptor Alpha: A New Frontier in Hormone-Responsive Breast Cancer Management
Abstract
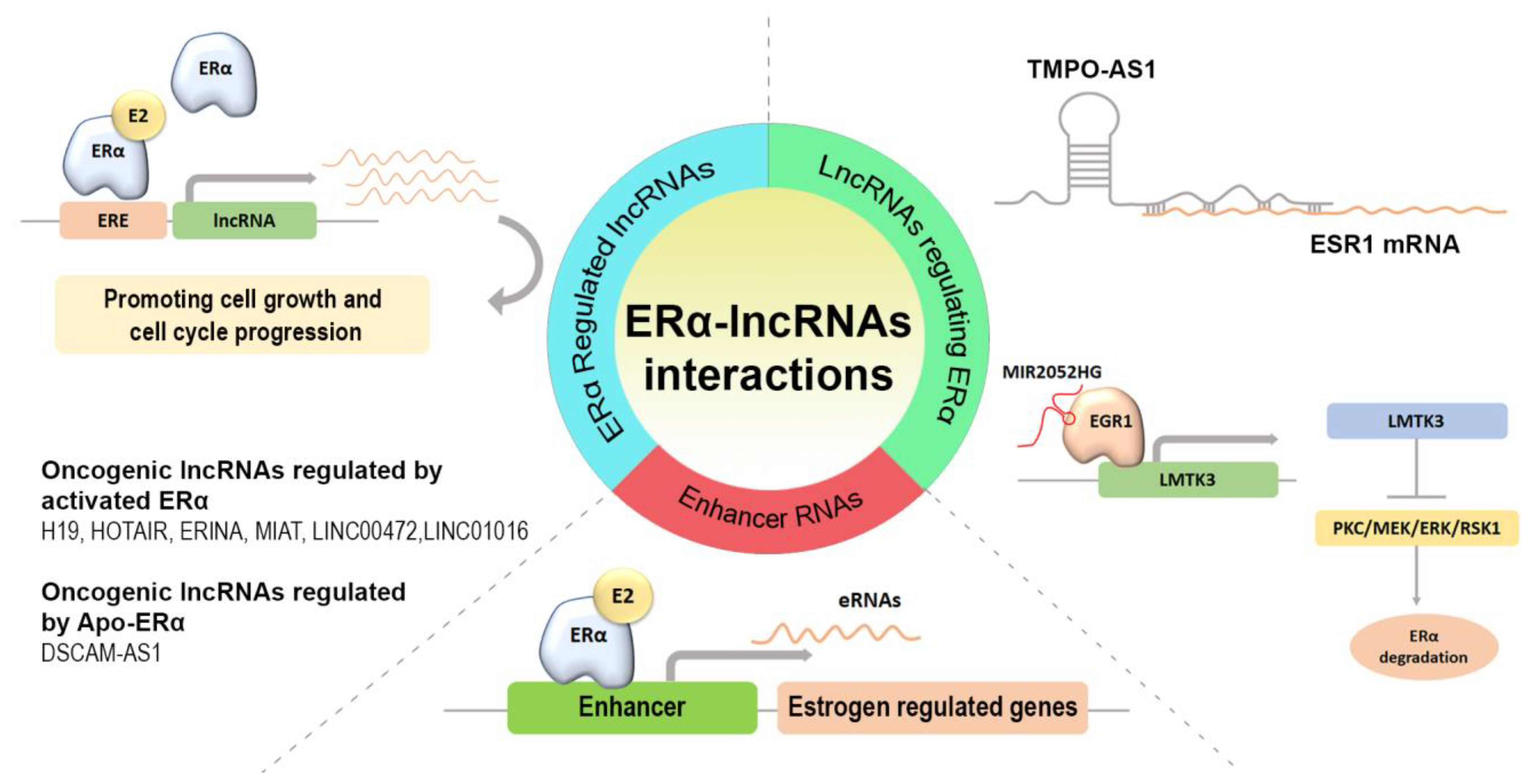
1. Introduction
2. Ligand-Induced and Constitutive Activity of ERα
3. lncRNAs and Cancer
4. Current Methods for the Functional Analysis of lncRNAs
| Method | Reference |
|---|---|
| Determination of the intracellular lncRNA localization | |
| Quantitative PCR (qPCR) | [34] |
| RNA-fluorescent in situ hybridization (RNA-FISH) | [35] |
| RNA-FISH combined with stochastic optical reconstruction microscopy (STORM) | [36] |
| lncRNA labeling with aptamers linked to fluorescent tags | [37,38] |
| lncRNA depletion or over-expression | |
| Small interfering RNA (siRNA) silencing | [78,79] |
| Short hairpin RNA (shRNA) silencing | [80] |
| Antisense oligonucleotide (ASO) silencing | [79] |
| CRISPR/Cas9 knock-out/knock-in | [81] |
| The establishment of secondary and three-dimensional lncRNA structures | |
| Shotgun secondary structure fragment analysis | [40] |
| Solution-state nuclear magnetic resonance (NMR) spectroscopy | [43,44] |
| Small-angle scattering (SAS) | [45] |
| X-ray diffraction and cryo-electron microscopy | [39] |
| Computational prediction | [41,42,46] |
| The determination of lncRNA-protein interactions (LPIs) | |
| Cross-linking immunoprecipitation (CLIP) | [49,50] |
| Targets of RNA-binding proteins identified by editing (TRIBE) | [51] |
| Digestion-optimized RNA immunoprecipitation cDNA library sequencing (DO-RIP-seq) | [52] |
| RNA-affinity purification followed by mass spectrometry | [53,54,55,56] |
| RNA-affinity purification followed by protein microarrays | [57] |
| The isolation of target RNA molecules by biotinylated antisense probes | [54,58] |
| The isolation of target RNA molecules by peptide nucleic acid oligomers | [55] |
| HB-tag-based affinity RNA purification | [56] |
| Chromatin isolation with RNA purification (ChIRP) | [59,60] |
| RNA chromosome conformation capture (R3C) | [61] |
| LPI computational prediction | [72] |
| The biophysical characterization of quantitative and qualitative LPIs | |
| Electrophoretic mobility shift assay (EMSA) | [62] |
| Filter-binding assays | [82] |
| Surface plasmon resonance | [63,64] |
| The evaluation of the coding capacity of lncRNAs | |
| Ribosome profiling | [83] |
| Mass spectrometry | [84] |
| Global translation initiation sequencing (GTI-seq) | [73] |
5. LncRNA Mechanisms in Breast Cancer
5.1. Estrogen-Inducible lncRNAs
5.1.1. lncRNA H19
5.1.2. HOX Transcript Antisense RNA (HOTAIR)
5.1.3. lncRNA ERINA
5.1.4. Myocardial Infarction-Associated Transcript (MIAT)
5.1.5. Long Intergenic Non-Protein Coding RNA 472 (LINC00472) and Long Intergenic Non-Protein Coding RNA 1016 (LINC01016)
5.1.6. lncRNA DSCAM-AS1 Regulation by Unliganded ERα
5.2. lncRNAs Able to Regulate ERα Expression
5.3. Enhancer RNAs (eRNAs)
| LncRNAs | Expression in BC | Regulation | Cellular Functions | References |
|---|---|---|---|---|
| lncRNA H19 | Up-regulation | Estrogen-dependent | Proliferation, tumorigenesis, migration, invasion, and EMT | [90,92,93] |
| HOTAIR (HOX transcript antisense RNA | Up-regulation | Estrogen-dependent | Proliferation, invasion, migration, survival, epigenetic regulation, and chemotherapy resistance | [94,97] |
| LncRNA ERINA | Up-regulation | Estrogen-dependent | Proliferation, survival, and chemotherapy resistance | [101] |
| MIAT (myocardial infarction-associated transcript) | Up-regulation | Estrogen-dependent | Proliferation, migration, invasion, chemotherapy resistance, and EMT | [102] |
| LINC00472 | Up-regulation | Estrogen-dependent | Proliferation, survival, migration, and invasion | [110,111] |
| LINC01016 | Up-regulation | Estrogen-dependent | Proliferation and survival | [114] |
| LncRNA DSCAM-AS1 | Up-regulation | Estrogen-independent | Tumorigenic processes, DNA replication, chromosome, segregation, survival, and EMT | [113,116] |
| TMPO-AS1 (TMPO antisense RNA1) | Up-regulation | Regulation of ERα expression | Proliferation and cell growth | [118] |
| MIR2052HG | Up-regulation | Regulation of ERα expression | Proliferation and cell growth | [120,121] |
6. Prognostic and Clinical Significance of lncRNAs in Hormone-Responsive BC Treatments
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Aromatase Inhibitor |
| AKT | Ak strain Transforming gene |
| ANRIL | Antisense Non-coding RNA in the INK4 Locus |
| ASOs | Antisense Oligonucleotides |
| BC | Breast Cancer |
| BrCSCs | Breast Cancer Stem-like Cells |
| c-MET | Mesenchymal Epithelial Transition cellular oncogene |
| CBP | cAMP-regulated-enhancer (CRE)-binding protein (CREB)-Binding Protein |
| CDK2 | Cyclin-dependent Kinase 2 |
| CDK4-6 | Cyclin-dependent Kinase 4-6 |
| ceRNA | Competing endogenous RNA |
| ChIP-Seq | Chromatin Immunoprecipitation Sequencing |
