Bacteriobiota of the Cave Church of Sts. Peter and Paul in Serbia—Culturable and Non-Culturable Communities’ Assessment in the Bioconservation Potential of a Peculiar Fresco Painting
Abstract
1. Introduction
2. Results
2.1. Diversity of the Cave Church of Sts. Peter and Paul Microbiome
2.1.1. The Analysis of Alpha and Beta Diversities
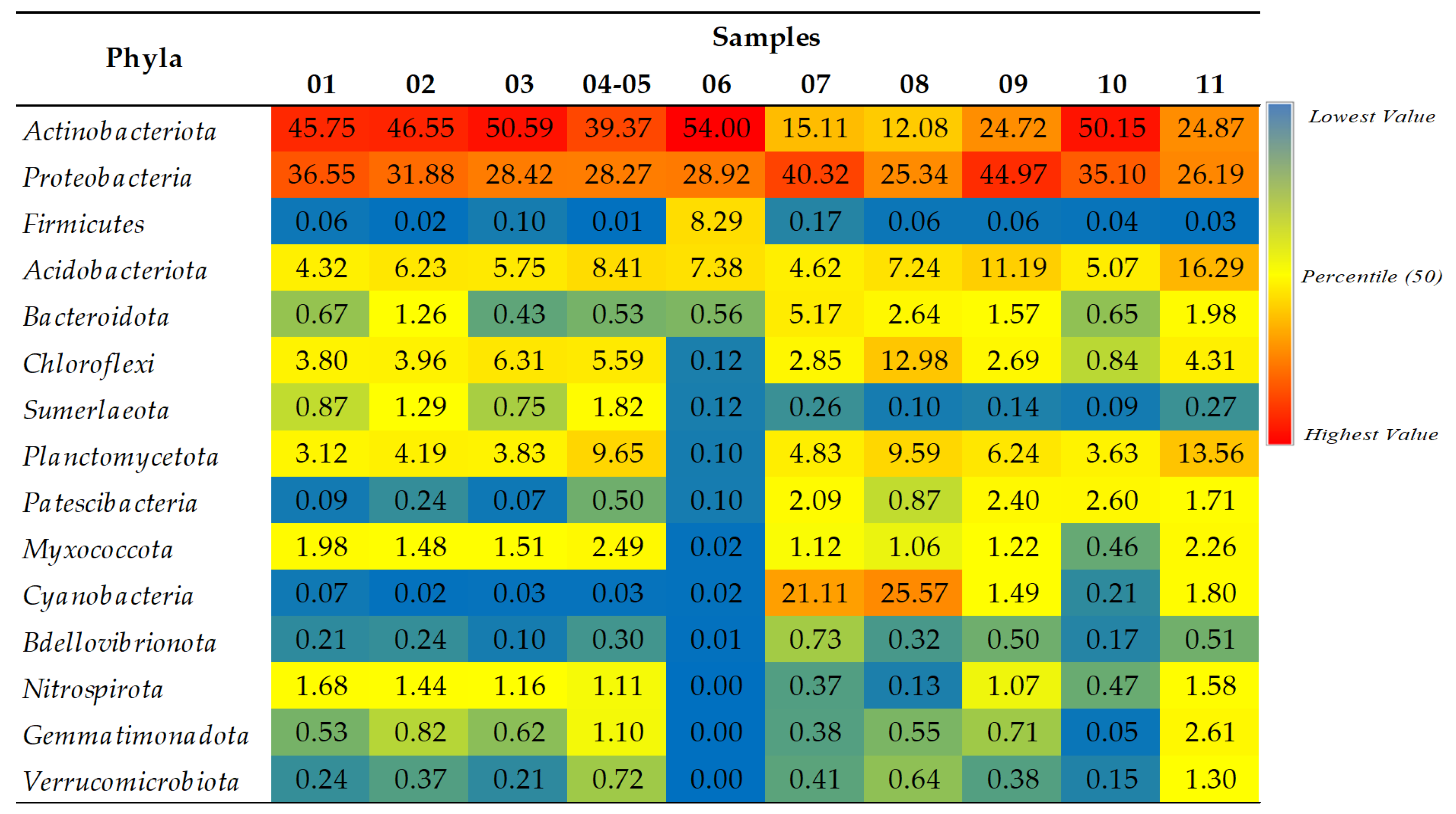
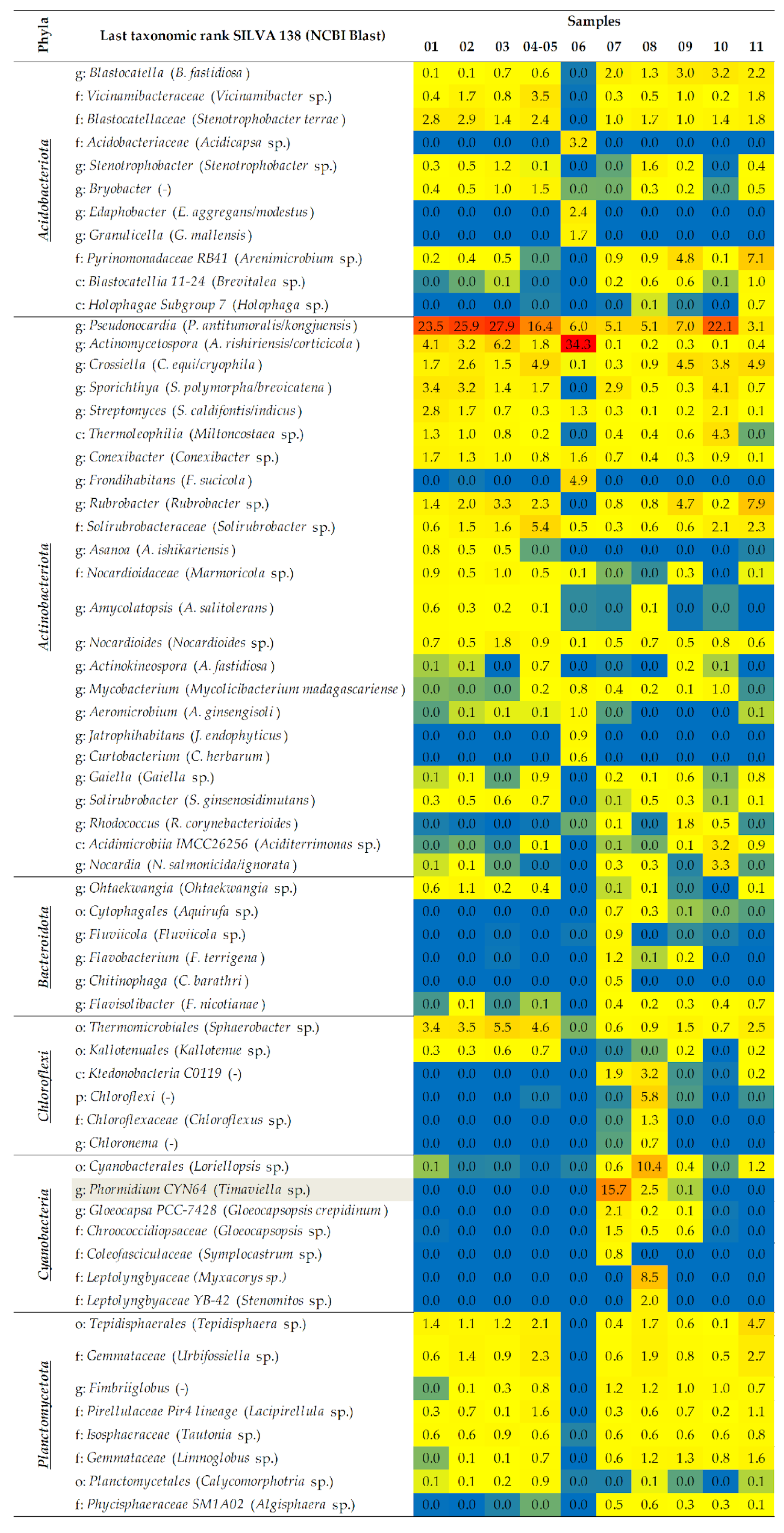
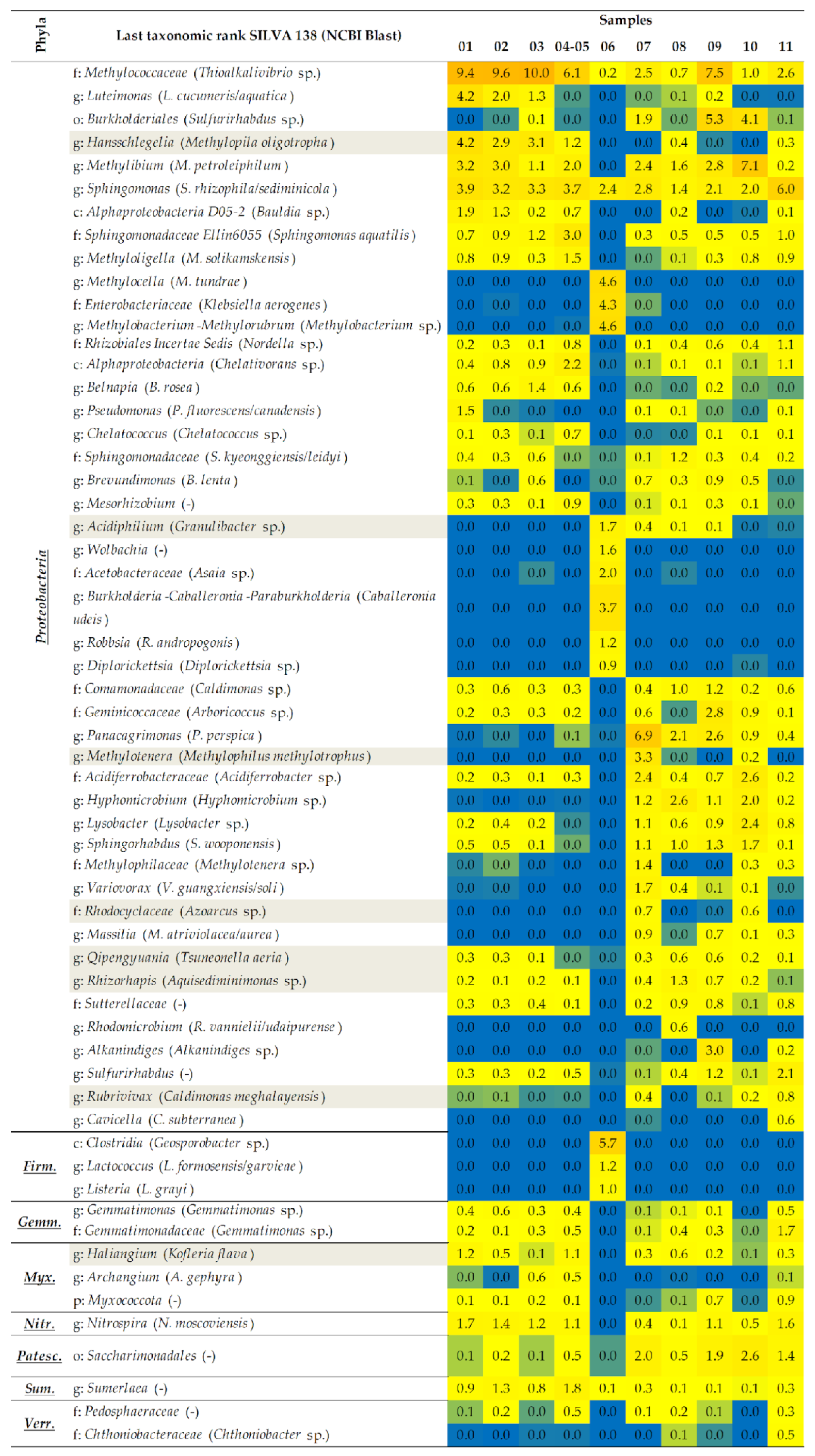
2.1.2. A Taxonomic Analysis of the Total Bacteriobiota
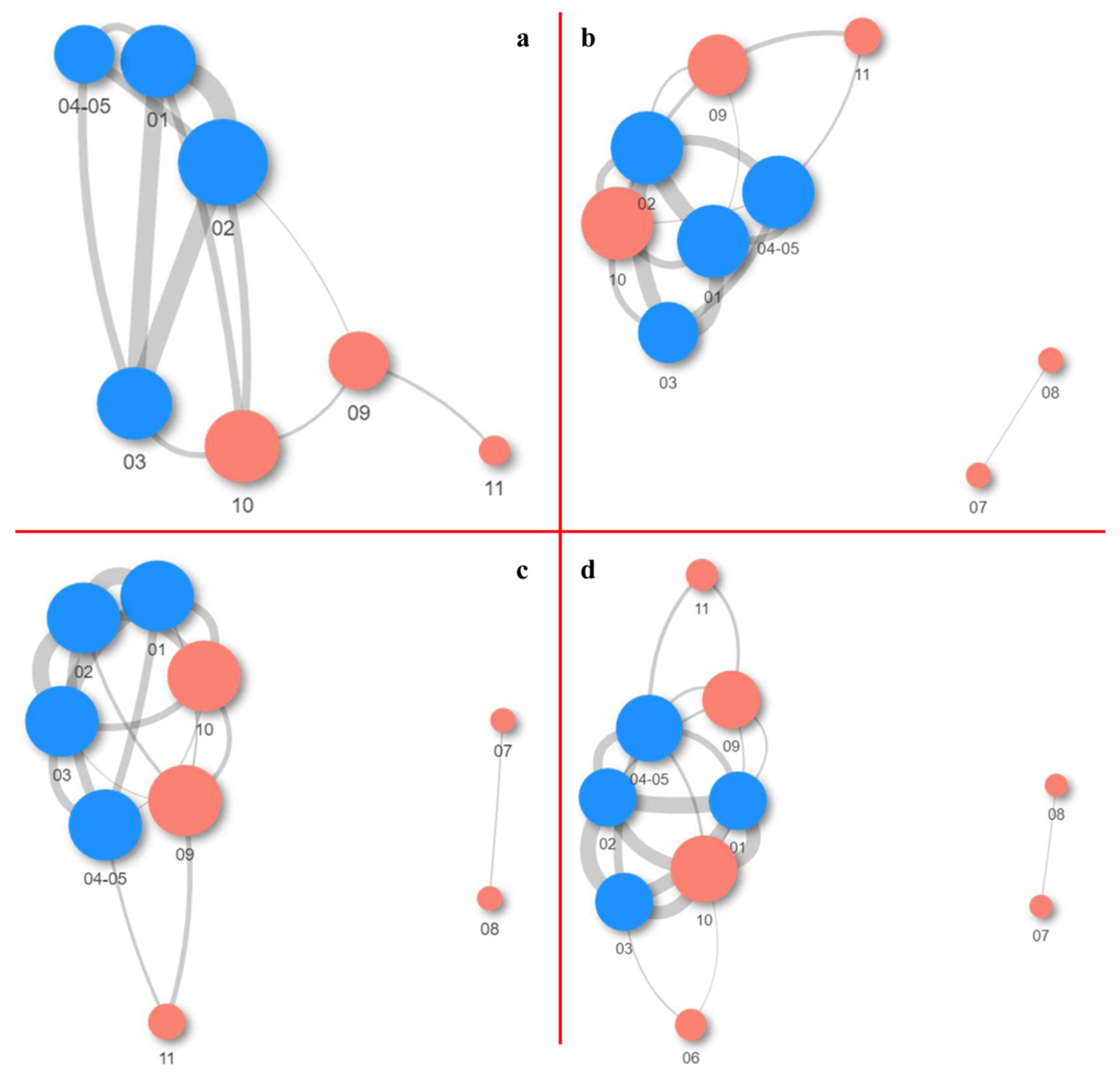
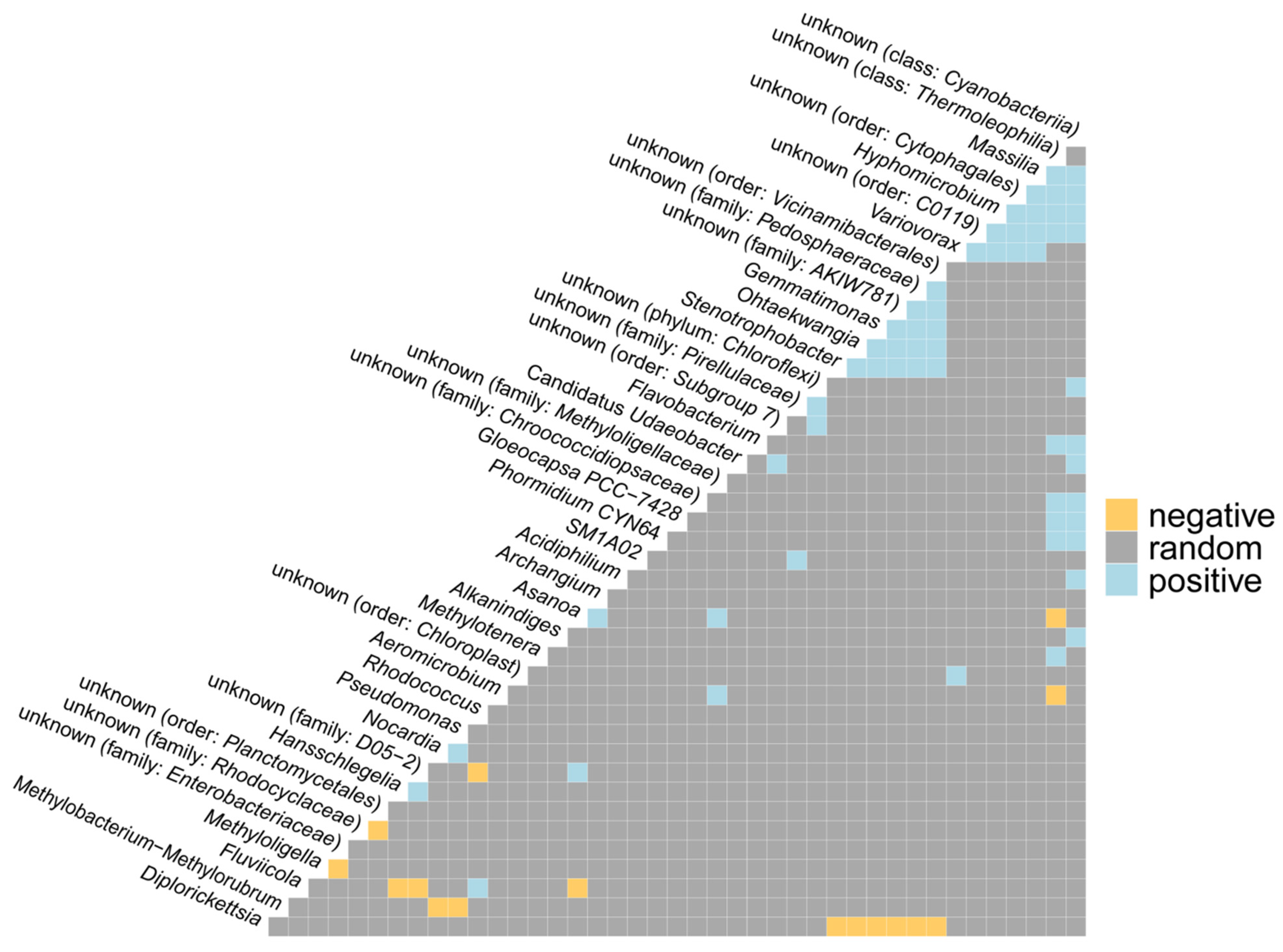
2.1.3. Co-Occurrence and Differential Abundance Analyses
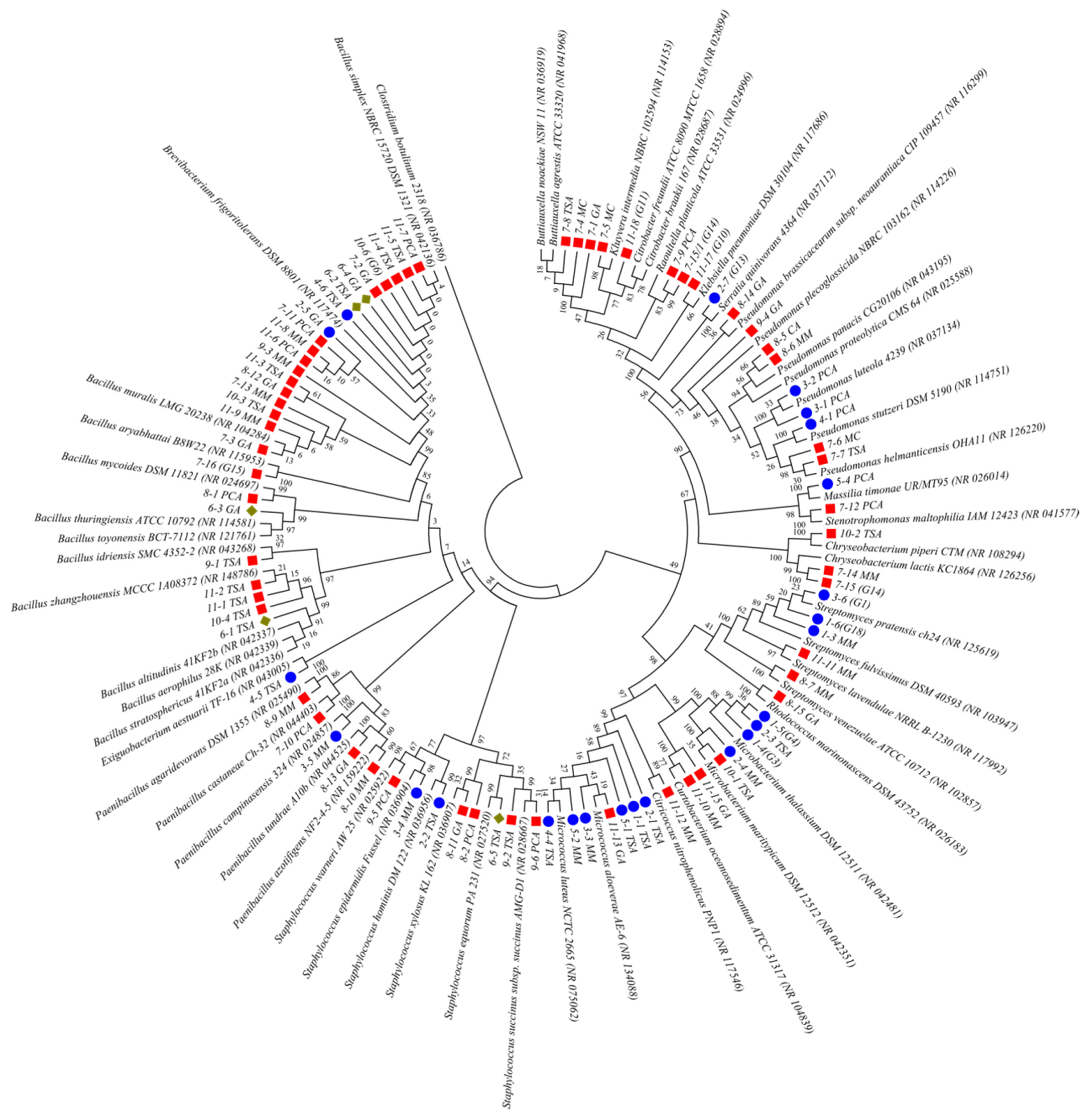
2.2. An Analysis of the Culturable Bacterial Communities
2.3. Antifungal Activity
2.4. Whole-Genome Sequencing
3. Discussion
4. Materials and Methods
4.1. Sample Location
4.2. Metabarcoding Analysis
4.2.1. DNA Extraction, Library Preparation, and NGS Sequencing
4.2.2. NGS Sequencing Data Processing and Taxonomic Annotation
