Streptozotocin-Induced Type 1 and 2 Diabetes Mellitus Mouse Models Show Different Functional, Cellular and Molecular Patterns of Diabetic Cardiomyopathy
Abstract
1. Introduction
2. Results
2.1. STZ-Based T1DM and T2DM Mouse Models Affected Global Left Ventricular Function Differently
2.2. STZ-Based T1DM and T2DM Mouse Models Affected Myocardial Performance Differently
2.3. STZ-Based T1DM and T2DM Mouse Models Affected Left Ventricular Remodeling Differently
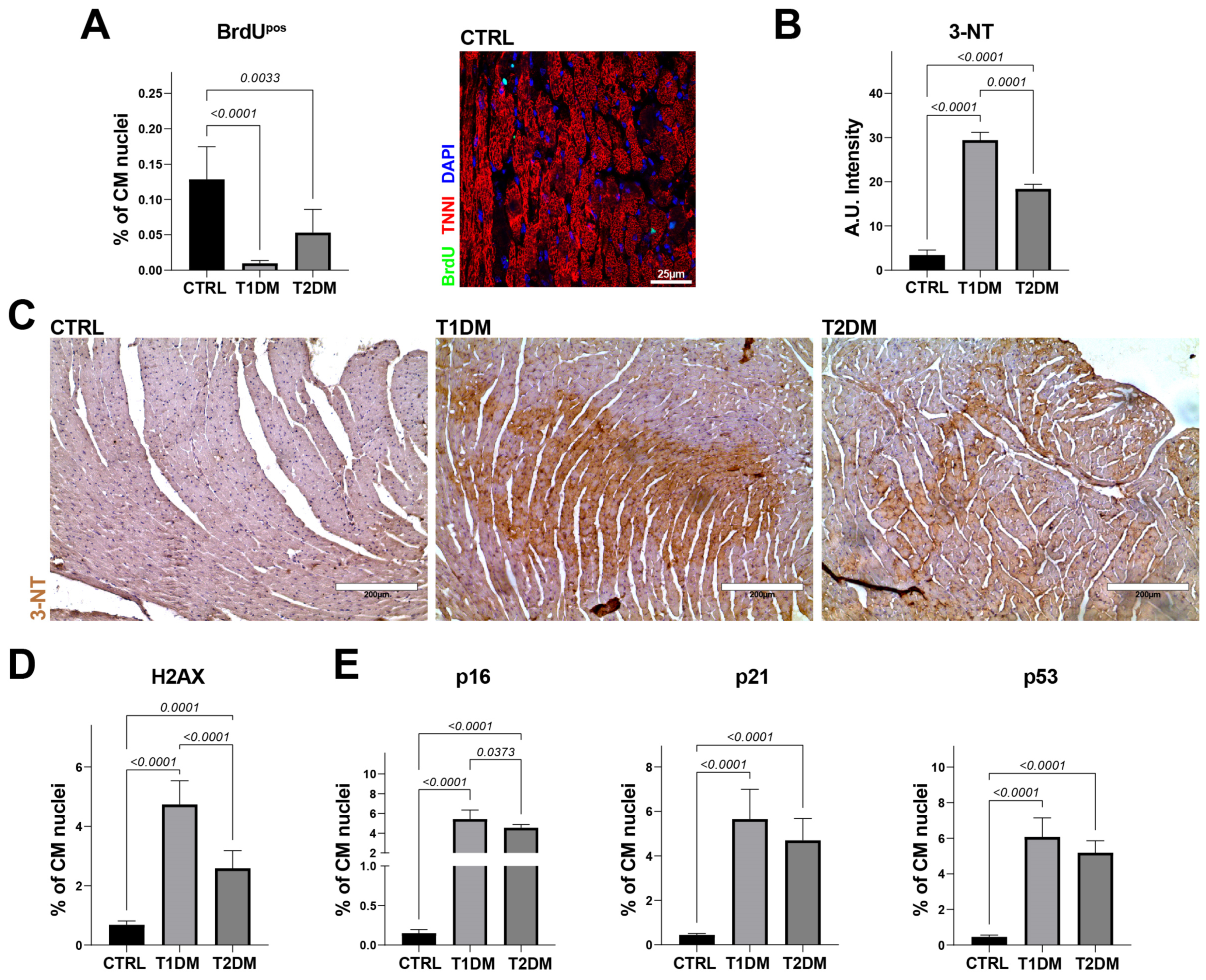
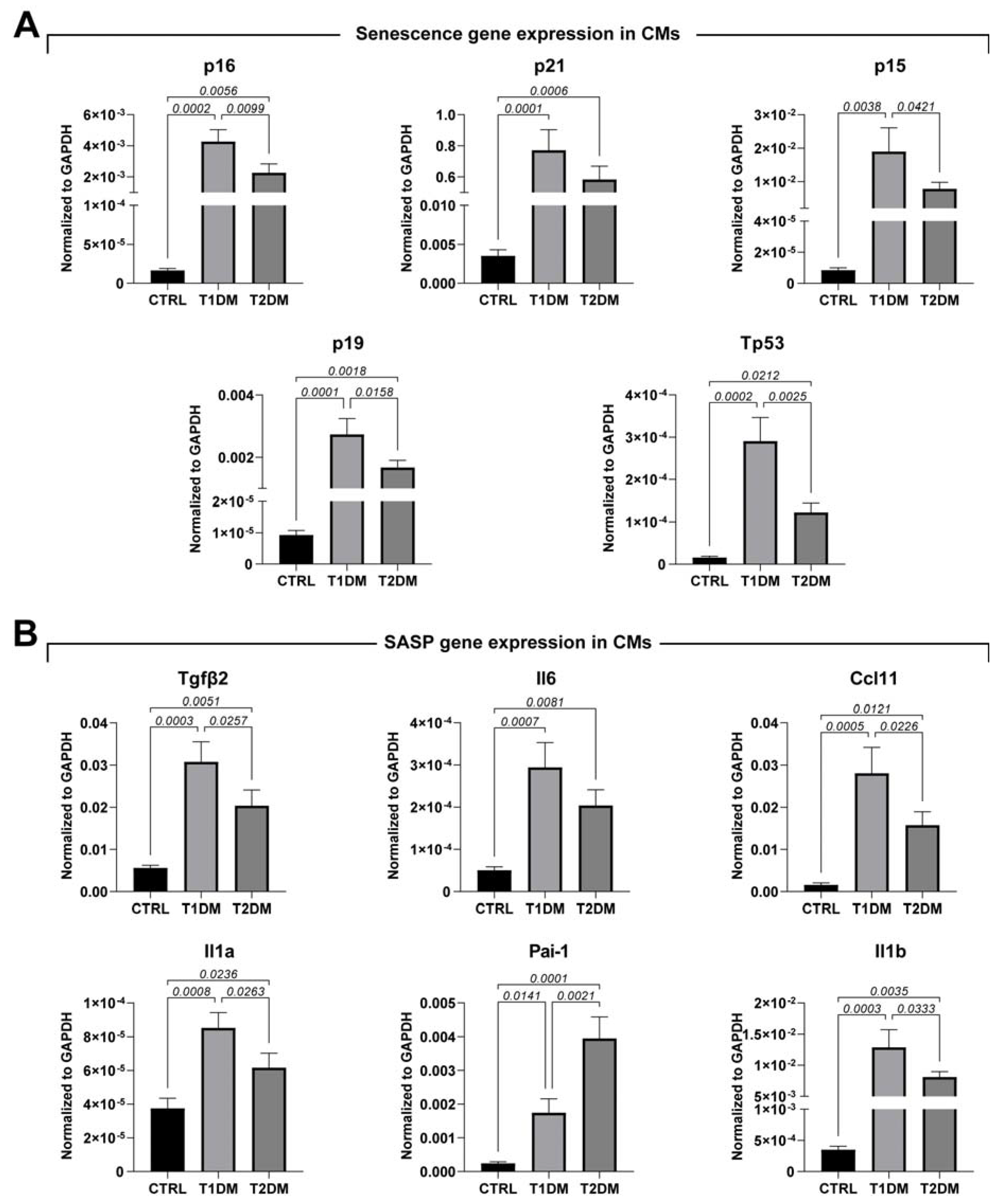
2.4. STZ-Based T1DM and T2DM Mouse Models Affected Oxidative Stress and Cell Senescence Differently
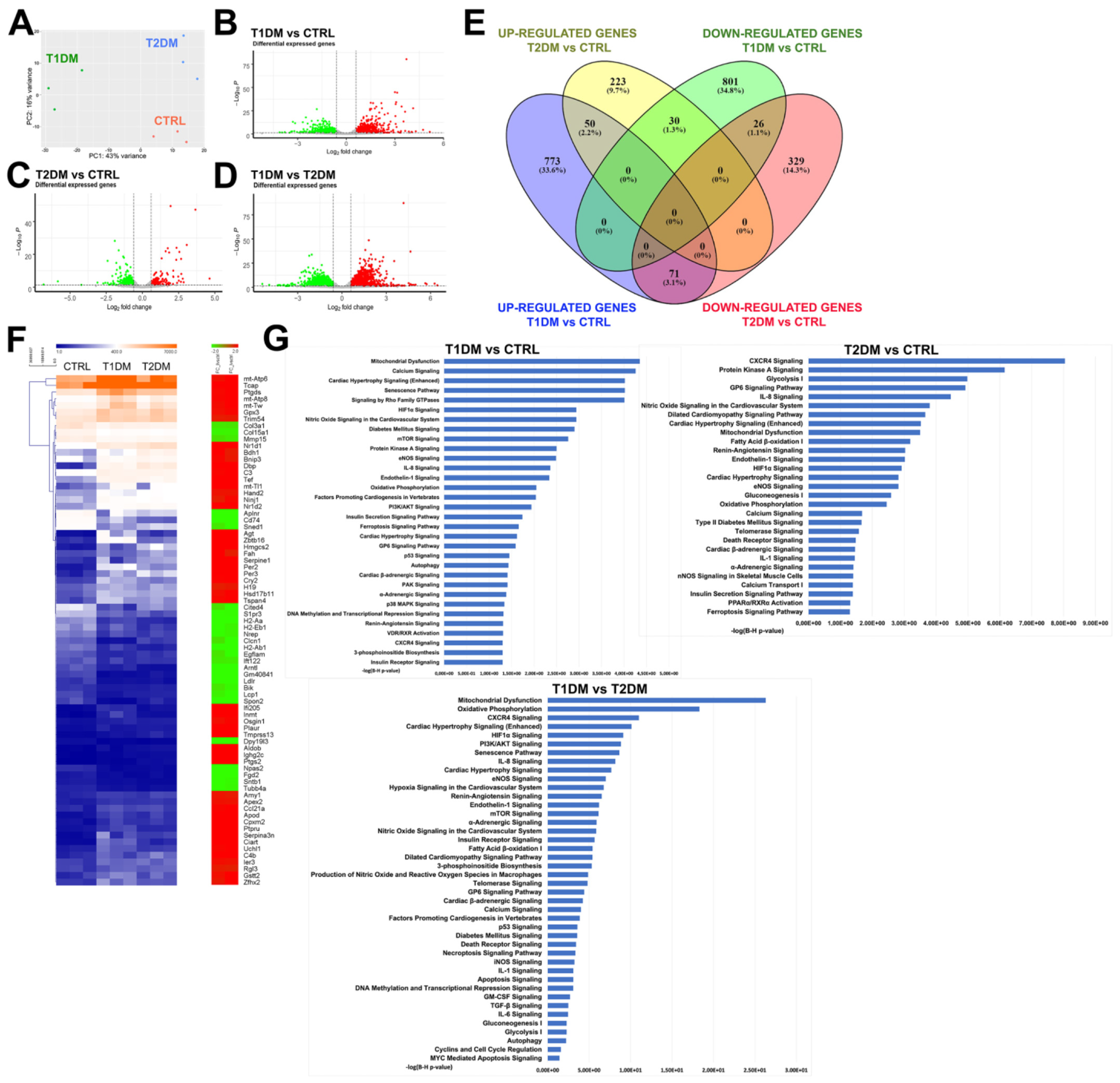
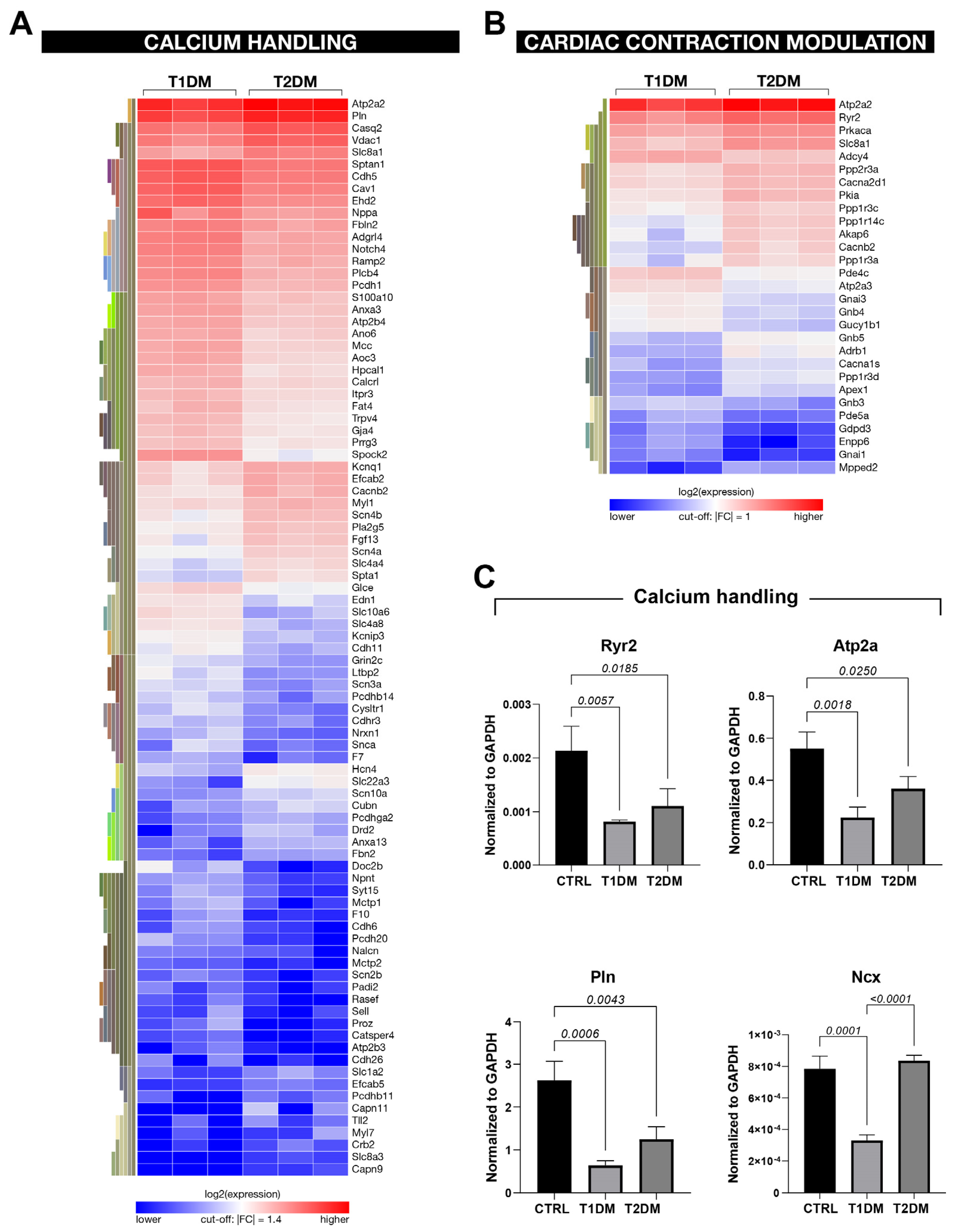
2.5. The Global Transcriptome Profile Showed Different Gene-Expression Signatures in the STZ-Based T1DM and T2DM Mouse Models
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Mouse Cardiomyocyte Isolation
4.3. Echocardiography
4.4. Tissue Harvesting, Histology and Immunohistochemistry
4.5. Quantitative RT-PCR (qPCR)
4.6. RNA Sequencing
4.6.1. RNA Extraction
4.6.2. Library Preparation
4.6.3. Sequencing
4.7. RNA-Seq Data Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- NCD Risk Factor Collaboration. Worldwide trends in diabetes since 1980: A pooled analysis of 751 population-based studies with 4.4 million participants. Lancet 2016, 387, 1513–1530. [Google Scholar] [CrossRef] [PubMed]
- Guariguata, L.; Whiting, D.; Weil, C.; Unwin, N. The International Diabetes Federation diabetes atlas methodology for estimating global and national prevalence of diabetes in adults. Diabetes Res. Clin. Pract. 2011, 94, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Rajbhandari, J.; Fernandez, C.J.; Agarwal, M.; Yeap, B.X.Y.; Pappachan, J.M. Diabetic heart disease: A clinical update. World J. Diabetes 2021, 12, 383–406. [Google Scholar] [CrossRef]
- Raghavan, S.; Vassy, J.L.; Ho, Y.L.; Song, R.J.; Gagnon, D.R.; Cho, K.; Wilson, P.W.F.; Phillips, L.S. Diabetes Mellitus-Related All-Cause and Cardiovascular Mortality in a National Cohort of Adults. J. Am. Heart Assoc. 2019, 8, e011295. [Google Scholar] [CrossRef]
- Leon, B.M.; Maddox, T.M. Diabetes and cardiovascular disease: Epidemiology, biological mechanisms, treatment recommendations and future research. World J. Diabetes 2015, 6, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.M.; Vitale, C.; Seferovic, P. Heart Failure in Patients with Diabetes Mellitus. Card. Fail. Rev. 2017, 3, 52–55. [Google Scholar] [CrossRef]