| ChIRP | Chromatin Isolation by RNA Purification |
| CLIP | Cross-Linking Immunoprecipitation |
| CRISPR/Cas9 | Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated protein 9 |
| DES | Diethylstilbestrol |
| DO-RIP-seq | Digestion-Optimized RNA Immunoprecipitation cDNA library sequencing |
| DSCAM-AS1 | Down Syndrome Cell Adhesion Molecule antisense1 |
| DUSP7 | Dual Specificity Phosphatase 7 |
| E2F1 | E2F transcription factor 1 |
| EED | Embryonic Ectoderm Development |
| EFEMP1 | EGF-containing fibulin-like extracellular matrix protein 1 |
| EGR1 | Early Growth Response 1 |
| EMSA | Electrophoretic mobility shift assay |
| EMT | Epithelial-mesenchymal transition |
| EREs | Estrogen Response Element |
| ERINA | Estrogen inducible lncRNA |
| ERK | Extracellular signal-Regulated Kinase |
| eRNAs | Enhancer RNAs |
| ERα | Estrogen Receptor α |
| ERα+ | Estrogen Receptor α positive |
| ESR1 | Estrogen Receptor 1 |
| ET | Endocrine Therapy |
| ETR | Endocrine Therapy Resistance |
| EZH2 | Enhancer of Zeste Homolog 2 |
| FOXO3 | Forkhead Box O3 |
| GRO-Seq | Global Run-On sequencing |
| GTI-seq | Global Translation Initiation sequencing |
| H3K27Ac | Histone 3 lysine 27 acetylation |
| H3K4 | Histone H3 lysine K4 |
| HB | Histidine-Biotin |
| HBD | Hormone Binding Domain |
| HDAC | Histone Deacetylase |
| HER2 | Human Epidermal growth factor Receptor 2 |
| HGF | Hepatocyte Growth Factor |
| hnRNPL | Heterogeneous nuclear Ribonucleoprotein L |
| HOTAIR | HOX Transcript Antisense RNA |
| ICI | Fulvestrant, ICI 182,780 |
| KDM2A | Lysine demethylase 2A |
| linc-ROR | Long Intergenic Non-Protein Coding RNA, Regulator Of Reprogramming |
| LINC00472 | Long Intergenic Non-Protein Coding RNA 472 |
| LINC01016 | Long Intergenic Non-Protein Coding RNA 1016 |
| LMTK3 | Lemur Tyrosine Kinase 3 |
| LNA | Locked nucleic acid-modified oligonucleotides |
| lncRNA H19 | Long non-coding RNA H19 |
| lncRNA ROR | Long non-coding RNA Regulator Of Reprogramming |
| lncRNAs | Long non-coding RNAs |
| LPI | LncRNA-Protein iInteraction |
| LSD1 | Lysine-Specific Demethylase 1 |
| MAFG-AS1 | MAF BZIP Transcription Factor G -AS1 |
| MAPK | Mitogen-activated protein kinase |
| MEK | Mitogen-activated protein kinase kinase |
| MIAT | Myocardial Infarction-Associated Transcript |
| miR-155-5p | microRNA-155-5p |
| miR-339-5p | microRNA-339-5p |
| MIR2052HG | MIR2052 Host Gene |
| MLL1 | Mixed Lineage Leukemiaprotein-1 |
| MLL3 | Mixed Lineage Leukemiaprotein-3 |
| MLLs | Mixed Lineage Leukemias |
| mRNA | messenger RNA |
| mTOR | Mammalian Target Of Rapamycin |
| ncRNAs | Non-coding RNAs |
| NGS | Next-generation sequencing |
| NMR | Nuclear Magnetic Resonance |
| ORF | Open Reading Frame |
| PI3K | Phosphatidylinositol-3 Kinase |
| PKC | Protein Kinase C |
| PR | Progesterone Receptor |
| PRC2 | Polycomb Repressive Complex 2 |
| PTM | Post-Translational Modification |
| qPCR | Quantitative PCR |
| R3C | RNA chromosome conformation capture |
| RB1 | Retinoblastoma protein 1 |
| RNA Pol II | RNA Polymerase II |
| RNA-FISH | RNA-Fluorescent In Situ Hybridization |
| RNA-Seq | RNA Sequencing |
| RSK1 | Ribosomal S6 Kinase 1 |
| SAS | Small-angle Scattering |
| SERDs | Selective Estrogen Receptor Down-regulators |
| SERMs | Selective Estrogen Receptor Modulators |
| shRNAs | Short hairpin RNAs |
| siRNAs | Small interfering RNAs |
| sncRNAs | Small non-coding RNAs |
| STORM | Stochastic Optical Reconstruction mMicroscopy |
| SUZ12 | Suppressor of Zeste 12 homolog |
| TDG | Thymine DNA glycosylase |
| TM4SF1 | Transmembrane-4 L-six family member-1 |
| TMPO | Thymopoietin |
| TMPO-AS1 | TMPO antisense RNA1 |
| TRIBE | Targets of RNA-binding proteins Identified By Editing |
| UCA1 | Urothelial Carcinoma-Associated protein1 |
| XIST | X-Inactive Specific Transcript |
| ZEB1 | Zinc Finger E-Box Binding Homeobox 1 |
| ZEB2 | Zinc Finger E-Box Binding Homeobox 2 |
References
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar]
- Skibinski, A.; Kuperwasser, C. The origin of breast tumor heterogeneity. Oncogene 2015, 34, 5309–5316. [Google Scholar] [CrossRef]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical management of breast cancer heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381–394. [Google Scholar] [CrossRef] [PubMed]
- McKenna, N.J.; Nawaz, Z.; Tsai, S.Y.; Tsai, M.-J.; O’Malley, B.W. Distinct steady-state nuclear receptor coregulator complexes exist in vivo. Proc. Natl. Acad. Sci. USA 1998, 95, 11697–11702. [Google Scholar] [CrossRef] [PubMed]
- Dobrzycka, K.M.; Townson, S.M.; Jiang, S.; Oesterreich, S. Estrogen receptor corepressors—A role in human breast cancer? Endocr. Relat. Cancer 2003, 10, 517–536. [Google Scholar] [CrossRef]
