Mutational Landscape of Bladder Cancer in Mexican Patients: KMT2D Mutations and chr11q15.5 Amplifications Are Associated with Muscle Invasion
Abstract
1. Introduction
2. Results
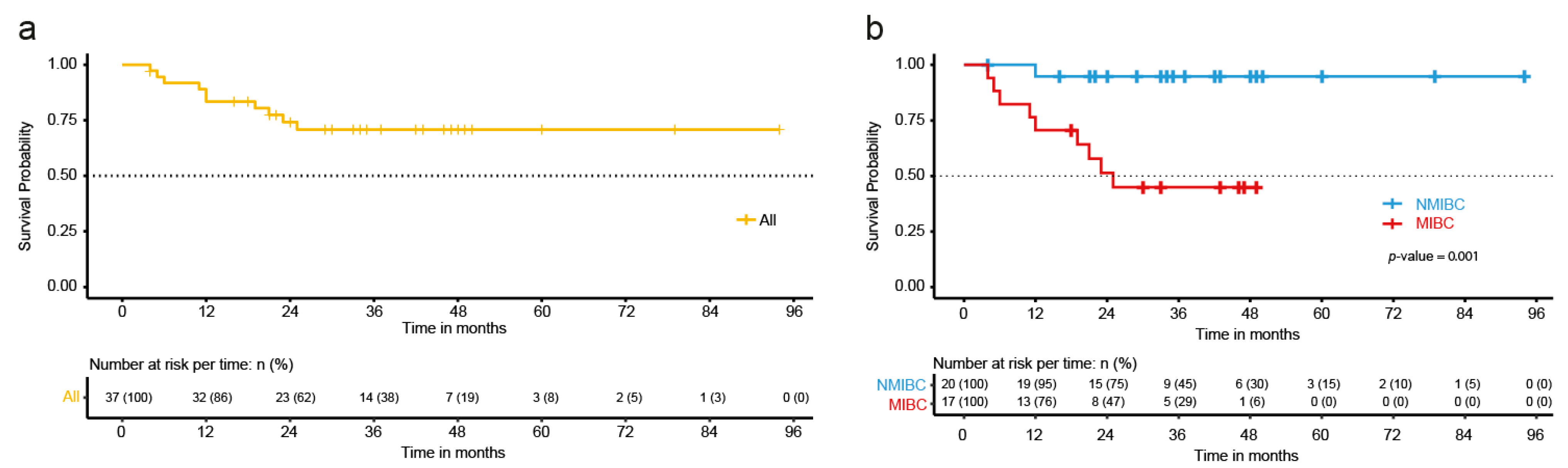
2.1. Clinical-Pathological Characteristics of the BC Patients
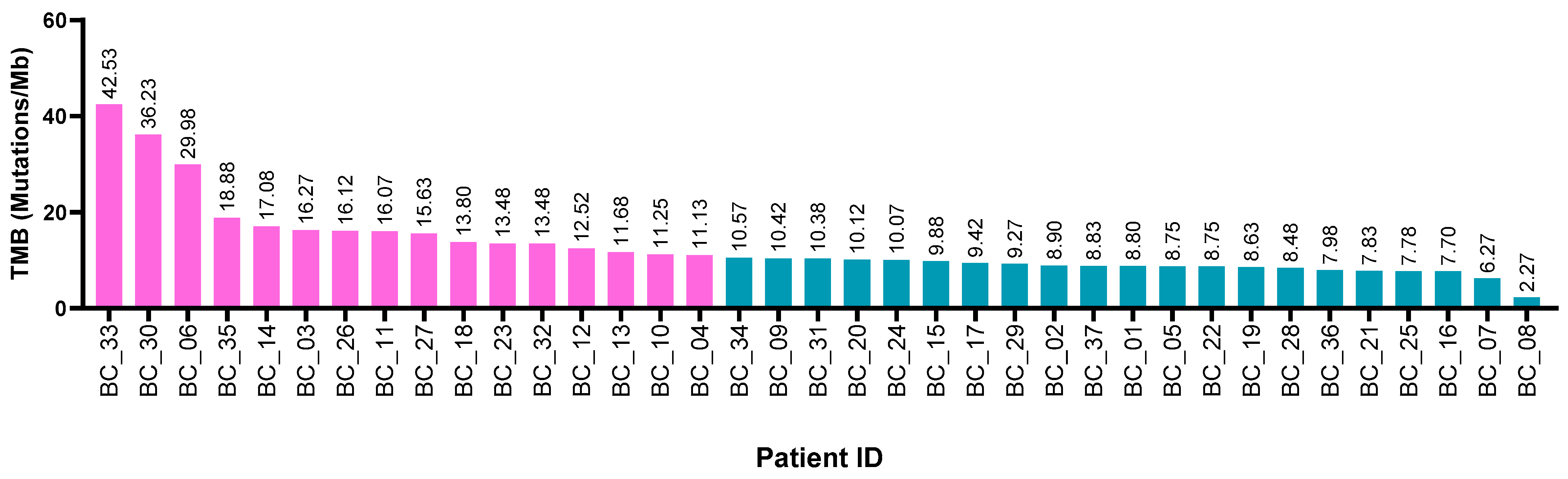
2.2. Tumor Mutational Burden and Clinical-Pathological Characteristics
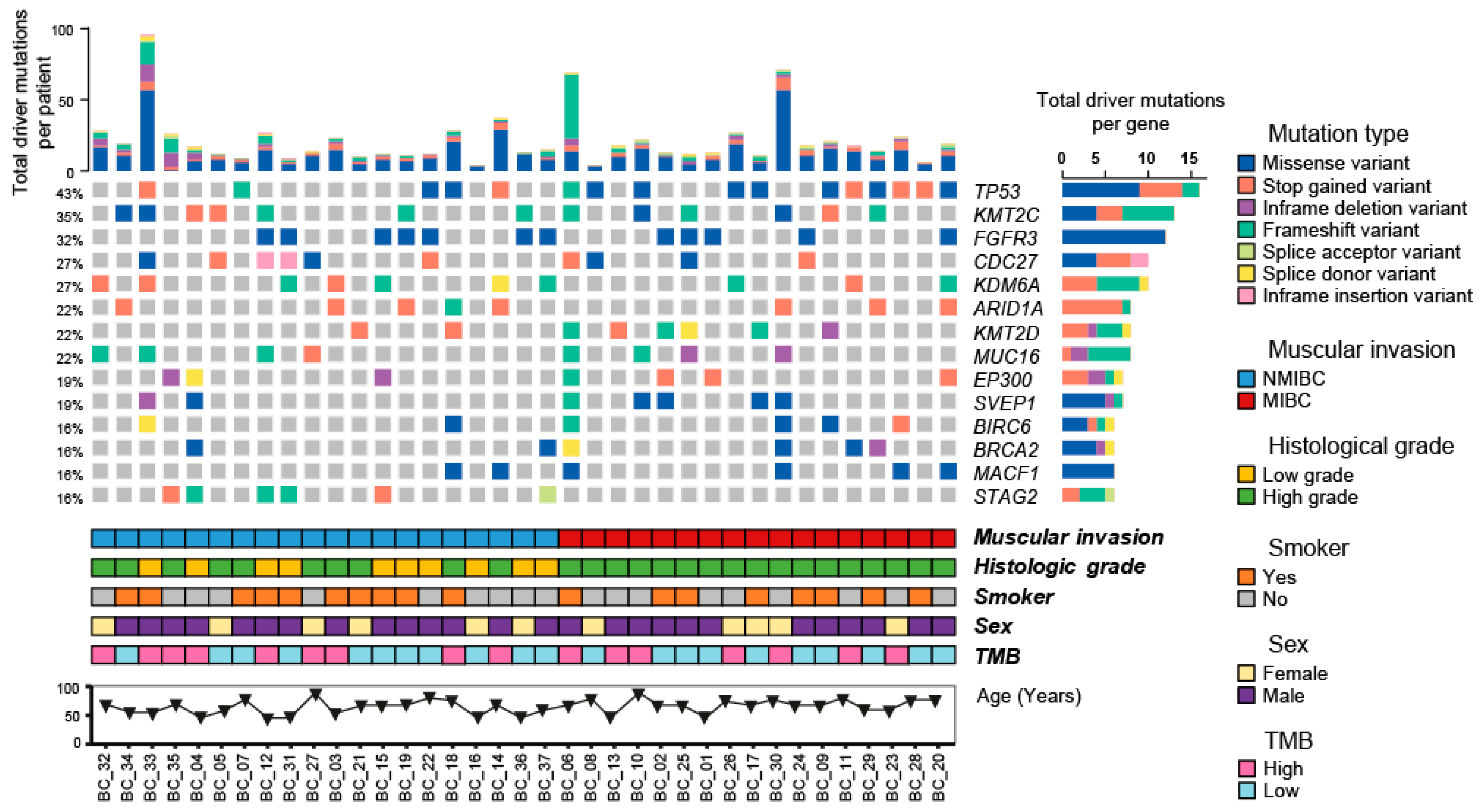
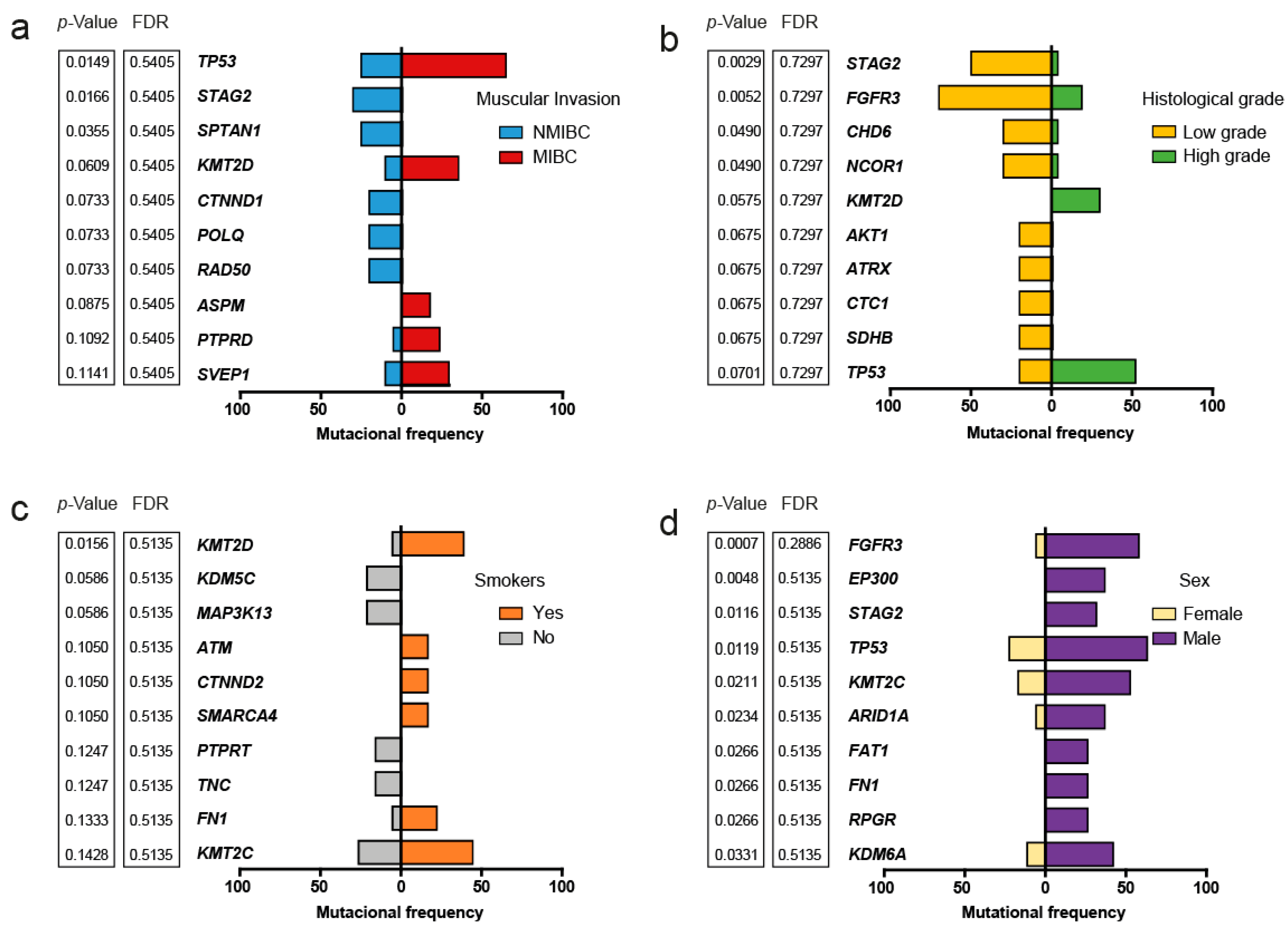
2.3. Somatic Variants and Clinical-Pathological Characteristics of the Patients
2.4. Structural Variants and Clinical-Pathological Characteristics of the Patients
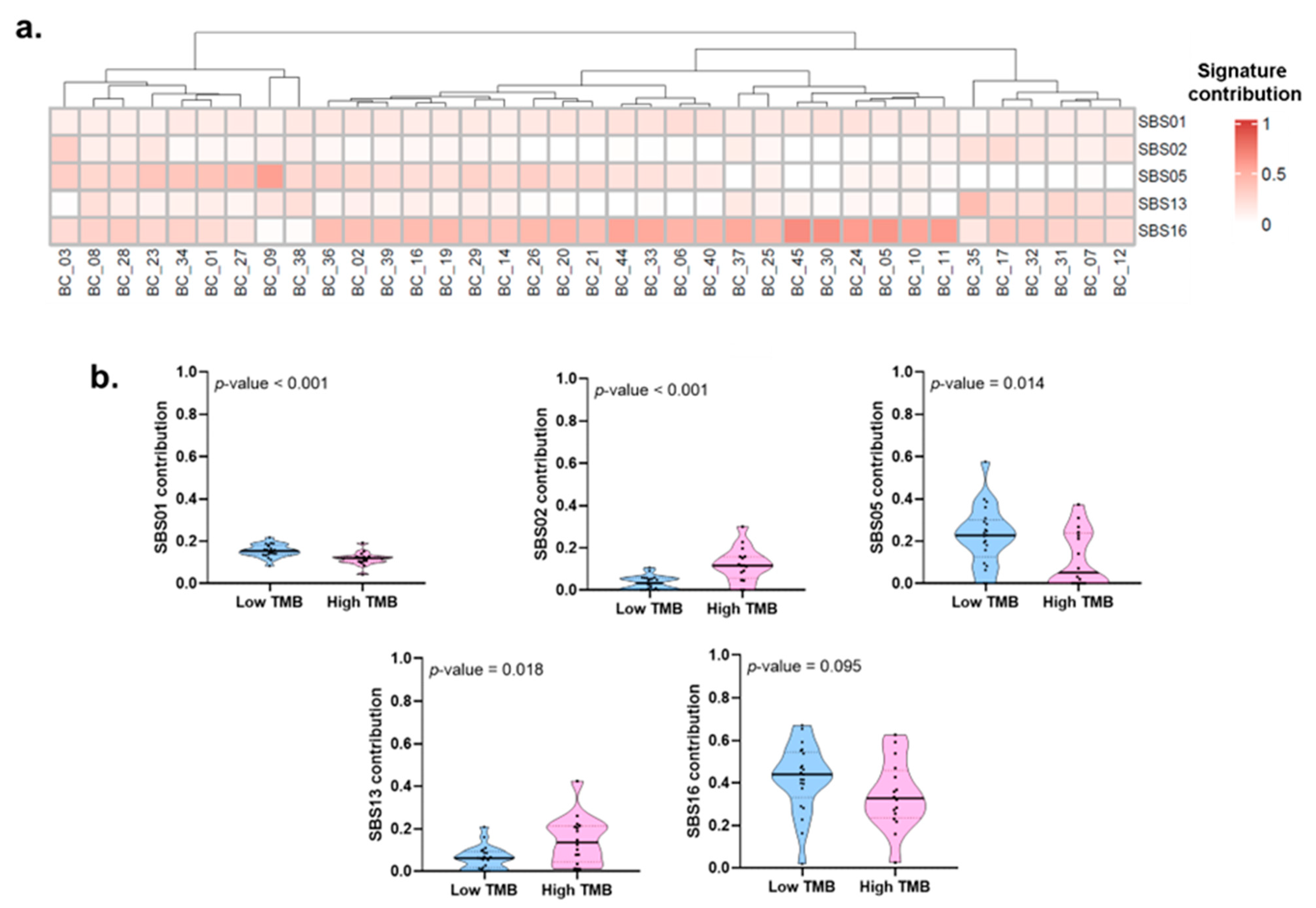
2.5. Mutational Processes and Clinical-Pathological Characteristics
3. Discussion
4. Material and Methods
4.1. Population
4.2. Clinical Data Collection
4.3. DNA Extraction and Quality Control
4.4. Library Preparation, Hybridation Capture, and WES
4.5. Bioinformatics Pipeline
4.6. Mutational Signature Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dobruch, J.; Oszczudłowski, M. Bladder Cancer: Current Challenges and Future Directions. Medicina 2021, 57, 749. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Freedman, N.D.; Silverman, D.T.; Hollenbeck, A.R.; Schatzkin, A.; Abnet, C.C. Association between smoking and risk of bladder cancer among men and women. JAMA 2011, 306, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Kirkali, Z.; Chan, T.; Manoharan, M.; Algaba, F.; Busch, C.; Cheng, L.; Kiemeney, L.; Kriegmair, M.; Montironi, R.; Murphy, W.M.; et al. Bladder cancer: Epidemiology, staging and grading, and diagnosis. Urology 2005, 66, 4–34. [Google Scholar] [CrossRef] [PubMed]