- Gómez-Perez, A.M.; Damas-Fuentes, M.; Cornejo-Pareja, I.; Tinahones, F.J. Heart Failure in Type 1 Diabetes: A Complication of Concern? A Narrative Review. J. Clin. Med. 2021, 10, 4497. [Google Scholar] [CrossRef] [PubMed]
- Palazzuoli, A.; Iacoviello, M. Diabetes leading to heart failure and heart failure leading to diabetes: Epidemiological and clinical evidence. Heart Fail. Rev. 2022. [Google Scholar] [CrossRef] [PubMed]
- Trimarco, B.; Barbato, E.; Izzo, R.; Morisco, C. Microvascular Disease and the Pathogenesis of Heart Failure in Diabetes: A Tiny Piece of the Tricky Puzzle. Diabetes Care 2022, 45, 2817–2819. [Google Scholar] [CrossRef]
- Shaw, J.A.; Cooper, M.E. Contemporary Management of Heart Failure in Patients With Diabetes. Diabetes Care 2020, 43, 2895–2903. [Google Scholar] [CrossRef]
- Jia, G.; Hill, M.A.; Sowers, J.R. Diabetic Cardiomyopathy: An Update of Mechanisms Contributing to This Clinical Entity. Circ. Res. 2018, 122, 624–638. [Google Scholar] [CrossRef]
- Gulsin, G.S.; Athithan, L.; McCann, G.P. Diabetic cardiomyopathy: Prevalence, determinants and potential treatments. Ther. Adv. Endocrinol. Metab. 2019, 10, 2042018819834869. [Google Scholar] [CrossRef]
- Abdellatif, Y.A.; Nazmy, N.E.M.; Bastawy, I.M.; Raafat, S.S. A Subtle Decline in Cardiac Mechanics is correlated with Albuminuria in Asymptomatic Normotensive Patients with Type 2 Diabetes Mellitus: A Two Dimensional Strain Echocardiography Study. J. Cardiovasc. Echogr. 2021, 31, 220–226. [Google Scholar] [CrossRef]
- Salerno, N.; Salerno, L.; Marino, F.; Scalise, M.; Chiefalo, A.; Panuccio, G.; De Angelis, A.; Cianflone, E.; Urbanek, K.; Torella, D. Myocardial regeneration protocols towards the routine clinical scenario: An unseemly path from bench to bedside. EClinicalMedicine 2022, 50, 101530. [Google Scholar] [CrossRef] [PubMed]
- Furman, B.L. Streptozotocin-Induced Diabetic Models in Mice and Rats. Curr. Protoc. Pharmacol. 2015, 70, 5–47. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yan, L.J. Streptozotocin-induced type 1 diabetes in rodents as a model for studying mitochondrial mechanisms of diabetic β cell glucotoxicity. Diabetes Metab. Syndr. Obes. 2015, 8, 181–188. [Google Scholar] [CrossRef]
- Akinlade, O.M.; Owoyele, B.V.; Soladoye, A.O. Streptozotocin-induced type 1 and 2 diabetes in rodents: A model for studying diabetic cardiac autonomic neuropathy. Afr. Health Sci. 2021, 21, 719–727. [Google Scholar] [CrossRef]