- Selli, C.; Dixon, J.M.; Sims, A.H. Accurate prediction of response to endocrine therapy in breast cancer patients: Current and future biomarkers. Breast Cancer Res. 2016, 18, 118. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Stebbing, J.; Giamas, G.; Murphy, J. Endocrine Resistance in Hormone Receptor Positive Breast Cancer—From Mechanism to Therapy. Front. Endocrinol. (Lausanne) 2019, 10, 245. [Google Scholar] [CrossRef] [PubMed]
- Rugo, H.S.; Vidula, N.; Ma, C. Improving Response to Hormone Therapy in Breast Cancer: New Targets, New Therapeutic Options. Am. Soc. Clin. Oncol. Educ. Book 2016, 35, e40–e54. [Google Scholar] [CrossRef]
- Du, T.; Shi, Y.; Xu, S.; Wan, X.; Sun, H.; Liu, B. Long Non-Coding RNAs in Drug Resistance of Breast Cancer. Onco Targets Ther. 2020, 13, 7075–7087. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef]
- Harlen, K.M.; Churchman, L.S. The code and beyond: Transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 2017, 18, 263–273. [Google Scholar] [CrossRef]
- Yaşar, P.; Ayaz, G.; User, S.D.; Güpür, G.; Muyan, M. Molecular mechanism of estrogen-estrogen receptor signaling. Reprod. Med. Biol. 2017, 16, 4–20. [Google Scholar] [CrossRef]
- Maggi, A. Liganded and unliganded activation of estrogen receptor and hormone replacement therapies. Biochim. Biophys. Acta 2011, 1812, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Caizzi, L.; Ferrero, G.; Cutrupi, S.; Cordero, F.; Ballaré, C.; Miano, V.; Reineri, S.; Ricci, L.; Friard, O.; Testori, A.; et al. Genome-wide activity of unliganded estrogen receptor-α in breast cancer cells. Proc. Natl. Acad. Sci. USA 2014, 111, 4892–4897. [Google Scholar] [CrossRef] [PubMed]
- Stellato, C.; Porreca, I.; Cuomo, D.; Tarallo, R.; Nassa, G.; Ambrosino, C. The “busy life” of unliganded estrogen receptors. Proteomics 2016, 16, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Elhasnaoui, J.; Ferrero, G.; Miano, V.; Cutrupi, S.; De Bortoli, M. The Estrogen Receptor α Signaling Pathway Controls Alternative Splicing in the Absence of Ligands in Breast Cancer Cells. Cancers 2021, 13, 6261. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huangyang, P.; Wang, Y.; Xue, L.; Devericks, E.; Nguyen, H.G.; Yu, X.; Oses-Prieto, J.A.; Burlingame, A.L.; Miglani, S.; et al. ERα is an RNA-binding protein sustaining tumor cell survival and drug resistance. Cell 2021, 184, 5215–5229.e17. [Google Scholar] [CrossRef]
- Salvati, A.; Gigantino, V.; Nassa, G.; Cappa, V.M.; Ventola, G.M.; Cracas, D.G.C.; Mastrocinque, R.; Rizzo, F.; Tarallo, R.; Weisz, A.; et al. Global View of Candidate Therapeutic Target Genes in Hormone-Responsive Breast Cancer. Int. J. Mol. Sci. 2020, 21, 4068. [Google Scholar] [CrossRef]
- Nassa, G.; Salvati, A.; Tarallo, R.; Gigantino, V.; Alexandrova, E.; Memoli, D.; Sellitto, A.; Rizzo, F.; Malanga, D.; Mirante, T.; et al. Inhibition of histone methyltransferase DOT1L silences ERα gene and blocks proliferation of antiestrogen-resistant breast cancer cells. Sci. Adv. 2019, 5, eaav5590. [Google Scholar] [CrossRef]
- Gigantino, V.; Salvati, A.; Giurato, G.; Palumbo, D.; Strianese, O.; Rizzo, F.; Tarallo, R.; Nyman, T.A.; Weisz, A.; Nassa, G. Identification of Antiestrogen-Bound Estrogen Receptor α Interactomes in Hormone-Responsive Human Breast Cancer Cell Nuclei. Proteomics 2020, 20, e2000135. [Google Scholar] [CrossRef]
- Nassa, G.; Giurato, G.; Salvati, A.; Gigantino, V.; Pecoraro, G.; Lamberti, J.; Rizzo, F.; Nyman, T.A.; Tarallo, R.; Weisz, A. The RNA-mediated estrogen receptor α interactome of hormone-dependent human breast cancer cell nuclei. Sci. Data 2019, 6, 173. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Xu, K.; Jin, X.; Li, J.; Shi, Y.; Zhang, M.; Jin, X.; Li, Y.; Xu, J.; Li, X. LncSpA: LncRNA Spatial Atlas of Expression across Normal and Cancer Tissues. Cancer Res. 2020, 80, 2067–2071. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Sun, M.; Kraus, W.L. Minireview: Long noncoding RNAs: New “links” between gene expression and cellular outcomes in endocrinology. Mol. Endocrinol. 2013, 27, 1390–1402. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Lee, J.T. Epigenetic regulation by long noncoding RNAs. Science 2012, 338, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Lu, X.; Yuan, L. LncRNA: A link between RNA and cancer. Biochim. Biophys. Acta 2014, 1839, 1097–1109. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, W.; Levy, D.E. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods 2006, 39, 356–362. [Google Scholar] [CrossRef]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015, 16, 20. [Google Scholar] [CrossRef]
- Sunwoo, H.; Wu, J.Y.; Lee, J.T. The Xist RNA-PRC2 complex at 20-nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proc. Natl. Acad. Sci. USA 2015, 112, E4216–E4225. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.S.; Sunwoo, H.; Zhang, B.; Spector, D.L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011, 13, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA mimics of green fluorescent protein. Science 2011, 333, 642–646. [Google Scholar] [CrossRef]