- Siracusano, S.; Rizzetto, R.; Porcaro, A.B. Bladder cancer genomics. Urologia 2020, 87, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, A.; de Reyniès, A.; Allory, Y.; Sjödahl, G.; Robertson, A.G.; Seiler, R.; Hoadley, K.A.; Groeneveld, C.S.; Al-Ahmadie, H.; Choi, W.; et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur. Urol. 2020, 77, 420–433. [Google Scholar] [CrossRef]
- Miyake, M.; Hirao, S.; Mibu, H.; Tanaka, M.; Takashima, K.; Shimada, K.; Hirao, K. Clinical significance of subepithelial growth patterns in non-muscle invasive bladder cancer. BMC Urol. 2011, 11, 17. [Google Scholar] [CrossRef]
- Gofrit, O.N.; Shapiro, A.; Pode, D.; Katz, R.; Yutkin, V.; Zorn, K.C.; Pizov, G. Subepithelial growth patterns in urothelial carcinoma-frequency and prognostic significance. Urol. Oncol. 2012, 30, 49–54. [Google Scholar] [CrossRef]
- Hedegaard, J.; Lamy, P.; Nordentoft, I.; Algaba, F.; Høyer, S.; Ulhøi, B.P.; Vang, S.; Reinert, T.; Hermann, G.G.; Mogensen, K.; et al. Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer Cell 2016, 30, 27–42. [Google Scholar] [CrossRef]
- Sjödahl, G.; Lauss, M.; Lövgren, K.; Chebil, G.; Gudjonsson, S.; Veerla, S.; Patschan, O.; Aine, M.; Fernö, M.; Ringnér, M.; et al. A molecular taxonomy for urothelial carcinoma. Clin. Cancer Res. 2012, 18, 3377–3386. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2018, 174, 1033. [Google Scholar] [CrossRef]