4.2.3. Bioinformatic and Statistical Analyses
4.3. Culturable Bacteriobiota
4.4. Whole-Genome Sequencing
4.5. In vitro Determination of Antifungal Activity
5. Conclusions
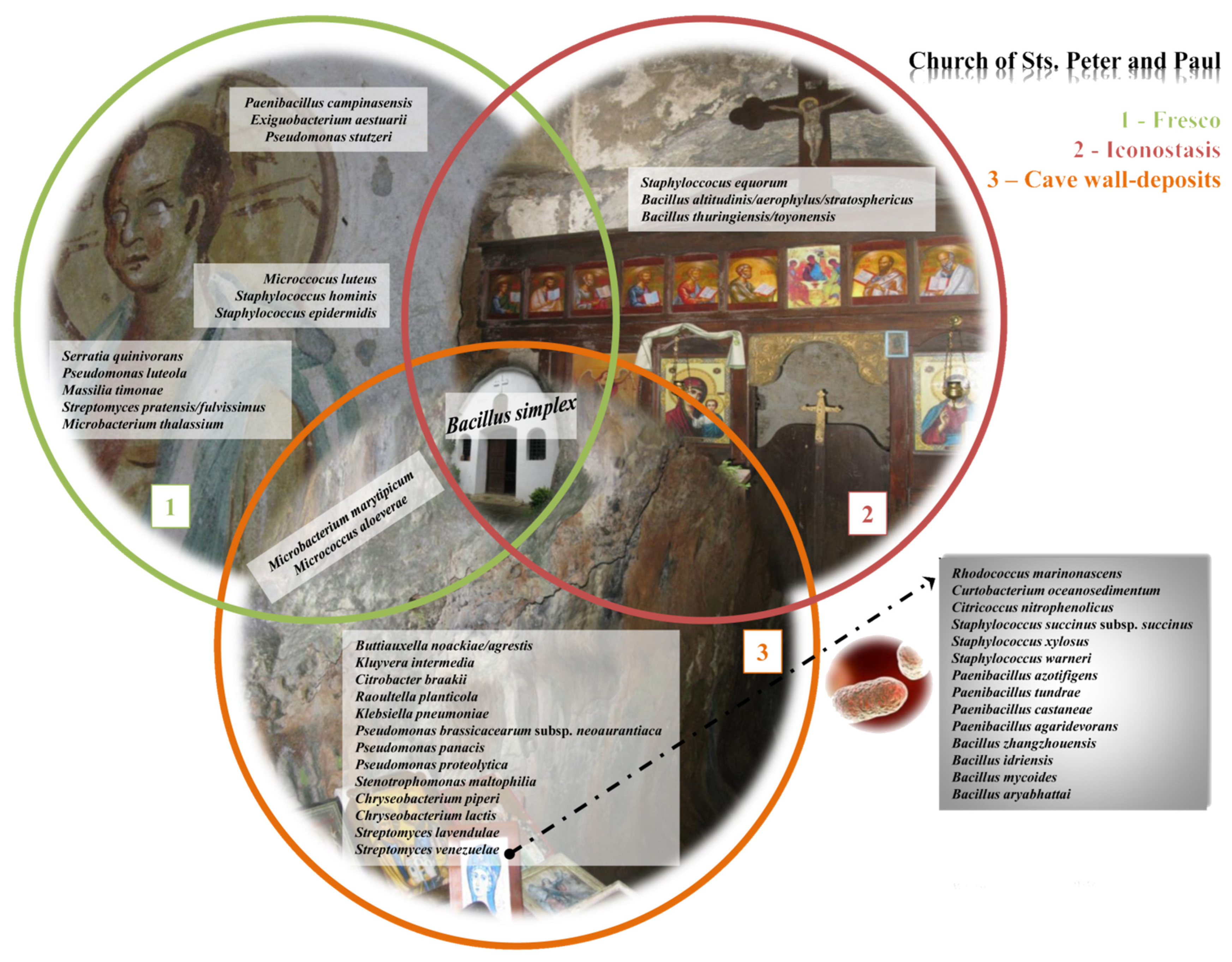
- The highest abundance of species was documented on the side walls of the church, while the lowest abundance was noted on the iconostasis.
- Samples from the fresco are mutually similar, as are the samples obtained from the side walls of the church.
- Actinobacteriota and Proteobacteria are dominant phyla in the majority of studied samples.
- The dominant bacterial taxa belonged to families Methylococcaceae, Blastocatellaceae, and Nocardioidaceae of Thermomicrobiales class.
- The most prevalent genera linked to the various kinds of wall deposits were Blastocatella and Crossiella.
- The number of positive correlations between bacterial genera far outnumbered the number of negative correlations.
- A total of 44 bacteria were identified, out of 96 obtained isolates, with the dominance of species from genus Bacillus. Bacillus simplex was the only isolated species simultaneously present in all investigated substrata within the church.
- The most promising antagonistic bacteria, with the potential to suppress growth of deteriogenic fungi and which represent excellent candidates for developing biocontrol strategies, are Streptomyces anulatus (1-3 TSA), Bacillus altitudinis (6-1 TSA), Chryseobacterium viscerum (7-15/G14), and Streptomyces sp. (11-11MM).
- Streptomyces sp. (11-11MM) represents a new species for science.
- Further research is necessary to design the most appropriate biocontrol formulation, taking into account the environment and sensitivity of the substrate, so that the desired effect of sustainable and long-term elimination of biodeteriogenic fungi can be achieved after in situ application.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bosnić, D. Amazing Serbia, 1st ed.; Mladinska knjiga: Belgrade, Serbia, 2020. [Google Scholar]
- Rakocija, M. Manastiri i Crkve Južne i Istočne Srbije; Zavod za zaštitu spomenika kulture Niš: Niš, Serbia, 2013.