- McFadden, E.J.; Hargrove, A.E. Biochemical Methods to Investigate lncRNA and the Influence of lncRNA: Protein Complexes on Chromatin. Biochemistry 2016, 55, 1615–1630. [Google Scholar] [CrossRef]
- Novikova, I.V.; Dharap, A.; Hennelly, S.P.; Sanbonmatsu, K.Y. 3S: Shotgun secondary structure determination of long non-coding RNAs. Methods 2013, 63, 170–177. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef]
- Foster, M.P.; McElroy, C.A.; Amero, C.D. Solution NMR of large molecules and assemblies. Biochemistry 2007, 46, 331–340. [Google Scholar] [CrossRef]
- Lu, K.; Miyazaki, Y.; Summers, M.F. Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR 2010, 46, 113–125. [Google Scholar] [CrossRef]
- Neylon, C. Small angle neutron and X-ray scattering in structural biology: Recent examples from the literature. Eur. Biophys. J. 2008, 37, 531–541. [Google Scholar] [CrossRef]
- Freier, S.M.; Kierzek, R.; Jaeger, J.A.; Sugimoto, N.; Caruthers, M.H.; Neilson, T.; Turner, D.H. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. USA 1986, 83, 9373–9377. [Google Scholar] [CrossRef]
- Wu, M.; McDowell, J.A.; Turner, U.H. A periodic table of symmetric tandem mismatches in RNA. Biochemistry 1995, 34, 3204–3211. [Google Scholar] [CrossRef]
- Ramanathan, M.; Porter, D.F.; Khavari, P.A. Methods to study RNA-protein interactions. Nat. Methods 2019, 16, 225–234. [Google Scholar] [CrossRef]
- Ule, J.; Jensen, K.B.; Ruggiu, M.; Mele, A.; Ule, A.; Darnell, R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science 2003, 302, 1212–1215. [Google Scholar] [CrossRef]
- Kim, B.; Kim, V.N. fCLIP-seq for transcriptomic footprinting of dsRNA-binding proteins: Lessons from DROSHA. Methods 2019, 152, 3–11. [Google Scholar] [CrossRef]
- McMahon, A.C.; Rahman, R.; Jin, H.; Shen, J.L.; Fieldsend, A.; Luo, W.; Rosbash, M. TRIBE: Hijacking an RNA-Editing Enzyme to Identify Cell-Specific Targets of RNA-Binding Proteins. Cell 2016, 165, 742–753. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, C.O.; Friedersdorf, M.; Keene, J.D. Quantifying RNA binding sites transcriptome-wide using DO-RIP-seq. RNA 2017, 23, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Faoro, C.; Ataide, S.F. Ribonomic approaches to study the RNA-binding proteome. FEBS Lett. 2014, 588, 3649–3664. [Google Scholar] [CrossRef]
- McHugh, C.A.; Guttman, M. RAP-MS: A Method to Identify Proteins that Interact Directly with a Specific RNA Molecule in Cells. Methods Mol. Biol. 2018, 1649, 473–488. [Google Scholar] [CrossRef]
- Zeng, F.; Peritz, T.; Kannanayakal, T.J.; Kilk, K.; Eiríksdóttir, E.; Langel, U.; Eberwine, J. A protocol for PAIR: PNA-assisted identification of RNA binding proteins in living cells. Nat. Protoc. 2006, 1, 920–927. [Google Scholar] [CrossRef]
- Tsai, B.P.; Wang, X.; Huang, L.; Waterman, M.L. Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol. Cell. Proteom. 2011, 10, M110.007385. [Google Scholar] [CrossRef]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef]
- Simon, M.D. Capture hybridization analysis of RNA targets (CHART). Curr. Protoc. Mol. Biol. 2013, 101, 21–25. [Google Scholar] [CrossRef]
- Chu, C.; Quinn, J.; Chang, H.Y. Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. 2012, 61, 3912. [Google Scholar] [CrossRef]
- Quinn, J.J.; Ilik, I.A.; Qu, K.; Georgiev, P.; Chu, C.; Akhtar, A.; Chang, H.Y. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat. Biotechnol. 2014, 32, 933–940. [Google Scholar] [CrossRef]
- Zhang, H.; Zeitz, M.J.; Wang, H.; Niu, B.; Ge, S.; Li, W.; Cui, J.; Wang, G.; Qian, G.; Higgins, M.J.; et al. Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J. Cell Biol. 2014, 204, 61–75. [Google Scholar] [CrossRef]
- Rio, D.C. 5′-end labeling of RNA with [γ-32P]ATP and T4 polynucleotide kinase. Cold Spring Harb. Protoc. 2014, 2014, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Katsamba, P.; Park, S.; Laird-Offringa, I.A. Kinetic studies of RNA-protein interactions using surface plasmon resonance. Methods 2002, 26, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Di Primo, C.; Dausse, E.; Toulmé, J.-J. Surface plasmon resonance investigation of RNA aptamer-RNA ligand interactions. Methods Mol. Biol. 2011, 764, 279–300. [Google Scholar] [CrossRef]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Liu, T.; Cui, T.; Wang, Z.; Zhang, Y.; Tan, P.; Huang, Y.; Yu, J.; Wang, D. RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res. 2020, 48, D189–D197. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Yang, Y.; Huang, Y.; Zhu, Y.; Lu, Z.J. POSTAR: A platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucleic Acids Res. 2017, 45, D104–D114. [Google Scholar] [CrossRef] [PubMed]