- Chaudhry, Z.Z.; Morris, D.L.; Moss, D.R.; Sims, E.K.; Chiong, Y.; Kono, T.; Evans-Molina, C. Streptozotocin is equally diabetogenic whether administered to fed or fasted mice. Lab. Anim. 2013, 47, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Eleazu, C.O.; Eleazu, K.C.; Chukwuma, S.; Essien, U.N. Review of the mechanism of cell death resulting from streptozotocin challenge in experimental animals, its practical use and potential risk to humans. J. Diabetes Metab. Disord. 2013, 12, 60. [Google Scholar] [CrossRef]
- Heydemann, A. An Overview of Murine High Fat Diet as a Model for Type 2 Diabetes Mellitus. J. Diabetes Res. 2016, 2016, 2902351. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, Z.; Zheng, C.; Wintergerst, K.A.; Keller, B.B.; Cai, L. Mechanisms of diabetic cardiomyopathy and potential therapeutic strategies: Preclinical and clinical evidence. Nat. Rev. Cardiol. 2020, 17, 585–607. [Google Scholar] [CrossRef]
- Marino, F.; Scalise, M.; Salerno, N.; Salerno, L.; Molinaro, C.; Cappetta, D.; Torella, M.; Greco, M.; Foti, D.; Sasso, F.C.; et al. Diabetes-Induced Cellular Senescence and Senescence-Associated Secretory Phenotype Impair Cardiac Regeneration and Function Independently of Age. Diabetes 2022, 71, 1081–1098. [Google Scholar] [CrossRef]
- Salerno, N.; Marino, F.; Scalise, M.; Salerno, L.; Molinaro, C.; Filardo, A.; Chiefalo, A.; Panuccio, G.; De Angelis, A.; Urbanek, K.; et al. Pharmacological clearance of senescent cells improves cardiac remodeling and function after myocardial infarction in female aged mice. Mech. Ageing Dev. 2022, 208, 111740. [Google Scholar] [CrossRef]
- Dobrin, J.S.; Lebeche, D. Diabetic cardiomyopathy: Signaling defects and therapeutic approaches. Expert Rev. Cardiovasc. Ther. 2010, 8, 373–391. [Google Scholar] [CrossRef]
- Cai, L.; Li, W.; Wang, G.; Guo, L.; Jiang, Y.; Kang, Y.J. Hyperglycemia-induced apoptosis in mouse myocardium: Mitochondrial cytochrome C-mediated caspase-3 activation pathway. Diabetes 2002, 51, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Frustaci, A.; Kajstura, J.; Chimenti, C.; Jakoniuk, I.; Leri, A.; Maseri, A.; Nadal-Ginard, B.; Anversa, P. Myocardial cell death in human diabetes. Circ. Res. 2000, 87, 1123–1132. [Google Scholar] [CrossRef]
- Aquila, I.; Cianflone, E.; Scalise, M.; Marino, F.; Mancuso, T.; Filardo, A.; Smith, A.J.; Cappetta, D.; De Angelis, A.; Urbanek, K.; et al. c-kit Haploinsufficiency impairs adult cardiac stem cell growth, myogenicity and myocardial regeneration. Cell Death Dis. 2019, 10, 436. [Google Scholar] [CrossRef] [PubMed]