- Seok Ju, Y. The mutational signatures and molecular alterations of bladder cancer. Transl. Cancer Res. 2017, 6, S689–S701. [Google Scholar]
- Poon, S.L.; Huang, M.N.; Choo, Y.; McPherson, J.R.; Yu, W.; Heng, H.L.; Gan, A.; Myint, S.S.; Siew, E.Y.; Ler, L.D.; et al. Mutation signatures implicate aristolochic acid in bladder cancer development. Genome Med. 2015, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Ariza, V.P.; Solares, S.M.E.; Chanona, V.J.G.; Martínez, C.P.; Jiménez-Ríos, M.A. Adenocarcinoma primario de vejiga. Experiencia de 20 años en el Instituto Nacional de Cancerología. Rev. Mex. Urol. 2007, 67, 256–260. [Google Scholar]
- Mayorga, G.E.; Ibarra, O.I.; Sedano, B.J.; Trujillo, O.L.; Cornejo, D.V.; Palmeros, R.A.; Uberetagoyena, T.I.; Garza, S.G.; Osornio, S.V.; Camacho, C.A.; et al. Aplicación de nomogramas en México para cáncer de vejiga en pacientes del Hospital General “Dr. Manuel Gea González”. Rev. Mex. Urol. 2014, 74, 3–8. [Google Scholar] [CrossRef]
- Nyame, Y.A.; Baker, K.K.; Montgomery, R.B.; Grivas, P.; Redman, M.W.; Wright, J.L. Racial and sex differences in somatic mutations in bladder cancer patients: An analysis of the cBioPortal for Cancer Genomics. J. Clin. Oncol. 2020, 38, 556. [Google Scholar] [CrossRef]