- Teng, X.; Chen, X.; Xue, H.; Tang, Y.; Zhang, P.; Kang, Q.; Hao, Y.; Chen, R.; Zhao, Y.; He, S. NPInter v4.0: An integrated database of ncRNA interactions. Nucleic Acids Res. 2020, 48, D160–D165. [Google Scholar] [CrossRef]
- Junge, A.; Refsgaard, J.C.; Garde, C.; Pan, X.; Santos, A.; Alkan, F.; Anthon, C.; von Mering, C.; Workman, C.; Jensen, L.J.; et al. RAIN: RNA-protein Association and Interaction Networks. Database 2017, 2017, baw167. [Google Scholar] [CrossRef]
- Giudice, G.; Sanchez-Cabo, F.; Torroja, C.; Lara-Pezzi, E. ATtRACT—A database of RNA-binding proteins and associated motifs. Database 2016, 2016, baw035. [Google Scholar] [CrossRef]
- Bouvrette, L.P.B.; Bovaird, S.; Blanchette, M.; Lécuyer, E. oRNAment: A database of putative RNA binding protein target sites in the transcriptomes of model species. Nucleic Acids Res. 2020, 48, D166–D173. [Google Scholar] [CrossRef]
- Philip, M.; Chen, T.; Tyagi, S. A Survey of Current Resources to Study lncRNA-Protein Interactions. Noncoding RNA 2021, 7, 33. [Google Scholar] [CrossRef]
- Choi, S.-W.; Kim, H.-W.; Nam, J.-W. The small peptide world in long noncoding RNAs. Brief Bioinform. 2019, 20, 1853–1864. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Liu, H.; Jiang, W.; Wang, L. LncRNA-Encoded Peptide: Functions and Predicting Methods. Front. Oncol. 2020, 10, 622294. [Google Scholar] [CrossRef]
- Guo, B.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Wang, Y.; Zhang, S.; Wu, R.; Lu, J.; et al. Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO J. 2020, 39, e102190. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Guo, B.; Zhang, S.; Wu, R.; Zhang, Z.; et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J. Exp. Med. 2020, 217, jem20190950. [Google Scholar] [CrossRef]
- Giambruno, R.; Mihailovich, M.; Bonaldi, T. Mass Spectrometry-Based Proteomics to Unveil the Non-coding RNA World. Front. Mol. Biosci. 2018, 5, 90. [Google Scholar] [CrossRef]
- Thijssen, M.S.; Bintz, J.; Arnes, L. In Vitro Silencing of lncRNA Expression Using siRNAs. Methods Mol. Biol. 2021, 2348, 141–156. [Google Scholar] [CrossRef]
- Watts, J.K.; Corey, D.R. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012, 226, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Goyal, A.; Myacheva, K.; Groß, M.; Klingenberg, M.; Arqué, B.D.; Diederichs, S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017, 45, e12. [Google Scholar] [CrossRef] [PubMed]
- Rio, D.C. Filter-binding assay for analysis of RNA-protein interactions. Cold Spring Harb. Protoc. 2012, 2012, 1078–1081. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Xie, S.; Liu, Y.; Xie, Z. Global and cell-type specific properties of lincRNAs with ribosome occupancy. Nucleic Acids Res. 2017, 45, 2786–2796. [Google Scholar] [CrossRef] [PubMed]
- Verheggen, K.; Volders, P.-J.; Mestdagh, P.; Menschaert, G.; Van Damme, P.; Gevaert, K.; Martens, L.; Vandesompele, J. Noncoding after All: Biases in Proteomics Data Do Not Explain Observed Absence of lncRNA Translation Products. J. Proteome Res. 2017, 16, 2508–2515. [Google Scholar] [CrossRef]
- Tian, T.; Gong, Z.; Wang, M.; Hao, R.; Lin, S.; Liu, K.; Guan, F.; Xu, P.; Deng, Y.; Song, D.; et al. Identification of long non-coding RNA signatures in triple-negative breast cancer. Cancer Cell Int. 2018, 18, 103. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, H.; Yan, G.; Wu, T.; Liu, S.; Chen, W.; Ning, Y.; Lu, Z. Long Non-Coding RNA and Breast Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819843889. [Google Scholar] [CrossRef]
- Choo, S.-W.; Zhong, Y.; Sendler, E.; Goustin, A.-S.; Cai, J.; Ju, D.; Kosir, M.A.; Giordo, R.; Lipovich, L. Estrogen distinctly regulates transcription and translation of lncRNAs and pseudogenes in breast cancer cells. Genomics 2022, 114, 110421. [Google Scholar] [CrossRef]
- Raveh, E.; Matouk, I.J.; Gilon, M.; Hochberg, A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis—A proposed unifying theory. Mol. Cancer 2015, 14, 184. [Google Scholar] [CrossRef]
- Han, J.; Han, B.; Wu, X.; Hao, J.; Dong, X.; Shen, Q.; Pang, H. Knockdown of lncRNA H19 restores chemo-sensitivity in paclitaxel-resistant triple-negative breast cancer through triggering apoptosis and regulating Akt signaling pathway. Toxicol. Appl. Pharmacol. 2018, 359, 55–61. [Google Scholar] [CrossRef]
- Adriaenssens, E.; Dumont, L.; Lottin, S.; Bolle, D.; Leprêtre, A.; Delobelle, A.; Bouali, F.; Dugimont, T.; Coll, J.; Curgy, J.-J. H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am. J. Pathol. 1998, 153, 1597–1607. [Google Scholar] [CrossRef]