- Cianflone, E.; Cappetta, D.; Mancuso, T.; Sabatino, J.; Marino, F.; Scalise, M.; Albanese, M.; Salatino, A.; Parrotta, E.I.; Cuda, G.; et al. Statins Stimulate New Myocyte Formation After Myocardial Infarction by Activating Growth and Differentiation of the Endogenous Cardiac Stem Cells. Int. J. Mol. Sci. 2020, 21, 7927. [Google Scholar] [CrossRef]
- Murakami, T.; Inagaki, N.; Kondoh, H. Cellular Senescence in Diabetes Mellitus: Distinct Senotherapeutic Strategies for Adipose Tissue and Pancreatic β Cells. Front. Endocrinol. 2022, 13, 869414. [Google Scholar] [CrossRef]
- Molinaro, C.; Salerno, L.; Marino, F.; Scalise, M.; Salerno, N.; Pagano, L.; De Angelis, A.; Cianflone, E.; Torella, D.; Urbanek, K. Unraveling and Targeting Myocardial Regeneration Deficit in Diabetes. Antioxidants 2022, 11, 208. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.; Lagnado, A.; Maggiorani, D.; Walaszczyk, A.; Dookun, E.; Chapman, J.; Birch, J.; Salmonowicz, H.; Ogrodnik, M.; Jurk, D.; et al. Length-independent telomere damage drives post-mitotic cardiomyocyte senescence. EMBO J. 2019, 38, e100492. [Google Scholar] [CrossRef] [PubMed]
- Cosentino, F.; Grant, P.J.; Aboyans, V.; Bailey, C.J.; Ceriello, A.; Delgado, V.; Federici, M.; Filippatos, G.; Grobbee, D.E.; Hansen, T.B.; et al. 2019 ESC Guidelines on diabetes, pre-diabetes, and cardiovascular diseases developed in collaboration with the EASD. Eur. Heart J. 2020, 41, 255–323. [Google Scholar] [CrossRef] [PubMed]
- Falcão-Pires, I.; Leite-Moreira, A.F. Diabetic cardiomyopathy: Understanding the molecular and cellular basis to progress in diagnosis and treatment. Heart Fail. Rev. 2012, 17, 325–344. [Google Scholar] [CrossRef] [PubMed]
- Richter, C.; Hinkel, R. Research(’s) Sweet Hearts: Experimental Biomedical Models of Diabetic Cardiomyopathy. Front. Cardiovasc. Med. 2021, 8, 703355. [Google Scholar] [CrossRef]
- Harmancey, R.; Taegtmeyer, H. The complexities of diabetic cardiomyopathy: Lessons from patients and animal models. Curr. Diabetes Rep. 2008, 8, 243–248. [Google Scholar] [CrossRef]
- Kottaisamy, C.P.D.; Raj, D.S.; Prasanth Kumar, V.; Sankaran, U. Experimental animal models for diabetes and its related complications-a review. Lab. Anim. Res. 2021, 37, 23. [Google Scholar] [CrossRef]
- Abudureyimu, M.; Luo, X.; Wang, X.; Sowers, J.R.; Wang, W.; Ge, J.; Ren, J.; Zhang, Y. Heart failure with preserved ejection fraction (HFpEF) in type 2 diabetes mellitus: From pathophysiology to therapeutics. J. Mol. Cell Biol. 2022, 14, mjac028. [Google Scholar] [CrossRef]