- Sterflinger, K.; Piñar, G. Microbial deterioration of cultural heritage and works of art—Tilting at windmills? Appl. Microbiol. Biotechnol. 2013, 97, 9637–9646. [Google Scholar] [CrossRef]
- Beata, G. The use of-omics tools for assessing biodeterioration of cultural heritage: A review. J. Cult. Herit 2020, 45, 351–361. [Google Scholar] [CrossRef]
- Ciferri, O. Microbial degradation of paintings. Appl. Environ. Microbiol. 1999, 65, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Rölleke, S.; Witte, A.; Wanner, G.; Lubitz, W. Medieval wall paintings—A habitat for archaea: Identification of archaea by denaturing gradient gel electrophoresis (DGGE) of PCR-amplified gene fragments coding for 16S rRNA in a medieval wall painting. Int. Biodeterior. Biodegrad. 1998, 41, 85–92. [Google Scholar] [CrossRef]
- Unković, N. Diversity and Role of Micromycetes in Wall Painting Biodeterioration Process: Church of the Holy Ascension in Veliki Krčimir. Ph.D. Thesis, University of Belgrade, Faculty of Biology, Belgrade, Serbia, 2018. [Google Scholar]
- Zucconi, L.; Gagliardi, M.; Isola, D.; Onofri, S.; Andaloro, M.C.; Pelosi, C.; Selbmann, L. Biodeterioration agents dwelling in or on the wall paintings of the Holy Saviour’s cave (Vallerano, Italy). Int. Biodeterior. Biodegrad. 2012, 70, 40–46. [Google Scholar] [CrossRef]
- Saarela, M.; Alakomi, H.L.; Suihko, M.L.; Maunuksela, L.; Raaska, L.; Mattila-Sandholm, T. Heterotrophic microorganisms in air and biofilm samples from Roman catacombs, with special emphasis on actinobacteria and fungi. Int. Biodeterior. Biodegrad. 2004, 54, 27–37. [Google Scholar] [CrossRef]
- Mitchell, R.; Gu, J.D. Changes in the biofilm microflora of limestone caused by atmospheric pollutants. Int. Biodeterior. Biodegrad. 2000, 46, 299–303. [Google Scholar] [CrossRef]
- Miller, A.Z.; Leal, N.; Laiz, L.; Rogerio-Candelera, M.A.; Silva, R.J.; Dionísio, A.; Saiz-Jimenez, C. Primary bioreceptivity of limestones used in southern European monuments. Geol. Soc. Spec. Publ. 2010, 331, 79–92. [Google Scholar] [CrossRef]
- Gaylarde, C.C.; Morton, L.H.G.; Loh, K.; Shirakawa, M.A. Biodeterioration of external architectural paint films–A review. Int. Biodeterior. Biodegrad. 2011, 65, 1189–1198. [Google Scholar] [CrossRef]
- Kusumi, A.; Li, X.S.; Katayama, Y. Mycobacteria isolated from Angkor monument sandstones grow chemolithoautotrophically by oxidizing elemental sulfur. Front. Microbiol. 2011, 2, 104. [Google Scholar] [CrossRef]
- Nugari, M.P.; Pietrini, A.M.; Caneva, G.; Imperi, F.; Visca, P. Biodeterioration of mural paintings in a rocky habitat: The Crypt of the Original Sin (Matera, Italy). Int. Biodeterior. Biodegrad. 2009, 63, 705–711. [Google Scholar] [CrossRef]
- Gorbushina, A.A.; Heyrman, J.; Dornieden, T.; Gonzalez-Delvalle, M.; Krumbein, W.E.; Laiz, L.; Swings, J. Bacterial and fungal diversity and biodeterioration problems in mural painting environments of St. Martins church (Greene–Kreiensen, Germany). Int. Biodeterior. Biodegrad. 2004, 53, 13–24. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, B.; He, Z.; Yang, X. Distribution and diversity of bacteria and fungi colonization in stone monuments analyzed by high-throughput sequencing. PLoS ONE 2016, 11, e0163287. [Google Scholar] [CrossRef]
- Trovão, J.; Portugal, A.; Soares, F.; Paiva, D.S.; Mesquita, N.; Coelho, C.; Tiago, I. Fungal diversity and distribution across distinct biodeterioration phenomena in limestone walls of the old cathedral of Coimbra, UNESCO World Heritage Site. Int. Biodeterior. Biodegrad. 2019, 142, 91–102. [Google Scholar] [CrossRef]
- Paiva, D.S.; Fernandes, L.; Trovão, J.; Mesquita, N.; Tiago, I.; Portugal, A. Uncovering the Fungal Diversity Colonizing Limestone Walls of a Forgotten Monument in the Central Region of Portugal by High-Throughput Sequencing and Culture-Based Methods. Appl. Sci. 2022, 12, 10650. [Google Scholar] [CrossRef]
- Ortega-Morales, O.; Montero-Muñoz, J.L.; Neto, J.A.B.; Beech, I.B.; Sunner, J.; Gaylarde, C. Deterioration and microbial colonization of cultural heritage stone buildings in polluted and unpolluted tropical and subtropical climates: A meta-analysis. Int. Biodeterior. Biodegrad. 2019, 143, 104734. [Google Scholar] [CrossRef]
- Dziurzynski, M.; Ciuchcinski, K.; Dyda, M.; Szych, A.; Drabik, P.; Laudy, A.; Dziewit, L. Assessment of bacterial contamination of air at the museum of King John III’s palace at wilanow (Warsaw, Poland): Selection of an optimal growth medium for analyzing airborne bacteria diversity. Appl. Sci. 2020, 10, 7128. [Google Scholar] [CrossRef]
- Bosch-Roig, P.; Ranalli, G. The safety of biocleaning technologies for cultural heritage. Front. Microbiol. 2014, 5, 155. [Google Scholar] [CrossRef][Green Version]
- Caldeira, A.T. Green Mitigation Strategy for Cultural Heritage Using Bacterial Biocides. In Microorganisms in the Deterioration and Preservation of Cultural Heritage; Springer: Berlin, Germany, 2021; p. 137. [Google Scholar]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Dimkić, I.; Janakiev, T.; Petrović, M.; Degrassi, G.; Fira, D. Plant-associated Bacillus and Pseudomonas antimicrobial activities in plant disease suppression via biological control mechanisms—A review. Physiol. Mol. Plant. Pathol. 2022, 117, 101754. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, H.; Du, Y.; Tian, T.; Xiang, T.; Liu, X.; Feng, H. The community distribution of bacteria and fungi on ancient wall paintings of the Mogao Grottoes. Sci. Rep. 2015, 5, 7752. [Google Scholar] [CrossRef] [PubMed]
- Pfendler, S.; Karimi, B.; Maron, P.A.; Ciadamidaro, L.; Valot, B.; Bousta, F.; Aleya, L. Biofilm biodiversity in French and Swiss show caves using the metabarcoding approach: First data. Sci. Total Environ. 2018, 615, 1207–1217. [Google Scholar] [CrossRef] [PubMed]
- Cennamo, P.; De Luca, D.A. Metabarcoding approach for the study of biodeterioration of ancient wall paintings in an Italian cave. In Proceedings of the Journal of Physics: Conference Series, Volume 2204, International Conference on Metrology for Archaeology and Cultural Heritage, Milan, Italy, 20–22 October 2021. [Google Scholar]
- Alaoui-Sosse, B.; Ozaki, S.; Barriquand, L.; De Luca, D.; Cennamo, P.; Valot, B.; Pfendler, S. Assessment of microbial communities colonizing the Azé prehistoric cave. J. Cult. Herit 2023, 59, 1–9. [Google Scholar] [CrossRef]
- Ljaljević Grbić, M.; Dimkić, I.; Savković, Ž.; Stupar, M.; Knežević, A.; Jelikić, A.; Unković, N. Mycobiome diversity of the Cave Church of Sts. Peter and Paul in Serbia—Risk assessment implication for the conservation of rare cavern habitat housing a peculiar fresco. J. Fungi 2022, 8, 1263. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M. Analysis of bacterial communities and characterization of antimicrobial strains from cave microbiota. Braz J. Microbiol. 2018, 49, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Lopez, C.; Rodriguez-Navarro, C.; Piñar, G.; Carrillo-Rosúa, F.J.; Rodriguez-Gallego, M.; Gonzalez-Muñoz, M.T. Consolidation of degraded ornamental porous limestone stone by calcium carbonate precipitation induced by the microbiota inhabiting the stone. Chemosphere 2007, 68, 1929–1936. [Google Scholar] [CrossRef] [PubMed]
- Stomeo, F.; Portillo, M.C.; Gonzalez, J.M.; Laiz, L.; Sáiz-Jiménez, C. Pseudonocardia in white colonizations in two caves with Paleolithic paintings. Int. Biodeterior. Biodegrad. 2008, 62, 483–486. [Google Scholar] [CrossRef]