- Sun, H.; Wang, G.; Peng, Y.; Zeng, Y.; Zhu, Q.-N.; Li, T.-L.; Cai, J.-Q.; Zhou, H.-H.; Zhu, Y.-S. H19 lncRNA mediates 17β-estradiol-induced cell proliferation in MCF-7 breast cancer cells. Oncol. Rep. 2015, 33, 3045–3052. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, Y.; Xiao, G.-D.; Zheng, X.-Q.; Wang, J.-C.; Xu, C.-W.; Qin, S.; Ren, H.; Tang, S.-C.; Sun, X. H19 regulation of oestrogen induction of symmetric division is achieved by antagonizing Let-7c in breast cancer stem-like cells. Cell Prolif. 2019, 52, e12534. [Google Scholar] [CrossRef]
- Bhan, A.; Hussain, I.; Ansari, K.I.; Kasiri, S.; Bashyal, A.; Mandal, S.S. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J. Mol. Biol. 2013, 425, 3707–3722. [Google Scholar] [CrossRef] [PubMed]
- Hajjari, M.; Salavaty, A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 2015, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sedano, M.J.; Harrison, A.L.; Zilaie, M.; Das, C.; Choudhari, R.; Ramos, E.; Gadad, S.S. Emerging Roles of Estrogen-Regulated Enhancer and Long Non-Coding RNAs. Int. J. Mol. Sci. 2020, 21, 3711. [Google Scholar] [CrossRef] [PubMed]
- Davidovich, C.; Zheng, L.; Goodrich, K.J.; Cech, T.R. Promiscuous RNA binding by Polycomb repressive complex. Nat. Struct. Mol. Biol. 2013, 20, 1250–1257. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Song, H.K.; Kim, S.Y. The Role of Sex-specific Long Non-coding RNAs in Cancer Prevention and Therapy. J. Cancer Prev. 2021, 26, 98–109. [Google Scholar] [CrossRef]
- Fang, Z.; Wang, Y.; Wang, Z.; Xu, M.; Ren, S.; Yang, D.; Hong, M.; Xie, W. Is an Estrogen-Responsive LncRNA That Drives Breast Cancer through the E2F1/RB1 Pathway. Cancer Res. 2020, 80, 4399–4413. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, B.; Wu, X.; Huang, Q.; Chen, W.; Zhu, H.; Qu, X.; Xie, L.; Ma, X.; Huang, G. Long non-coding RNA MIAT is estrogen-responsive and promotes estrogen-induced proliferation in ER-positive breast cancer cells. Biochem. Biophys. Res. Commun. 2018, 503, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Saito, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006, 51, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Vausort, M.; Wagner, D.R.; Devaux, Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 2014, 115, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.-Q.; Hu, H.-L.; Ye, N.; Shen, Y.; Xu, Q. Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr. Res. 2015, 166, 125–130. [Google Scholar] [CrossRef]
- Sun, C.; Huang, L.; Li, Z.; Leng, K.; Xu, Y.; Jiang, X.; Cui, Y. Long non-coding RNA MIAT in development and disease: A new player in an old game. J. Biomed. Sci. 2018, 25, 23. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, H.; Li, S.; Lv, B.; Shi, J.; Yan, H.; Zhang, H.; He, Y. Long Non-coding RNA MIAT Mediates Non-small Cell Lung Cancer Development through Regulating the miR-128-3p/PELI3 Axis. Biochem. Genet. 2020, 58, 867–882. [Google Scholar] [CrossRef]
- Wang, Y.; Fu, L.; Lu, T.; Zhang, G.; Zhang, J.; Zhao, Y.; Jin, H.; Yang, K.; Cai, H. Clinicopathological and Prognostic Significance of Long Non-coding RNA MIAT in Human Cancers: A Review and Meta-Analysis. Front. Genet. 2021, 12, 729768. [Google Scholar] [CrossRef]
- Luan, T.; Zhang, X.; Wang, S.; Song, Y.; Zhou, S.; Lin, J.; An, W.; Yuan, W.; Yang, Y.; Cai, H.; et al. Long non-coding RNA MIAT promotes breast cancer progression and functions as ceRNA to regulate DUSP7 expression by sponging miR-155-5p. Oncotarget 2017, 8, 76153–76164. [Google Scholar] [CrossRef]
- Shen, Y.; Wang, Z.; Loo, L.W.M.; Ni, Y.; Jia, W.; Fei, P.; Risch, H.A.; Katsaros, D.; Yu, H. LINC00472 expression is regulated by promoter methylation and associated with disease-free survival in patients with grade 2 breast cancer. Breast Cancer Res. Treat. 2015, 154, 473–482. [Google Scholar] [CrossRef]
- Wang, Z.; Katsaros, D.; Biglia, N.; Shen, Y.; Loo, L.; Yu, X.; Lin, H.; Fu, Y.; Chu, W.-M.; Fei, P.; et al. ERα upregulates the expression of long non-coding RNA LINC00472 which suppresses the phosphorylation of NF-κB in breast cancer. Breast Cancer Res. Treat. 2019, 175, 353–368. [Google Scholar] [CrossRef]
- Pan, X.; Li, D.; Huo, J.; Kong, F.; Yang, H.; Ma, X. LINC01016 promotes the malignant phenotype of endometrial cancer cells by regulating the miR-302a-3p/miR-3130-3p/NFYA/SATB1 axis. Cell Death. Dis. 2018, 9, 303. [Google Scholar] [CrossRef] [PubMed]
- Miano, V.; Ferrero, G.; Reineri, S.; Caizzi, L.; Annaratone, L.; Ricci, L.; Cutrupi, S.; Castellano, I.; Cordero, F.; De Bortoli, M. Luminal long non-coding RNAs regulated by estrogen receptor alpha in a ligand-independent manner show functional roles in breast cancer. Oncotarget 2016, 7, 3201–3216. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, P.; Coarfa, C.; Mesmar, F.; Raz, T.; Rajapakshe, K.; Thompson, J.; Gunaratne, P.H.; Williams, C. Single-Molecule Sequencing Reveals Estrogen-Regulated Clinically Relevant lncRNAs in Breast Cancer. Mol. Endocrinol. 2015, 29, 1634–1645. [Google Scholar] [CrossRef] [PubMed]