- McHugh, K.; DeVore, A.D.; Wu, J.; Matsouaka, R.A.; Fonarow, G.C.; Heidenreich, P.A.; Yancy, C.W.; Green, J.B.; Altman, N.; Hernandez, A.F. Heart Failure With Preserved Ejection Fraction and Diabetes: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2019, 73, 602–611. [Google Scholar] [CrossRef]
- Giacco, F.; Brownlee, M. Oxidative stress and diabetic complications. Circ. Res. 2010, 107, 1058–1070. [Google Scholar] [CrossRef]
- Kaludercic, N.; Di Lisa, F. Mitochondrial ROS Formation in the Pathogenesis of Diabetic Cardiomyopathy. Front. Cardiovasc. Med. 2020, 7, 12. [Google Scholar] [CrossRef]
- Newsholme, P.; Haber, E.P.; Hirabara, S.M.; Rebelato, E.L.; Procopio, J.; Morgan, D.; Oliveira-Emilio, H.C.; Carpinelli, A.R.; Curi, R. Diabetes associated cell stress and dysfunction: Role of mitochondrial and non-mitochondrial ROS production and activity. J. Physiol. 2007, 583, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Marino, F.; Scalise, M.; Cianflone, E.; Salerno, L.; Cappetta, D.; Salerno, N.; De Angelis, A.; Torella, D.; Urbanek, K. Physical Exercise and Cardiac Repair: The Potential Role of Nitric Oxide in Boosting Stem Cell Regenerative Biology. Antioxidants 2021, 10, 1002. [Google Scholar] [CrossRef]
- Shah, M.S.; Brownlee, M. Molecular and Cellular Mechanisms of Cardiovascular Disorders in Diabetes. Circ. Res. 2016, 118, 1808–1829. [Google Scholar] [CrossRef]
- D’Oria, R.; Schipani, R.; Leonardini, A.; Natalicchio, A.; Perrini, S.; Cignarelli, A.; Laviola, L.; Giorgino, F. The Role of Oxidative Stress in Cardiac Disease: From Physiological Response to Injury Factor. Oxid. Med. Cell. Longev. 2020, 2020, 5732956. [Google Scholar] [CrossRef]
- Zhou, B.; Tian, R. Mitochondrial dysfunction in pathophysiology of heart failure. J. Clin. Investig. 2018, 128, 3716–3726. [Google Scholar] [CrossRef] [PubMed]
- Pospelova, T.V.; Demidenko, Z.N.; Bukreeva, E.I.; Pospelov, V.A.; Gudkov, A.V.; Blagosklonny, M.V. Pseudo-DNA damage response in senescent cells. Cell Cycle 2009, 8, 4112–4118. [Google Scholar] [CrossRef]
- Schäfer, K.; Fujisawa, K.; Konstantinides, S.; Loskutoff, D.J. Disruption of the plasminogen activator inhibitor 1 gene reduces the adiposity and improves the metabolic profile of genetically obese and diabetic ob/ob mice. FASEB J. 2001, 15, 1840–1842. [Google Scholar] [CrossRef]