- Burgess, E.F.; Sanders, J.A.; Livasy, C.; Symanowski, J.; Gatalica, Z.; Steuerwald, N.M.; Arguello, D.; Brouwer, C.R.; Korn, W.M.; Grigg, C.M.; et al. Identification of potential biomarkers and novel therapeutic targets through genomic analysis of small cell bladder carcinoma and associated clinical outcomes. Urol. Oncol. 2022, 40, 383.e1–383.e10. [Google Scholar] [CrossRef]
- Dhar, S.S.; Lee, M.G. Cancer-epigenetic function of the histone methyltransferase KMT2D and therapeutic opportunities for the treatment of KMT2D-deficient tumors. Oncotarget 2021, 12, 1296–1308. [Google Scholar] [CrossRef]
- Ding, B.; Yan, L.; Zhang, Y.; Wang, Z.; Xia, D.; Ye, Z.; Xu, H. Analysis of the role of mutations in the KMT2D histone lysine methyltransferase in bladder cancer. FEBS Open Bio 2019, 9, 693–706. [Google Scholar] [CrossRef]
- Sun, P.; Wu, T.; Sun, X.; Cui, Z.; Zhang, H.; Xia, Q.; Zhang, D. KMT2D inhibits the growth and metastasis of bladder Cancer cells by maintaining the tumor suppressor genes. Biomed. Pharm. 2019, 115, 108924. [Google Scholar] [CrossRef] [PubMed]
- Alam, H.; Tang, M.; Maitituoheti, M.; Dhar, S.S.; Kumar, M.; Han, C.Y.; Ambati, C.R.; Amin, S.B.; Gu, B.; Chen, T.-Y.; et al. KMT2D Deficiency Impairs Super-Enhancers to Confer a Glycolytic Vulnerability in Lung Cancer. Cancer Cell 2020, 37, 599–617.e7. [Google Scholar] [CrossRef]
- Laukhtina, E.; Lemberger, U.; Bruchbacher, A.; Ilijazi, D.; Korn, S.; Berndl, F.; D’Andrea, D.; Susani, M.; Enikeev, D.; Compérat, E.; et al. Expression Analysis and Mutational Status of Histone Methyltransferase. J. Pers. Med. 2021, 11, 1147. [Google Scholar] [CrossRef] [PubMed]