- Schabereiter-Gurtner, C.; Saiz-Jimenez, C.; Piñar, G.; Lubitz, W.; Rölleke, S. Altamira cave Paleolithic paintings harbor partly unknown bacterial communities. FEMS Microbiol. Lett. 2002, 211, 7–11. [Google Scholar] [CrossRef]
- Heyrman, J.; Mergaert, J.; Denys, R.; Swings, J. The use of fatty acid methyl ester analysis (FAME) for the identification of heterotrophic bacteria present on three mural paintings showing severe damage by microorganisms. FEMS Microbiol. Lett. 1999, 181, 55–62. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Wu, F.; Ma, W.; Zhang, Y.; Gu, J.D.; Duan, Y.; Li, S.W. Insights into the bacterial and fungal communities and microbiome that causes a microbe outbreak on ancient wall paintings in the Maijishan Grottoes. Int. Biodeterior. Biodegrad. 2021, 163, 105250. [Google Scholar] [CrossRef]
- Schabereiter-Gurtner, C.; Piñar, G.; Lubitz, W.; Rölleke, S. An advanced molecular strategy to identify bacterial communities on art objects. J. Microbiol. Methods 2001, 45, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Haliem, M.E.F.; Sakr, A.A.; Ali, M.F.; Ghaly, M.F.; Sohlenkamp, C. Characterization of Streptomyces isolates causing colour changes of mural paintings in ancient Egyptian tombs. Microbiol. Res. 2013, 168, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Sorokovikova, E.; Belykh, O.; Krasnopeev, A.; Potapov, S.; Tikhonova, I.; Khanaev, I.; Timoshkin, O. First data on cyanobacterial biodiversity in benthic biofilms during mass mortality of endemic sponges in Lake Baikal. J. Great Lakes Res. 2020, 46, 75–84. [Google Scholar] [CrossRef]
- Sciuto, K.; Moschin, E.; Moro, I. Cryptic cyanobacterial diversity in the Giant Cave (Trieste, Italy): The new genus Timaviella (Leptolyngbyaceae). Cryptogam. Algol. 2017, 38, 285–323. [Google Scholar] [CrossRef]
- Vinogradova, O.M.; Mikhailyuk, T.I. On the taxonomy and nomenclature of some terrestrial taxa of plectonema s. I.(cyanophyceae). 1. The case of Plectonema edaphicum. Int. J. Algae 2018, 20, 211–224. [Google Scholar] [CrossRef]
- Mai, T.; Johansen, J.R.; Pietrasiak, N.; Bohunická, M.; Martin, M.P. Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 2018, 365, 1–59. [Google Scholar] [CrossRef]
- Mulec, J.; Kosi, G.; Vrhovšek, D. Characterization of cave aerophytic algal communities and effects of irradiance levels on production of pigments. J. Caves Karst Stud. 2008, 70, 3–12. [Google Scholar]
- Martinez, A.; Asencio, A.D. Distribution of Cyanobacteria at the Gelada Cave (Spain) by physical parameters. J. Caves Karst Stud. 2010, 72, 11–20. [Google Scholar] [CrossRef]
- Lamprinou, V.; Pantazidou, A.; Papadogiannaki, G.; Radea, C.; Economou-Amilli, A. Cyanobacteria and associated invertebrates in Leontari Cave, Attica (Greece). Fottea 2009, 9, 155–164. [Google Scholar] [CrossRef]
- Whitton, B.A.; Potts, M. Ecology of Cyanobacteria II, 1st ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 1–13. [Google Scholar]
- Keshari, N.; Adhikary, S.P. Diversity of cyanobacteria on stone monuments and building facades of India and their phylogenetic analysis. Int. Biodeterior. Biodegrad. 2014, 90, 45–51. [Google Scholar] [CrossRef]
- Cappitelli, F.; Abbruscato, P.; Foladori, P.; Zanardini, E.; Ranalli, G.; Principi, P.; Sorlini, C. Detection and elimination of cyanobacteria from frescoes: The case of the St. Brizio Chapel (Orvieto Cathedral, Italy). Microb. Ecol. 2009, 57, 633–639. [Google Scholar] [CrossRef]
- Albertano, P.; Bruno, L.; Bellezza, S. New strategies for the monitoring and control of cyanobacterial films on valuable lithic faces. Plant. Biosyst. 2005, 139, 311–322. [Google Scholar] [CrossRef]
- Popović, S. Diversity of Aerophytic Cyanobacteria and Algae in Biofilm from Selected Caves in Serbia. Ph.D. Thesis, University of Belgrade, Faculty of Biology, Belgrade, Serbia, 2018. [Google Scholar]
- Ortega-Calvo, J.J.; Ariño, X.; Hernandez-Marine, M.; Saiz-Jimenez, C. Factors affecting the weathering and colonization of monuments by phototrophic microorganisms. Sci. Total Environ. 1995, 167, 329–341. [Google Scholar] [CrossRef]
- Golubic, S.; Friedmann, E.I.; Schneider, J. The lithobiontic ecological niche, with special reference to microorganisms. J. Sediment. Res. 1981, 51, 475–478. [Google Scholar] [CrossRef]
- Pepe, O.; Sannino, L.; Palomba, S.; Anastasio, M.; Blaiotta, G.; Villani, F.; Moschetti, G. Heterotrophic microorganisms in deteriorated medieval wall paintings in southern Italian churches. Microbiol. Res. 2010, 165, 21–32. [Google Scholar] [CrossRef]
- Capodicasa, S.; Fedi, S.; Porcelli, A.M.; Zannoni, D. The microbial community dwelling on a biodeteriorated 16th century painting. Int. Biodeterior. Biodegrad. 2010, 64, 727–733. [Google Scholar] [CrossRef]
- Fajardo-Cavazos, P.; Nicholson, W. Bacillus endospores isolated from granite: Close molecular relationships to globally distributed Bacillus spp. from endolithic and extreme environments. Appl. Environ. Microbiol. 2006, 72, 2856–2863. [Google Scholar] [CrossRef]
- Lin, L.; Kan, X.; Yan, H.; Wang, D. Characterization of extracellular cellulose-degrading enzymes from Bacillus thuringiensis strains. Electron. J. Biotechnol. 2012, 15, 2. [Google Scholar] [CrossRef]
- Garza-González, E.; Morfin-Otero, R.; Martínez-Vázquez, M.A.; Gonzalez-Diaz, E.; González-Santiago, O.; Rodríguez-Noriega, E. Microbiological and molecular characterization of human clinical isolates of Staphylococcus cohnii, Staphylococcus hominis, and Staphylococcus sciuri. Scand. J. Infect. Dis. 2011, 43, 930–936. [Google Scholar] [CrossRef]
- Mudgil, D.; Baskar, S.; Baskar, R.; Paul, D.; Shouche, Y.S. Biomineralization potential of Bacillus subtilis, Rummeliibacillus stabekisii and Staphylococcus epidermidis strains in vitro isolated from Speleothems, Khasi Hill Caves, Meghalaya, India. GeoMicrobiol. J. 2018, 35, 675–694. [Google Scholar] [CrossRef]
- Steinman, R.; Zuniga, R. Towards the isolation and identification of antimicrobial compounds produced by Curtobacterium oceanosedimentum and the initial screening of compounds isolated from Proteus penneri. In Proceedings of the JCCC Science and Mathematics Poster Symposium, Johnson County Community College, Overland Park, KS, USA, 28 April 2017. [Google Scholar]
- Faramarzi, M.A.; Fazeli, M.; Yazdi, M.T.; Adrangi, S.; Al-Ahmadi, K.J.; Tasharrofi, N.; Mohseni, F.A. Optimization of cultural conditions for production of chitinase by a soil isolate of Massilia timonae. Biotechnology 2009, 8, 93–99. [Google Scholar] [CrossRef]
- Lee, B.; Farag, M.A.; Park, H.B.; Kloepper, J.W.; Lee, S.H.; Ryu, C.M. Induced resistance by a long-chain bacterial volatile: Elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS ONE 2012, 7, e48744. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Illueca, F.; Vila-Donat, P.; Calpe, J.; Luz, C.; Meca, G.; Quiles, J.M. Antifungal activity of biocontrol agents in vitro and potential application to reduce mycotoxins (aflatoxin B1 and ochratoxin A). Toxins 2021, 13, 752. [Google Scholar] [CrossRef] [PubMed]
- Sakr, A.A.; Ghaly, M.F.; Edwards, H.G.M.; Ali, M.F.; Abdel-Haliem, M.E. Involvement of Streptomyces in the deterioration of cultural heritage materials through biomineralization and bio-pigment production pathways: A review. Geomicrobiol. J. 2020, 37, 653–662. [Google Scholar] [CrossRef]