- Elhasnaoui, J.; Miano, V.; Ferrero, G.; Doria, E.; Leon, A.E.; Fabricio, A.S.C.; Annaratone, L.; Castellano, I.; Sapino, A.; De Bortoli, M. DSCAM-AS1-Driven Proliferation of Breast Cancer Cells Involves Regulation of Alternative Exon Splicing and 3′-End Usage. Cancers 2020, 12, 1453. [Google Scholar] [CrossRef]
- Niknafs, Y.S.; Han, S.; Ma, T.; Speers, C.; Zhang, C.; Wilder-Romans, K.; Iyer, M.K.; Pitchiaya, S.; Malik, R.; Hosono, Y.; et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat. Commun. 2016, 7, 12791. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Shi, Y.; Liu, J.-B.; Wu, T.-M.; Jia, C.-Y.; Yang, H.-Q.; Zhang, D.-D.; Yang, X.-L.; Wang, H.-M.; Ma, Y.-S. Targeting Long Non-coding RNA to Therapeutically Regulate Gene Expression in Cancer. Mol. Ther. Nucleic Acids. 2020, 21, 712–724. [Google Scholar] [CrossRef] [PubMed]
- Mitobe, Y.; Ikeda, K.; Suzuki, T.; Takagi, K.; Kawabata, H.; Horie-Inoue, K.; Inoue, S. ESR1-Stabilizing Long Noncoding RNA. Mol. Cell Biol. 2019, 39, e00261-19. [Google Scholar] [CrossRef]
- Liu, G.; Yang, H.; Cao, L.; Han, K.; Li, G. LncRNA TMPO-AS1 Promotes Proliferation and Invasion by Sponging miR-383-5p in Glioma Cells. Cancer Manag. Res. 2020, 12, 12001. [Google Scholar] [CrossRef]
- Cairns, J.; Ingle, J.N.; Kalari, K.R.; Shepherd, L.E.; Kubo, M.; Goetz, M.P.; Weinshilboum, R.M.; Wang, L. The lncRNA MIR2052HG regulates ERα levels and aromatase inhibitor resistance through LMTK3 by recruiting EGR1. Breast Cancer Res. 2019, 21, 47. [Google Scholar] [CrossRef]
- Ingle, J.N.; Xie, F.; Ellis, M.J.; Goss, P.E.; Shepherd, L.E.; Chapman, J.-A.W.; Chen, B.E.; Kubo, M.; Furukawa, Y.; Momozawa, Y.; et al. Genetic Polymorphisms in the Long Noncoding RNA MIR2052HG Offer a Pharmacogenomic Basis for the Response of Breast Cancer Patients to Aromatase Inhibitor Therapy. Cancer Res. 2016, 76, 7012–7023. [Google Scholar] [CrossRef]
- Arner, E.; Daub, C.O.; Vitting-Seerup, K.; Andersson, R.; Lilje, B.; Drabløs, F.; Lennartsson, A.; Rönnerblad, M.; Hrydziuszko, O.; Vitezic, M.; et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 2015, 347, 1010–1014. [Google Scholar] [CrossRef]
- Hsieh, C.-L.; Fei, T.; Chen, Y.; Li, T.; Gao, Y.; Wang, X.; Sun, T.; Sweeney, C.J.; Lee, G.-S.M.; Chen, S.; et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. USA 2014, 111, 7319–7324. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-K.; Hemberg, M.; Gray, J.M. Enhancer RNAs: A class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb. Perspect. Biol. 2015, 7, a018622. [Google Scholar] [CrossRef] [PubMed]
- Andersson, R.; Gebhard, C.; Miguel-Escalada, I.; Hoof, I.; Bornholdt, J.; Boyd, M.; Chen, Y.; Zhao, X.; Schmidl, C.; Suzuki, T.; et al. An atlas of active enhancers across human cell types and tissues. Nature 2014, 507, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Liu, Y.; Liao, X.; Zhan, H.; Liu, Y.; Huang, W. Enhancer RNAs (eRNAs): New Insights into Gene Transcription and Disease Treatment. J. Cancer 2018, 9, 2334–2340. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Ma, Q.; Tanasa, B.; Nunez, E.; Chen, A.Y.; Merkurjev, D.; Zhang, J.; Ohgi, K.; Song, X.; et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013, 498, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Schaukowitch, K.; Joo, J.-Y.; Liu, X.; Watts, J.K.; Martinez, C.; Kim, T.-K. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 2014, 56, 29–42. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, L.; Ren, S.; Wang, L.; Blackburn, P.R.; McNulty, M.S.; Gao, X.; Qiao, M.; Vessella, R.L.; Kohli, M.; et al. Activation of P-TEFb by Androgen Receptor-Regulated Enhancer RNAs in Castration-Resistant Prostate Cancer. Cell Rep. 2016, 15, 599–610. [Google Scholar] [CrossRef]
- Carroll, J.S. Mechanisms of oestrogen receptor (ER) gene regulation in breast cancer. Eur. J. Endocrinol. 2016, 175, R41–R49. [Google Scholar] [CrossRef]
- Carroll, J.S.; Brown, M. Estrogen receptor target gene: An evolving concept. Mol. Endocrinol. 2006, 20, 1707–1714. [Google Scholar] [CrossRef]
- Yang, M.; Lee, J.H.; Zhang, Z.; De La Rosa, R.; Bi, M.; Tan, Y.; Liao, Y.; Hong, J.; Du, B.; Wu, Y.; et al. Enhancer RNAs Mediate Estrogen-Induced Decommissioning of Selective Enhancers by Recruiting ERα and Its Cofactor. Cell Rep. 2020, 31, 107803. [Google Scholar] [CrossRef]
- Kolendowski, B.; Hassan, H.; Krstic, M.; Isovic, M.; Thillainadesan, G.; Chambers, A.F.; Tuck, A.B.; Torchia, J. Genome-wide analysis reveals a role for TDG in estrogen receptor-mediated enhancer RNA transcription and 3-dimensional reorganization. Epigenet. Chromatin 2018, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Basak, P.; Chatterjee, S.; Bhat, V.; Su, A.; Jin, H.; Lee-Wing, V.; Liu, Q.; Hu, P.; Murphy, L.C.; Raouf, A. Long Non-Coding RNA H19 Acts as an Estrogen Receptor Modulator that is Required for Endocrine Therapy Resistance in ER+ Breast Cancer Cells. Cell Physiol. Biochem. 2018, 51, 1518–1532. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Hao, G.; Sun, Y.; Li, L.; Wang, Y. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. Onco Targets Ther. 2018, 11, 8001–8012. [Google Scholar] [CrossRef]