- Ma, L.J.; Mao, S.L.; Taylor, K.L.; Kanjanabuch, T.; Guan, Y.; Zhang, Y.; Brown, N.J.; Swift, L.L.; McGuinness, O.P.; Wasserman, D.H.; et al. Prevention of obesity and insulin resistance in mice lacking plasminogen activator inhibitor 1. Diabetes 2004, 53, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Samad, F.; Pandey, M.; Bell, P.A.; Loskutoff, D.J. Insulin continues to induce plasminogen activator inhibitor 1 gene expression in insulin-resistant mice and adipocytes. Mol. Med. 2000, 6, 680–692. [Google Scholar] [CrossRef]
- Fukui, S.; Fukumoto, Y.; Suzuki, J.; Saji, K.; Nawata, J.; Shinozaki, T.; Kagaya, Y.; Watanabe, J.; Shimokawa, H. Diabetes mellitus accelerates left ventricular diastolic dysfunction through activation of the renin-angiotensin system in hypertensive rats. Hypertens. Res. 2009, 32, 472–480. [Google Scholar] [CrossRef]
- Kalyanasundaram, A.; Lacombe, V.A.; Belevych, A.E.; Brunello, L.; Carnes, C.A.; Janssen, P.M.; Knollmann, B.C.; Periasamy, M.; Gyørke, S. Up-regulation of sarcoplasmic reticulum Ca2+ uptake leads to cardiac hypertrophy, contractile dysfunction and early mortality in mice deficient in CASQ2. Cardiovasc. Res. 2013, 98, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Sciarretta, S.; Boppana, V.S.; Umapathi, M.; Frati, G.; Sadoshima, J. Boosting autophagy in the diabetic heart: A translational perspective. Cardiovasc. Diagn. Ther. 2015, 5, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Sciarretta, S.; Maejima, Y.; Zablocki, D.; Sadoshima, J. The Role of Autophagy in the Heart. Annu. Rev. Physiol. 2018, 80, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Cianflone, E.; Torella, M.; Biamonte, F.; De Angelis, A.; Urbanek, K.; Costanzo, F.S.; Rota, M.; Ellison-Hughes, G.M.; Torella, D. Targeting Cardiac Stem Cell Senescence to Treat Cardiac Aging and Disease. Cells 2020, 9, 1558. [Google Scholar] [CrossRef]
- Scalise, M.; Marino, F.; Salerno, L.; Mancuso, T.; Cappetta, D.; Barone, A.; Parrotta, E.I.; Torella, A.; Palumbo, D.; Veltri, P.; et al. In vitro CSC-derived cardiomyocytes exhibit the typical microRNA-mRNA blueprint of endogenous cardiomyocytes. Commun. Biol. 2021, 4, 1146. [Google Scholar] [CrossRef]
- Martin, M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Vashishtha, K.; Gaud, C.; Andrews, S.; Krueger, C. Librarian: A quality control tool to analyse sequencing library compositions. F1000Research 2022, 11, 1122. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Salvati, A.; Gigantino, V.; Nassa, G.; Giurato, G.; Alexandrova, E.; Rizzo, F.; Tarallo, R.; Weisz, A. The Histone Methyltransferase DOT1L Is a Functional Component of Estrogen Receptor Alpha Signaling in Ovarian Cancer Cells. Cancers 2019, 11, 1720. [Google Scholar] [CrossRef] [PubMed]