- Pepe, O.; Palomba, S.; Sannino, L.; Blaiotta, G.; Ventorino, V.; Moschetti, G.; Villani, F. Characterization in the archaeological excavation site of heterotrophic bacteria and fungi of deteriorated wall painting of Herculaneum in Italy. J. Environ. Biol. 2011, 32, 241–250. [Google Scholar]
- Karpovich-Tate, N.; Rebrikova, N.L. Microbial communities on damaged frescoes and building materials in the cathedral of the nativity of the virgin in the Pafnutiy-Borovsky monastery, Russia. Int. Biodeterior. 1991, 27, 281–296. [Google Scholar] [CrossRef]
- Roig, P.B.; Ros, J.L.R.; Estellés, R.M. Biocleaning of nitrate alterations on wall paintings by Pseudomonas stutzeri. Int. Biodeterior. Biodegrad. 2013, 84, 266–274. [Google Scholar] [CrossRef]
- Ranalli, G.; Zanardini, E. Biocleaning on Cultural Heritage: New frontiers of microbial biotechnologies. J. Appl. Microbiol. 2021, 131, 583–603. [Google Scholar] [CrossRef]
- Enyedi, N.T.; Makk, J.; Kótai, L.; Berényi, B.; Klébert, S.; Sebestyén, Z.; Németh, P. Cave bacteria-induced amorphous calcium carbonate formation. Sci. Rep. 2020, 10, 8696. [Google Scholar] [CrossRef]
- Rautela, R.; Rawat, S. Analysis and optimization of process parameters for in vitro biomineralization of CaCO3 by Klebsiella pneumoniae, isolated from a stalactite from the Sahastradhara cave. RSC Adv. 2020, 10, 8470–8479. [Google Scholar] [CrossRef] [PubMed]
- Müller, H.E.; Brenner, D.J.; Fanning, G.R.; Grimont, P.A.; Kämpfer, P. Emended description of Buttiauxella agrestis with recognition of six new species of Buttiauxella and two new species of Kluyvera: Buttiauxella ferragutiae sp. nov., Buttiauxella gaviniae sp. nov., Buttiauxella brennerae sp. nov., Buttiauxella izardii sp. nov., Buttiauxella noackiae sp. nov., Buttiauxella warmboldiae sp. nov., Kluyvera cochleae sp. nov., and Kluyvera georgiana sp. nov. Int. J. Syst. Evol. Microbiol. 1996, 46, 50–63. [Google Scholar] [CrossRef]
- Ikeda, H.; Takada, Y.; Pang, C.H.; Tanaka, H.; Omura, S. Transposon mutagenesis by Tn4560 and applications with avermectin-producing Streptomyces avermitilis. J. Bacteriol. 1993, 175, 2077–2082. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Couillerot, O.; Loqman, S.; Toribio, A.; Hubert, J.; Gandner, L.; Nuzillard, J.M.; Renault, J.H. Purification of antibiotics from the biocontrol agent Streptomyces anulatus S37 by centrifugal partition chromatography. J. Chromatogr. B 2014, 944, 30–34. [Google Scholar] [CrossRef]
- Praveen, V.; Tripathi, C.K.M.; Bihari, V. Studies on optimum fermentation conditions for actinomycin-D production by two new strains of Streptomyces spp. Med. Chem. Res. 2008, 17, 114–122. [Google Scholar] [CrossRef]
- Laczeski, M.E.; Onetto, A.L.; Cortese, I.J.; Mallozzi, G.Y.; Castrillo, M.L.; Bich, G.A.; Otegui, M.B. Isolation and selection of endophytic spore-forming bacteria with plant growth promoting properties isolated from Ilex paraguariensis St. Hil. (Yerba mate). An. Acad. Bras. Ciênc 2022, 92. [Google Scholar] [CrossRef]
- Lu, X.; Zhou, D.; Chen, X.; Zhang, J.; Huang, H.; Wei, L. Isolation and characterization of Bacillus altitudinis JSCX-1 as a new potential biocontrol agent against Phytophthora sojae in soybean [Glycine max (L.) Merr.]. Plant. Soil. 2017, 416, 53–66. [Google Scholar] [CrossRef]
- Goswami, M.; Deka, S. Biosurfactant production by a rhizosphere bacteria Bacillus altitudinis MS16 and its promising emulsification and antifungal activity. Colloids Surf. B 2019, 178, 285–296. [Google Scholar] [CrossRef]
- Jeong, J.J.; Lee, Y.J.; Pathiraja, D.; Park, B.; Choi, I.G.; Kim, K.D. Draft genome sequences of Chryseobacterium lactis NCTC11390T isolated from milk, Chryseobacterium oncorhynchi 701B-08T from rainbow trout, and Chryseobacterium viscerum 687B-08T from diseased fish. Genome Announc. 2018, 6, e00628-18. [Google Scholar] [CrossRef]
- Kyndt, J.A.; Moore, T.C. Draft whole-genome sequence of a novel Chryseobacterium viscerum strain isolated from fresh water at dripping springs, New Mexico. Microbiol. Resour. Announc. 2019, 8, e01155-19. [Google Scholar] [CrossRef]
- Joseph, E. Microorganisms in the Deterioration and Preservation of Cultural Heritage, 1st ed.; Springer Nature: Cham, Switzerland, 2021. [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge–accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef] [PubMed]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Pavoine, S.; Dufour, A.B.; Chessel, D. From dissimilarities among species to dissimilarities among communities: A double principal coordinate analysis. J. Theor. Biol. 2004, 228, 523–537. [Google Scholar] [CrossRef]
- Wright, E.S. DECIPHER: Harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinform. 2015, 16, 322. [Google Scholar] [CrossRef]
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Griffith, D.M.; Veech, J.A.; Marsh, C.J. Cooccur: Probabilistic species co-occurrence analysis in R. J. Stat. Softw. 2016, 69, 1–17. [Google Scholar] [CrossRef]
- Foster, Z.S.; Sharpton, T.J.; Grünwald, N.J. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 2017, 13, e1005404. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef]
- Dimkić, I.; Živković, S.; Berić, T.; Ivanović, Ž.; Gavrilović, V.; Stanković, S.; Fira, D. Characterization and evaluation of two Bacillus strains, SS-12.6 and SS-13.1, as potential agents for the control of phytopathogenic bacteria and fungi. Biol. Control. 2013, 65, 312–321. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Pevzner, P.A. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Tanizawa, Y.; Fujisawa, T.; Kaminuma, E.; Nakamura, Y.; Arita, M. DFAST and DAGA: Web-based integrated genome annotation tools and resources. Biosci. Microbiota Food Health 2016, 35, 173–184. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Araujo, F.A.; Barh, D.; Silva, A.; Guimaraes, L.; Ramos, R.T.J. GO FEAT: A rapid web-based functional annotation tool for genomic and transcriptomic data. Sci. Rep. 2018, 8, 1794. [Google Scholar] [CrossRef] [PubMed]
- Asnicar, F.; Thomas, A.M.; Beghini, F.; Mengoni, C.; Manara, S.; Manghi, P.; Zhu, Q.; Bolzan, M.; Cumbo, F.; May, U.; et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020, 11, 2500. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Dimkić, I.; Berić, T.; Stević, T.; Pavlović, S.; Šavikin, K.; Fira, D.; Stanković, S. Additive and synergistic effects of Bacillus spp. isolates and essential oils on the control of phytopathogenic and saprophytic fungi from medicinal plants and marigold seeds. Biol. Control. 2015, 87, 6–13. [Google Scholar] [CrossRef]
- Korsten, L.; De Jager, E.S. Mode of action of Bacillus subtilis for control of avocado postharvest pathogens. S. Afr. Avocado Growers’ Assoc. Yearb. 1995, 18, 124–130. [Google Scholar]

| Indices | Description | Mean | St. Dev. | Min. | Max. | Test Statistic | Kruskal–Wallis Sign. (P < 0.05) |
|---|---|---|---|---|---|---|---|
| Phylum | |||||||
| OBS | Fresco | 19.0 | 0.8 | 18.0 | 20.0 | 2.927 | 0.087 |
| Around Fresco | 25.3 | 4.5 | 17.0 | 30.0 | |||
| Chao1 | Fresco | 20.0 | 1.8 | 18.0 | 22.0 | 2.909 | 0.088 |
| Around Fresco | 25.4 | 4.5 | 17.0 | 30.0 | |||
| ACE | Fresco | 21.4 | 2.2 | 19.1 | 24.1 | 2.909 | 0.088 |
| Around Fresco | 25.8 | 4.7 | 17.0 | 30.3 | |||
| Shannon | Fresco | 1.5 | 0.1 | 1.4 | 1.7 | 0.567 | 0.451 |