- Betge, J.; Schneider, N.I.; Harbaum, L.; Pollheimer, M.J.; Lindtner, R.A.; Kornprat, P.; Ebert, M.P.; Langner, C. MUC1, MUC2, MUC5AC, and MUC6 in colorectal cancer: Expression profiles and clinical significance. Virchows Arch. 2016, 469, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zuo, D.; Yin, L.; Lin, Y.; Liu, T.; Wang, L. Prognostic Value of MUC2 Expression in Colorectal Cancer: A Systematic Review and Meta-Analysis. Gastroenterol. Res. Pr. 2018, 2018, 6986870. [Google Scholar] [CrossRef]
- Gonul, I.I.; Cakir, A.; Sozen, S. Immunohistochemical expression profiles of MUC1 and MUC2 mucins in urothelial tumors of bladder. Indian J. Pathol. Microbiol. 2018, 61, 350–355. [Google Scholar] [CrossRef]
- Voutsadakis, I.A. Urothelial Bladder Carcinomas with High Tumor Mutation Burden Have a Better Prognosis and Targetable Molecular Defects beyond Immunotherapies. Curr. Oncol. 2022, 29, 1390–1407. [Google Scholar] [CrossRef]
- Natesan, D.; Zhang, L.; Martell, H.J.; Jindal, T.; Devine, P.; Stohr, B.; Espinosa-Mendez, C.; Grenert, J.; Van Ziffle, J.; Joseph, N.; et al. APOBEC Mutational Signature and Tumor Mutational Burden as Predictors of Clinical Outcomes and Treatment Response in Patients With Advanced Urothelial Cancer. Front. Oncol. 2022, 12, 816706. [Google Scholar] [CrossRef]
- Pena-Llopis, S.; Brugarolas, J. Simultaneous isolation of high-quality DNA, RNA, miRNA and proteins from tissues for genomic applications. Nat. Protoc. 2013, 8, 2240–2255. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Saunders, C.T.; Wong, W.S.; Swamy, S.; Becq, J.; Murray, L.J.; Cheetham, R.K. Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012, 28, 1811–1817. [Google Scholar] [CrossRef]