| CTRL (n = 10) | T1DM (n = 11) | T2DM (n = 12) | p-Value | |
|---|---|---|---|---|
| HR (bpm) | 446.67 ± 24.16 | 463.55 ± 49.13 | 442.25 ± 40.34 | 0.428 |
| LVEDD (mm) | 4.06 ± 0.08 | 4.40 ± 0.20 | 3.95 ± 0.23 | <0.001 |
| LVESD (mm) | 2.82 ± 0.07 | 3.41 ± 0.19 | 2.75 ± 0.30 | <0.001 |
| EF (%) | 58.33 ± 2.32 | 45.63 ± 3.84 | 58.41 ± 6.55 | <0.001 |
| FS (%) | 30.45 ± 1.63 | 22.60 ± 2.29 | 30.57 ± 4.49 | <0.001 |
| GLS (%) | −21.49 ± 1.33 | −13.93 ± 3.38 | −18.49 ± 2.99 | <0.001 |
| E (mm/sec) | 639.98 ± 88.09 | 702.06 ± 78.32 | 587.87 ± 73.18 | 0.008 |
| A (mm/sec) | 399.20 ± 54.74 | 422.69 ± 57.17 | 411.68 ± 80.27 | 0.756 |
| E/A | 1.62 ± 0.3 | 1.65 ± 0.19 | 1.47 ± 0.27 | 0.256 |
| E′ (mm/sec) | −25.18 ± 3.10 | −21.68 ± 3.97 | −18.44 ± 4.96 | 0.005 |
| E/E′ | 25.6 ± 3.5 | 33.10 ± 5.62 | 33.35 ± 7.24 | 0.009 |
| E′/A′ | 1.15 ± 0.26 | 0.77 ± 0.11 | 0.97 ± 0.23 | 0.002 |
| IVSd (mm) | 0.65 ± 0.06 | 0.62 ± 0.05 | 0.59 ± 0.04 | 0.067 |
| LVPWd (mm) | 0.63 ± 0.07 | 0.64 ± 0.07 | 0.62 ± 0.05 | 0.628 |
| Gene | Sequence (5′ -> 3′) |
|---|---|
| mGapdh | Fwd-CTCCACTCTTCCACCTTCG- |
| Rev-GCCTCTCTTGCTCAGTGTCC- | |
| mTgfb2 | Fwd-CCGCATCTCCTGCTAATGTTG- |
| Rev-AATAGGCGGCATCCAAAGC- | |
| mNppa | Fwd-CTGATGGATTTCAAGAACCTGCT- |
| Rev-TCTCAGAGGTGGGTTGACCT- | |
| mp21 | Fwd-AACATCTCAGGGCCGAAA- |
| Rev-TGCGCTTGGAGTGATAGAAA- | |
| mp16 | Fwd-GTGTGCATGACGTGCGGG- |
| Rev-GCAGTTCGAATCTGCACCGTAG- | |
| mp15 | Fwd-AGATCCCAACGCCCTGAAC- |
| Rev-CCCATCATCATCACCTGGATT- | |
| mp19 | Fwd-GCTCTGGCTTTCGTGAACATG- |
| Rev-TCGAATCTGCACCGTAGTTGAG- | |
| mMybpc2 | Fwd-CTGCTAGGGCCTGGTTAGAG- |
| Rev-CCTTTTTGGCCGCTGGTTTA- | |
| mIl-6 | Fwd-TGAGAAAAGAGTTGTGCAATGG- |
| Rev-GGTACTCCAGAAGACCAGAGG- | |
| mCcl11 | Fwd-TGCAGAGCTCCACAGCGCTT |
| Rev-GGGTGAGCCAGCACCTGGGA | |
| mPai-1 | Fwd-GGCCATTACTACGACATCCTG |
| Rev-GGTCATGTTGCCTTTCCAGT | |
| mIl1b | Fwd-TGCCACCTTTTGACAGTGATG |
| Rev-TGATGTGCTGCTGCGAGATT | |
| mGja1 | Fwd GGT GAT GAA CAG TCT GCC TTT CG |
| Rev GTG AGC CAA GTA CAG GAG TGT G | |
| mCol1a1 | Fwd CCT CAG GGT ATT GCT GGA CAA C |
| Rev CAG AAG GAC CTT GTT TGC CAG G | |
| mCol1a2 | Fwd TTC TGT GGG TCC TGC TGG GAA A |
| Rev TTG TCA CCT CGG ATG CCT TGA G | |
| mCol3a1 | Fwd GAC CAA AAG GTG ATG CTG GAC AG |
| Rev CAA GAC CTC GTG CTC CAG TTA G | |
| mMyl7 | Fwd AGG AAG CCA TCC TGA GTG CCT T |
| Rev CAT GGG TGT CAG CGC AAA CAG T | |
| mCasp3 | Fwd GGA GTC TGA CTG GAA AGC CGA A |
| Rev CTT CTG GCA AGC CAT CTC CTC A | |
| mBcl2 | Fwd CCT GTG GAT GAC TGA GTA CCT G |