| Around Fresco | 1.7 | 0.3 | 1.2 | 2.0 | |||
| Gini–Simpson | Fresco | 0.7 | 0.0 | 0.7 | 0.7 | 0.214 | 0.643 |
| Around Fresco | 0.7 | 0.1 | 0.6 | 0.8 | |||
| Family | |||||||
| OBS | Fresco | 146.75 | 10.14 | 137.00 | 161.00 | 2.909 | 0.088 |
| Around Fresco | 204.50 | 63.89 | 92.00 | 259.00 | |||
| Chao1 | Fresco | 154.33 | 10.29 | 146.20 | 169.30 | 2.909 | 0.088 |
| Around Fresco | 210.35 | 68.07 | 92.70 | 280.00 | |||
| ACE | Fresco | 153.05 | 10.34 | 142.60 | 167.30 | 2.909 | 0.088 |
| Around Fresco | 211.05 | 67.50 | 93.50 | 277.60 | |||
| Shannon | Fresco | 3.10 | 0.22 | 2.90 | 3.40 | 2.283 | 0.131 |
| Around Fresco | 3.52 | 0.61 | 2.40 | 3.90 | |||
| Gini–Simpson | Fresco | 0.90 | 0.00 | 0.90 | 0.90 | 2.000 | 0.157 |
| Around Fresco | 0.95 | 0.08 | 0.80 | 1.00 | |||
| Genus | |||||||
| OBS | Fresco | 223.25 | 8.66 | 215.00 | 233.00 | 2.909 | 0.088 |
| Around Fresco | 321.50 | 109.80 | 136.00 | 428.00 | |||
| Chao1 | Fresco | 230.40 | 11.83 | 220.00 | 242.00 | 2.909 | 0.088 |
| Around Fresco | 329.28 | 113.77 | 138.10 | 442.00 | |||
| ACE | Fresco | 231.18 | 10.15 | 221.90 | 240.40 | 2.909 | 0.088 |
| Around Fresco | 329.55 | 113.73 | 137.90 | 444.60 | |||
| Shannon | Fresco | 3.58 | 0.22 | 3.40 | 3.90 | 2.297 | 0.130 |
| Around Fresco | 3.98 | 0.61 | 2.90 | 4.40 | |||
| Gini–Simpson | Fresco | 0.93 | 0.05 | 0.90 | 1.00 | 1.500 | 0.221 |
| Around Fresco | 0.97 | 0.05 | 0.90 | 1.00 | |||
| ASV | |||||||
| OBS | Fresco | 735.00 | 47.63 | 689.00 | 790.00 | 0.727 | 0.394 |
| Around Fresco | 1209.17 | 616.55 | 254.00 | 1757.00 | |||
| Chao1 | Fresco | 755.53 | 49.62 | 710.20 | 814.60 | 0.727 | 0.394 |
| Around Fresco | 1253.83 | 644.91 | 254.30 | 1847.10 | |||
| ACE | Fresco | 752.38 | 47.74 | 712.50 | 808.10 | 0.727 | 0.394 |
| Around Fresco | 1246.60 | 638.87 | 254.70 | 1822.80 | |||
| Shannon | Fresco | 5.00 | 0.24 | 4.70 | 5.30 | 0.732 | 0.392 |
| Around Fresco | 5.23 | 1.04 | 3.40 | 6.30 | |||
| Gini–Simpson | Fresco | 1.00 | 0.00 | 1.00 | 1.00 | 0.667 | 0.414 |
| Around Fresco | 0.98 | 0.04 | 0.90 | 1.00 | |||
| Taxa | Taxa Rank | Fresco | Around Fresco | Test Statistic | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 02 | 03 | 04–05 | 06 | 07 | 08 | 09 | 10 | 11 | lfcSE | stat | Pvalue | Padj | ||
| Solimonadaceae | Family | 6 | 19 | 0 | 65 | 4 | 10111 | 2852 | 3491 | 1121 | 509 | 1.376 | 3.669 | 0.0002 | 0.0048 |
| Chroococcidiopsaceae | 5 | 0 | 0 | 0 | 0 | 4994 | 870 | 887 | 14 | 0 | 2.191 | 3.664 | 0.0002 | 0.0048 | |
| Hyphomicrobiaceae | 6 | 0 | 4 | 6 | 0 | 1701 | 3133 | 1378 | 2502 | 463 | 1.357 | 5.051 | 0.0000 | 0.0000 | |
| Rhodocyclaceae | 0 | 0 | 0 | 0 | 0 | 951 | 0 | 12 | 779 | 0 | 2.604 | 8.569 | 0.0000 | 0.0000 | |
| Intrasporangiaceae | 0 | 0 | 4 | 0 | 9 | 66 | 30 | 376 | 62 | 53 | 1.431 | 3.579 | 0.0003 | 0.0057 | |
| Oxalobacteraceae | 0 | 0 | 4 | 3 | 0 | 1377 | 49 | 877 | 280 | 414 | 1.511 | 4.125 | 0.0000 | 0.0011 | |
| Microcystaceae | 0 | 0 | 0 | 0 | 0 | 33 | 437 | 56 | 0 | 333 | 2.157 | 3.608 | 0.0003 | 0.0056 | |
| Chthonomonadaceae | 0 | 2 | 0 | 0 | 0 | 44 | 329 | 38 | 31 | 27 | 1.769 | 3.120 | 0.0018 | 0.0194 | |
| env.OPS 17 | 0 | 0 | 0 | 0 | 0 | 334 | 321 | 317 | 17 | 423 | 1.768 | 4.774 | 0.0000 | 0.0001 | |
| Cellvibrionaceae | 0 | 0 | 0 | 0 | 3 | 101 | 40 | 15 | 71 | 27 | 1.511 | 4.069 | 0.0000 | 0.0011 | |
| B1-7BS | 14 | 34 | 41 | 75 | 0 | 0 | 0 | 0 | 0 | 0 | 1.721 | −2.977 | 0.0029 | 0.0288 | |
| Acidobacteriae | 0 | 0 | 0 | 0 | 0 | 56 | 149 | 154 | 26 | 291 | 1.724 | 4.287 | 0.0000 | 0.0006 | |
| Saprospiraceae | 0 | 0 | 0 | 0 | 0 | 79 | 146 | 79 | 15 | 0 | 2.106 | 3.048 | 0.0023 | 0.0236 | |
| Micavibrionaceae | 0 | 0 | 0 | 0 | 0 | 4 | 23 | 138 | 7 | 19 | 1.940 | 2.950 | 0.0032 | 0.0308 | |
| Fimbriimonadaceae | 0 | 0 | 0 | 0 | 6 | 22 | 29 | 6 | 14 | 20 | 1.610 | 3.260 | 0.0011 | 0.0142 | |
| Phormidium CYN64 | Genus | 0 | 0 | 0 | 0 | 0 | 21581 | 3008 | 79 | 0 | 0 | 2.834 | 9.142 | 0.0000 | 0.0000 |
| Panacagrimonas | 6 | 19 | 0 | 65 | 4 | 9544 | 2565 | 3201 | 1089 | 509 | 1.371 | 3.612 | 0.0003 | 0.0056 | |
| Gloeocapsa PCC-7428 | 0 | 0 | 0 | 0 | 0 | 2921 | 198 | 156 | 8 | 0 | 2.354 | 9.806 | 0.0000 | 0.0000 | |
| Hyphomicrobium | 6 | 0 | 4 | 6 | 0 | 1692 | 3075 | 1352 | 2496 | 303 | 1.374 | 4.961 | 0.0000 | 0.0000 | |
| Asanoa | 1260 | 509 | 605 | 43 | 0 | 0 | 0 | 0 | 0 | 17 | 1.723 | −3.203 | 0.0014 | 0.0166 | |
| Rhodococcus | 16 | 5 | 0 | 6 | 20 | 180 | 12 | 2212 | 601 | 31 | 1.398 | 3.329 | 0.0009 | 0.0127 | |
| Afipia | 0 | 0 | 0 | 0 | 0 | 370 | 52 | 21 | 352 | 0 | 2.159 | 3.521 | 0.0004 | 0.0066 | |
| Knoellia | 0 | 0 | 0 | 0 | 0 | 66 | 0 | 342 | 62 | 34 | 2.132 | 3.315 | 0.0009 | 0.0130 | |
| Massilia | 0 | 0 | 0 | 0 | 0 | 1195 | 36 | 864 | 150 | 348 | 1.834 | 5.058 | 0.0000 | 0.0000 | |
| Chalicogloea CCALA 975 | 0 | 0 | 0 | 0 | 0 | 33 | 422 | 56 | 0 | 333 | 2.154 | 3.599 | 0.0003 | 0.0056 | |
| Chthonomonas | 0 | 2 | 0 | 0 | 0 | 44 | 329 | 38 | 31 | 27 | 1.769 | 3.120 | 0.0018 | 0.0194 | |
| Plantactinospora | 183 | 75 | 121 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.054 | −3.015 | 0.0026 | 0.0259 | |
| Cellvibrio | 0 | 0 | 0 | 0 | 3 | 101 | 40 | 15 | 71 | 27 | 1.511 | 4.069 | 0.0000 | 0.0011 | |
| Pseudofulvimonas | 0 | 0 | 0 | 0 | 0 | 11 | 47 | 150 | 34 | 48 | 1.769 | 3.555 | 0.0004 | 0.0060 | |
| Ferruginibacter | 0 | 0 | 0 | 0 | 0 | 36 | 59 | 197 | 0 | 65 | 2.089 | 3.139 | 0.0017 | 0.0194 | |
| Aureimonas | 0 | 0 | 0 | 0 | 17 | 109 | 37 | 102 | 41 | 0 | 1.841 | 3.761 | 0.0002 | 0.0035 | |
| Paludibaculum | 0 | 0 | 0 | 0 | 0 | 56 | 149 | 154 | 26 | 291 | 1.724 | 4.287 | 0.0000 | 0.0006 | |
| Rhodoferax | 0 | 0 | 0 | 0 | 5 | 80 | 14 | 138 | 0 | 0 | 2.218 | 2.799 | 0.0051 | 0.0447 | |
| Leptothrix | 0 | 0 | 0 | 0 | 0 | 109 | 196 | 153 | 23 | 21 | 1.769 | 3.995 | 0.0001 | 0.0015 | |
| Pedobacter | 0 | 0 | 0 | 0 | 0 | 139 | 7 | 20 | 105 | 0 | 2.157 | 2.799 | 0.0051 | 0.0447 | |
| Rhodopseudomonas | 0 | 0 | 0 | 0 | 21 | 29 | 11 | 0 | 81 | 0 | 2.259 | 2.878 | 0.0040 | 0.0381 | |
| P3OB-42 | 0 | 0 | 0 | 0 | 0 | 45 | 59 | 72 | 6 | 23 | 1.773 | 3.248 | 0.0012 | 0.0145 | |
| Sandaracinus | 0 | 0 | 0 | 0 | 5 | 58 | 18 | 36 | 4 | 25 | 1.599 | 3.482 | 0.0005 | 0.0075 | |
| Roseisolibacter | 0 | 0 | 0 | 0 | 0 | 7 | 120 | 179 | 0 | 64 | 2.152 | 3.090 | 0.0020 | 0.0210 | |
| Penicillium | Botryotrichum | Cladosporium | Aspergillus | Epicoccum | Parengyodontium | Botrytis | Beaueria | Mortierella | Trichoderma | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1-2 TSA | 0.0 ± 0.00 c | 2.0 ± 1.96 e | 46.1 ± 5.52 b | 18.2 ± 5.25 cde | 0.0 ± 0.00 e | 0.0 ± 0.00 d | 0.0 ± 0.00 e | 1.7 ± 1.67 e | 0.0 ± 0.00 e | 0.0 ± 0.00 b |
| 1-3 TSA | 70.2 ± 4.91 a | 70.6 ± 5.66 a | 80.9 ± 2.55 a | 51.5 ± 0.00 a | 66.0 ± 4.91 a | 0.0 ± 0.00 d | 0.0 ± 0.00 e | 27.5 ± 1.44 c | 0.0 ± 0.00 e | 0.0 ± 0.00 b |
| 6-1 TSA | 78.7 ± 0.00 a | 0.0 ± 0.00 e | 0.0 ± 0.00 d | 0.0 ± 0.00 e | 31.9 ± 2.46 bc | 16.1 ± 0.55 bc | 50.5 ± 0.00 b | 62.5 ± 4.33 a | 42.0 ± 0.00 a | 79.0 ± 0.58 a |
| 7-3 GA | 19.1 ± 2.46 b | 13.7 ± 4.53 de | 52.9 ± 6.79 b | 0.0 ± 0.00 e | 0.0 ± 0.00 e | 13.9 ± 4.81 c | 0.0 ± 0.00 e | 0.0 ± 0.00 e | 10.1 ± 1.67 d | 0.0 ± 0.00 b |
| 7-6 MC | 29.8 ± 6.14 b | 37.3 ± 0.00 b | 19.1 ± 7.64 cd | 30.3 ± 5.25 bc | 0.0 ± 0.00 e | 11.1 ± 3.21 c | 0.0 ± 0.00 e | 0.0 ± 0.00 e | 0.0 ± 0.00 e | 0.0 ± 0.00 b |