- Narzisi, G.; Corvelo, A.; Arora, K.; Bergmann, E.A.; Shah, M.; Musunuri, R.; Emde, A.K.; Robine, N.; Vacic, V.; Zody, M.C. Genome-wide somatic variant calling using localized colored de Bruijn graphs. Commun. Biol. 2018, 1, 20. [Google Scholar] [CrossRef] [PubMed]
- Wala, J.A.; Bandopadhayay, P.; Greenwald, N.F.; O’Rourke, R.; Sharpe, T.; Stewart, C.; Schumacher, S.; Li, Y.; Weischenfeldt, J.; Yao, X.; et al. SvABA: Genome-wide detection of structural variants and indels by local assembly. Genome Res. 2018, 28, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Seshan, V.E. FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016, 44, e131. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; Abecasis, G.R.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016, 11, 1–9. [Google Scholar] [CrossRef]
- Shihab, H.A.; Gough, J.; Mort, M.; Cooper, D.N.; Day, I.N.; Gaunt, T.R. Ranking non-synonymous single nucleotide polymorphisms based on disease concepts. Hum. Genom. 2014, 8, 11. [Google Scholar] [CrossRef]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S.; et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, J.R.; Ziman, R.; Yuen, R.K.; Feuk, L.; Scherer, S.W. The Database of Genomic Variants: A curated collection of structural variation in the human genome. Nucleic Acids Res. 2014, 42, D986–D992. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Stratton, M.R. Mutational signatures: The patterns of somatic mutations hidden in cancer genomes. Curr. Opin. Genet. Dev. 2014, 24, 52–60. [Google Scholar] [CrossRef]