| Rev AGC CAG GAG AAA TCA AAC AGA GG | |
| mBax | Fwd AGG ATG CGT CCA CCA AGA AGC T |
| Rev TCC GTG TCC ACG TCA GCA ATC A | |
| mFoxo3 | Fwd CCT ACT TCA AGG ATA AGG GCG AC |
| Rev GCC TTC ATT CTG AAC GCG CAT G | |
| mFoxo1 | Fwd CTA CGA GTG GAT GGT GAA GAG C |
| Rev CCA GTT CCT TCA TTC TGC ACT CG | |
| mIL1a | Fwd AGGGAGTCAACTCATTGGCG |
| Rev TGGCAGAACTGTAGTCTTCGT | |
| mTp53 | Fwd ATGGCCATCTACAAGAAGTCACAG |
| Rev ATCGGAGCAGCGCTCATG | |
| mMyh7 | Fwd GCTGGAAGATGAGTGCTCAGAG |
| Rev TCCAAACCAGCCATCTCCTCTG | |
| mRyr2 | Fwd ACCTACTCCGAAGGCTGGTGTT |
| Rev TTCTTCCGAGGCAGCACCAAAG | |
| mAtp2a | Fwd GTGAAGTGCCATCAGTATGACGG |
| Rev GTGAGAGCAGTCTCGGTAGCTT | |
| mPnl | Fwd GGACCAAAGGAACTTGCCAGCT |
| Rev CAACAGGCAGCCAAATGTGAGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marino, F.; Salerno, N.; Scalise, M.; Salerno, L.; Torella, A.; Molinaro, C.; Chiefalo, A.; Filardo, A.; Siracusa, C.; Panuccio, G.; et al. Streptozotocin-Induced Type 1 and 2 Diabetes Mellitus Mouse Models Show Different Functional, Cellular and Molecular Patterns of Diabetic Cardiomyopathy. Int. J. Mol. Sci. 2023, 24, 1132. https://doi.org/10.3390/ijms24021132
Marino F, Salerno N, Scalise M, Salerno L, Torella A, Molinaro C, Chiefalo A, Filardo A, Siracusa C, Panuccio G, et al. Streptozotocin-Induced Type 1 and 2 Diabetes Mellitus Mouse Models Show Different Functional, Cellular and Molecular Patterns of Diabetic Cardiomyopathy. International Journal of Molecular Sciences. 2023; 24(2):1132. https://doi.org/10.3390/ijms24021132
Chicago/Turabian StyleMarino, Fabiola, Nadia Salerno, Mariangela Scalise, Luca Salerno, Annalaura Torella, Claudia Molinaro, Antonio Chiefalo, Andrea Filardo, Chiara Siracusa, Giuseppe Panuccio, and et al. 2023. "Streptozotocin-Induced Type 1 and 2 Diabetes Mellitus Mouse Models Show Different Functional, Cellular and Molecular Patterns of Diabetic Cardiomyopathy" International Journal of Molecular Sciences 24, no. 2: 1132. https://doi.org/10.3390/ijms24021132
APA StyleMarino, F., Salerno, N., Scalise, M., Salerno, L., Torella, A., Molinaro, C., Chiefalo, A., Filardo, A., Siracusa, C., Panuccio, G., Ferravante, C., Giurato, G., Rizzo, F., Torella, M., Donniacuo, M., De Angelis, A., Viglietto, G., Urbanek, K., Weisz, A., ... Cianflone, E. (2023). Streptozotocin-Induced Type 1 and 2 Diabetes Mellitus Mouse Models Show Different Functional, Cellular and Molecular Patterns of Diabetic Cardiomyopathy. International Journal of Molecular Sciences, 24(2), 1132. https://doi.org/10.3390/ijms24021132