- Xue, X.; Yang, Y.A.; Zhang, A.; Fong, K.-W.; Kim, J.; Song, B.; Li, S.; Zhao, J.C.; Yu, J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 2016, 35, 2746–2755. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, D.; Li, H.; Lv, Y.; Li, S. Long non-coding RNA UCA1 confers tamoxifen resistance in breast cancer endocrinotherapy through regulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway. Int. J. Oncol. 2019, 54, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.G.; Yang, M.F.; Ren, Y.Q.; Wu, C.H.; Wang, L.Q. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4362–4368. [Google Scholar]
- Wu, C.; Luo, J. Long Non-Coding RNA (lncRNA) Urothelial Carcinoma-Associated 1 (UCA1) Enhances Tamoxifen Resistance in Breast Cancer Cells via Inhibiting mTOR Signaling Pathway. Med. Sci. Monit. 2016, 22, 3860–3867. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wen, T.; Li, Z.; Feng, L.; Zhou, L.; Yang, Z.; Xu, L.; Shi, S.; Hou, K.; Shen, J.; et al. Cross-talk between the ER pathway and the lncRNA MAFG-AS1/miR-339-5p/CDK2 axis promotes progression of ER+ breast cancer and confers tamoxifen resistance. Aging 2020, 12, 20658–20683. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, B.; Zhu, H.; Qu, X.; Zhao, L.; Tan, Y.; Jiang, Y.; Liao, M.; Wu, X. Inhibition of long non-coding RNA ROR reverses resistance to Tamoxifen by inducing autophagy in breast cancer. Tumour Biol. 2017, 39, 1010428317705790. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-Y.; Liang, F.; Zhang, J.-W.; Wang, F.; Wang, L.; Kang, X.-G. Effects of long noncoding RNA-ROR on tamoxifen resistance of breast cancer cells by regulating microRNA-205. Cancer Chemother. Pharmacol. 2017, 79, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.-X.; Huang, J.-G.; Yang, L.; Gong, A.-H.; Mo, Y.-Y. Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol. Cancer 2017, 16, 161. [Google Scholar] [CrossRef] [PubMed]
- Steiner, H.R.; Lammer, N.C.; Batey, R.T.; Wuttke, D.S. An Extended DNA Binding Domain of the Estrogen Receptor Alpha Directly Interacts with RNAs. Biochemistry 2022, 61, 2490–2494. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Du, X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod. Pathol. 2013, 26, 155–165. [Google Scholar] [CrossRef]
- Li, C.H.; Chen, Y. Targeting long non-coding RNAs in cancers: Progress and prospects. Int. J. Biochem. Cell Biol. 2013, 45, 1895–1910. [Google Scholar] [CrossRef]
- Haemmerle, M.; Gutschner, T. Long non-coding RNAs in cancer and development: Where do we go from here? Int. J. Mol. Sci. 2015, 16, 1395–1405. [Google Scholar] [CrossRef]
- Beylerli, O.; Gareev, I.; Sufianov, A.; Ilyasova, T.; Guang, Y. Long noncoding RNAs as promising biomarkers in cancer. Noncoding RNA Res. 2022, 7, 66–70. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melone, V.; Salvati, A.; Brusco, N.; Alexandrova, E.; D’Agostino, Y.; Palumbo, D.; Palo, L.; Terenzi, I.; Nassa, G.; Rizzo, F.; et al. Functional Relationships between Long Non-Coding RNAs and Estrogen Receptor Alpha: A New Frontier in Hormone-Responsive Breast Cancer Management. Int. J. Mol. Sci. 2023, 24, 1145. https://doi.org/10.3390/ijms24021145
Melone V, Salvati A, Brusco N, Alexandrova E, D’Agostino Y, Palumbo D, Palo L, Terenzi I, Nassa G, Rizzo F, et al. Functional Relationships between Long Non-Coding RNAs and Estrogen Receptor Alpha: A New Frontier in Hormone-Responsive Breast Cancer Management. International Journal of Molecular Sciences. 2023; 24(2):1145. https://doi.org/10.3390/ijms24021145
Chicago/Turabian StyleMelone, Viola, Annamaria Salvati, Noemi Brusco, Elena Alexandrova, Ylenia D’Agostino, Domenico Palumbo, Luigi Palo, Ilaria Terenzi, Giovanni Nassa, Francesca Rizzo, and et al. 2023. "Functional Relationships between Long Non-Coding RNAs and Estrogen Receptor Alpha: A New Frontier in Hormone-Responsive Breast Cancer Management" International Journal of Molecular Sciences 24, no. 2: 1145. https://doi.org/10.3390/ijms24021145
APA StyleMelone, V., Salvati, A., Brusco, N., Alexandrova, E., D’Agostino, Y., Palumbo, D., Palo, L., Terenzi, I., Nassa, G., Rizzo, F., Giurato, G., Weisz, A., & Tarallo, R. (2023). Functional Relationships between Long Non-Coding RNAs and Estrogen Receptor Alpha: A New Frontier in Hormone-Responsive Breast Cancer Management. International Journal of Molecular Sciences, 24(2), 1145. https://doi.org/10.3390/ijms24021145