- Rosenthal, R.; McGranahan, N.; Herrero, J.; Taylor, B.S.; Swanton, C. DeconstructSigs: Delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016, 17, 31. [Google Scholar] [CrossRef]


| Variable | Mean/n | SD/% |
|---|---|---|
| Age (Years) | 62.49 | 11.41 |
| BMI (kg/m2) | 26.75 | 3.99 |
| Time to follow-up (Months) | 39.91 | 19.44 |
| Sex | ||
| Women | 11 | 29.73% |
| Men | 26 | 70.73% |
| Education level | ||
| High school or less | 22 | 59.46% |
| College or vocational school | 5 | 13.51% |
| Grad school or higher | 10 | 27.03% |
| Smoking | 18 | 48.65% |
| Symptoms at diagnosis | ||
| Asymptomatic | 1 | 2.70% |
| Symptomatic 1 | 2 | 5.41% |
| Hematuria | 34 | 91.89% |
| Muscle invasion disease | ||
| Yes | 17 | 45.95% |
| Subepithelial infiltration | ||
| Yes | 22 | 59.46% |
| Histological grade | ||
| Low | 10 | 27.03% |
| High | 27 | 72.97% |
| Recurrence | 11 | 29.73% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Montiel, M.D.; Cerrato-Izaguirre, D.; Sánchez-Pérez, Y.; Diaz-Chavez, J.; Cortés-González, C.C.; Rubio, J.A.; Jiménez-Ríos, M.A.; Herrera, L.A.; Scavuzzo, A.; Meneses-García, A.; et al. Mutational Landscape of Bladder Cancer in Mexican Patients: KMT2D Mutations and chr11q15.5 Amplifications Are Associated with Muscle Invasion. Int. J. Mol. Sci. 2023, 24, 1092. https://doi.org/10.3390/ijms24021092
Pérez-Montiel MD, Cerrato-Izaguirre D, Sánchez-Pérez Y, Diaz-Chavez J, Cortés-González CC, Rubio JA, Jiménez-Ríos MA, Herrera LA, Scavuzzo A, Meneses-García A, et al. Mutational Landscape of Bladder Cancer in Mexican Patients: KMT2D Mutations and chr11q15.5 Amplifications Are Associated with Muscle Invasion. International Journal of Molecular Sciences. 2023; 24(2):1092. https://doi.org/10.3390/ijms24021092
Chicago/Turabian StylePérez-Montiel, María D., Dennis Cerrato-Izaguirre, Yesennia Sánchez-Pérez, Jose Diaz-Chavez, Carlo César Cortés-González, Jairo A. Rubio, Miguel A. Jiménez-Ríos, Luis A. Herrera, Anna Scavuzzo, Abelardo Meneses-García, and et al. 2023. "Mutational Landscape of Bladder Cancer in Mexican Patients: KMT2D Mutations and chr11q15.5 Amplifications Are Associated with Muscle Invasion" International Journal of Molecular Sciences 24, no. 2: 1092. https://doi.org/10.3390/ijms24021092
APA StylePérez-Montiel, M. D., Cerrato-Izaguirre, D., Sánchez-Pérez, Y., Diaz-Chavez, J., Cortés-González, C. C., Rubio, J. A., Jiménez-Ríos, M. A., Herrera, L. A., Scavuzzo, A., Meneses-García, A., Hernández-Martínez, R., Vaca-Paniagua, F., Ramírez, A., Orozco, A., Cantú-de-León, D., & Prada, D. (2023). Mutational Landscape of Bladder Cancer in Mexican Patients: KMT2D Mutations and chr11q15.5 Amplifications Are Associated with Muscle Invasion. International Journal of Molecular Sciences, 24(2), 1092. https://doi.org/10.3390/ijms24021092