| 7-10 PCA | 0.0 ± 0.00 c | 45.1 ± 2.26 b | 33.8 ± 4.25 bc | 39.4 ± 7.00 ab | 0.0 ± 0.00 e | 24.8 ± 1.46 ab | 0.0 ± 0.00 e | 0.0 ± 0.00 e | 0.0 ± 0.00 e | 0.0 ± 0.00 b |
| 7-15/G14 | 0.0 ± 0.00 c | 11.8 ± 1.13 de | 55.9 ± 0.00 ab | 18.2 ± 5.25 cde | 0.0 ± 0.00 e | 27.6 ± 3.21 a | 0.0 ± 0.00 e | 0.0 ± 0.00 e | 0.0 ± 0.00 e | 0.0 ± 0.00 b |
| 7-16/G15 | 0.0 ± 0.00 c | 33.3 ± 2.26 bc | 44.1 ± 5.10 bc | 39.4 ± 3.50 ab | 0.0 ± 0.00 e | 0.0 ± 0.00 d | 0.0 ± 0.00 e | 15.0 ± 0.00 d | 0.0 ± 0.00 e | 0.0 ± 0.00 b |
| 8-5 CA | 0.0 ± 0.00 c | 19.6 ± 7.92 cd | 55.9 ± 1.70 ab | 21.2 ± 3.50 bcd | 0.0 ± 0.00 e | 0.0 ± 0.00 d | 0.0 ± 0.00 e | 17.5 ± 1.44 cd | 37.7 ± 0.84 a | 0.0 ± 0.00 b |
| 8-7 MM | 26.3 ± 7.48 b | 0.0 ± 0.00 e | 0.0 ± 0.00 d | 0.0 ± 0.00 e | 25.5 ± 1.23 c | 0.0 ± 0.00 d | 46.2 ± 1.24 c | 15.0 ± 2.89 d | 0.0 ± 0.00 e | 0.0 ± 0.00 b |
| 8-10 MM | 0.0 ± 0.00 c | 15.7 ± 1.13 de | 42.6 ± 7.64 bc | 24.2 ± 1.75 bcd | 0.0 ± 0.00 e | 0.0 ± 0.00 d | 0.0 ± 0.00 e | 7.5 ± 4.33 de | 0.0 ± 0.00 e | 0.0 ± 0.00 b |
| 8-14 GA | 0.0 ± 0.00 c | 47.1 ± 3.39 b | 38.2 ± 8.49 bc | 36.4 ± 1.75 abc | 0.0 ± 0.00 e | 0.0 ± 0.00 d | 0.0 ± 0.00 e | 15.0 ± 2.89 d | 21.7 ± 0.00 c | 0.0 ± 0.00 b |
| 9-2 TSA | 0.0 ± 0.00 c | 0.0 ± 0.00 e | 50.0 ± 8.49 b | 9.1 ± 0.00 de | 0.0 ± 0.00 e | 0.0 ± 0.00 d | 0.0 ± 0.00 e | 7.5 ± 1.44 de | 29.0 ± 2.51 b | 0.0 ± 0.00 b |
| 10-4 TSA | 0.0 ± 0.00 c | 0.0 ± 0.00 e | 0.0 ± 0.00 d | 24.2 ± 1.75 bcd | 0.0 ± 0.00 e | 16.7 ± 0.00 bc | 0.0 ± 0.00 e | 40.0 ± 2.89 b | 20.3 ± 0.84 c | 0.0 ± 0.00 b |
| 11-10 MM | 0.0 ± 0.00 c | 39.2 ± 1.13 b | 0.0 ± 0.00 d | 18.2 ± 5.25 cde | 36.2 ± 2.46 b | 16.7 ± 0.00 bc | 35.5 ± 2.48 d | 1.7 ± 1.67 e | 30.4 ± 3.35 ab | 0.0 ± 0.00 b |
| 11-11 MM | 0.0 ± 0.00 c | 37.3 ± 2.27 b | 0.0 ± 0.00 d | 18.2 ± 1.75 cde | 61.7 ± 2.46 a | 27.6 ± 3.21 a | 63.4 ± 1.24 a | 10.0 ± 2.89 de | 36.2 ± 0.00 a | 0.0 ± 0.00 b |
| 1-3 TSA | 6-1 TSA | 7-15/G14 | 11-11MM | |
|---|---|---|---|---|
| Total Sequence Length (bp): | 8,497,050 | 3,834,536 | 5,315,248 | 9,378,728 |
| Number of Sequences: | 53 | 52 | 38 | 161 |
| Longest Sequences (bp): | 1,277,654 | 677,502 | 1,550,329 | 615,461 |
| N50 (bp): | 373,280 | 420,792 | 1,252,813 | 151,700 |
| Gap Ratio (%): | 0.015652 | 0.005216 | 0.005832 | 0.00853 |
| GCcontent (%): | 71.6 | 41.2 | 36.2 | 71.8 |
| Number of CDSs: | 7529 | 3923 | 4672 | 8549 |
| Average Protein Length: | 332.3 | 286.6 | 327.9 | 322.4 |
| Coding Ratio (%): | 88.3 | 88 | 86.5 | 88.2 |
| Number of rRNAs: | 2 | 3 | 3 | 4 |
| Number of tRNAs: | 85 | 75 | 80 | 94 |
| Number of CRISPRs: | 4 | 0 | 0 | 2 |
| Assembly according to the closest reference strain | ||||
| # contigs (≥0 bp) | 71 | 78 | 45 | |
| # contigs (≥1000 bp) | 38 | 24 | 16 | |
| # contigs (≥5000 bp) | 33 | 22 | 11 | |
| # contigs (≥10,000 bp) | 33 | 22 | 10 | |
| # contigs (≥25,000 bp) | 32 | 19 | 9 | |
| # contigs (≥50,000 bp) | 28 | 15 | 9 | |
| Total length (≥0 bp) | 8,499,567 | 3,838,568 | 5,316,373 | |
| Total length (≥1000 bp) | 8,492,054 | 3,824,321 | 5,308,269 | |
| Total length (≥5000 bp) | 8,483,321 | 3,818,604 | 5,297,689 | |
| Total length (≥10,000 bp) | 8,483,321 | 3,818,604 | 5,292,390 | |
| Total length (≥25,000 bp) | 8,464,247 | 3,769,813 | 5,281,078 | |
| Total length (≥50,000 bp) | 8,322,500 | 3,615,438 | 5,281,078 | |
| # contigs | 38 | 24 | 16 | |
| Largest contig | 1,277,654 | 677,502 | 1,550,329 | |
| Total length | 8,492,054 | 3824321 | 5,308,269 | |
| Reference length | 8,855,698 | 3,812,514 | 5,693,782 | |
| GC (%) | 71.61 | 41.24 | 36.15 | |
| Reference GC (%) | 71.71 | 41.11 | 36.16 | |
| N50 | 373,280 | 420,792 | 1,252,813 | |
| NG50 | 373,280 | 420,792 | 773,259 | |
| N75 | 237,595 | 268,237 | 598,143 | |
| NG75 | 237,278 | 268,237 | 407,868 | |
| L50 | 7 | 4 | 2 | |
| LG50 | 7 | 4 | 3 | |
| L75 | 14 | 7 | 4 | |
| LG75 | 15 | 7 | 5 | |
| # misassemblies | 251 | 71 | 149 | |
| # misassembled contigs | 29 | 14 | 8 | |
| Misassembled contigs length | 8,168,390 | 3,470,420 | 5,209,481 | |
| # local misassemblies | 570 | 69 | 329 | |
| # scaffold gap ext. mis. | 5 | 0 | 1 | |
| # scaffold gap loc. mis. | 59 | 0 | 2 | |
| # unaligned mis. contigs | 2 | 1 | 1 | |
| # unaligned contigs | 1 + 35 part | 2 + 14 part | 5 + 10 part | |
| Unaligned length | 1,427,995 | 306,517 | 1,225,761 | |
| Genome fraction (%) | 79.357 | 92.168 | 71.617 | |
| Duplication ratio | 1.005 | 1.001 | 1.001 | |
| # N’s per 100 kbp | 15.66 | 5.23 | 5.84 | |
| # mismatches per 100 kbp | 2167.9 | 1198.66 | 3504.82 | |
| # indels per 100 kbp | 111.29 | 24.9 | 85.42 | |
| # genomic features | 5912 + 606 part | 3670 + 107 part | 3986 + 304 part | |
| Largest alignment | 208,439 | 269,167 | 139,195 | |
| Total aligned length | 7,060,676 | 3,515,775 | 4,081,960 | |
| NA50 | 38,599 | 86510 | 30,553 | |
| NGA50 | 36,902 | 86510 | 27,840 | |
| NA75 | 11,869 | 39,117 | 6088 | |
| NGA75 | 8663 | 39,911 | - | |
| LA50 | 63 | 13 | 45 | |
| LGA50 | 68 | 13 | 52 | |
| LA75 | 154 | 30 | 126 | |
| LGA75 | 180 | 29 | - | |
| Species Name | Strain No. | Accession No. | NCBI Taxonomy IDs | Type Strain | Validated | ANI (%) | Folded Fragments | Total Fragments |
|---|---|---|---|---|---|---|---|---|
| 1-3 TSA | ||||||||
| Streptomyces anulatus | strain=JCM 4721 | GCA_014650675.1 | 1892 | type | TRUE | 97.0620 | 2441 | 2811 |
| Streptomyces anulatus | strain=ATCC 11523 | GCA_001434355.1 | 1892 | syntype | TRUE | 97.0122 | 2443 | 2811 |
| Streptomyces griseus | strain=NCTC13033 | GCA_900460065.1 | 1911 | type | TRUE | 93.1318 | 2171 | 2811 |
| Streptomyces griseus | strain=DSM 40236 | GCA_900105705.1 | 1911 | syntype | TRUE | 93.1183 | 2214 | 2811 |
| Streptomyces anulatus | strain=NRRL B-3362 | GCA_000717105.1 | 1892 | syntype | TRUE | 92.8788 | 1974 | 2811 |
| Streptomyces anulatus | strain=NRRL B-2873 | GCA_000721175.1 | 1892 | syntype | TRUE | 92.8387 | 1967 | 2811 |
| Streptomyces griseus | strain=JCM 4516 | GCA_014655335.1 | 1911 | syntype | FALSE | 92.5514 | 2195 | 2811 |
| 6-1 TSA | ||||||||
| Bacillus altitudinis | strain=DSM 26896 | GCA_000949525.1 | 293387 | syntype | TRUE | 98.4941 | 1160 | 1263 |
| Bacillus altitudinis | strain=NIO-1130 | GCA_900094985.1 | 293387 | syntype | TRUE | 97.9919 | 1150 | 1263 |
| Bacillus altitudinis | strain=NIO-1130 | GCA_001457015.1 | 293387 | type | TRUE | 97.9919 | 1150 | 1263 |
| Bacillus altitudinis | strain=41KF2b | GCA_000691145.1 | 293387 | type | TRUE | 97.9481 | 1154 | 1263 |
| Bacillus xiamenensis | strain=HYC-10 | GCA_000300535.1 | 1178537 | type | TRUE | 91.4340 | 1029 | 1263 |
| Bacillus zhangzhouensis | strain=DW5-4 | GCA_000715205.1 | 1178540 | type | TRUE | 89.9647 | 1085 | 1263 |
| 7-15/G14 | ||||||||
| Chryseobacterium viscerum | strain=687B-08 | GCA_002899945.2 | 1037377 | type | TRUE | 95.4107 | 1497 | 1760 |
| Chryseobacterium sediminis | strain=IMT-174 | GCA_008386505.1 | 1679494 | type | TRUE | 87.2802 | 1354 | 1760 |
| Chryseobacterium rhizoplanae | strain=DSM 29371 | GCA_900182655.1 | 1609531 | type | TRUE | 87.2214 | 1367 | 1760 |
| Chryseobacterium cucumeris | strain=GSE06 | GCA_001593385.1 | 1813611 | type | TRUE | 86.4695 | 1357 | 1760 |
| Chryseobacterium candidae | strain=JC507 | GCA_004916905.1 | 1978493 | type | TRUE | 86.3433 | 1199 | 1760 |
| Chryseobacterium gleum | strain=ATCC 35910 | GCA_000143785.1 | 250 | type | TRUE | 85.8473 | 1296 | 1760 |
| Chryseobacterium aureum | strain=17S1E7 | GCA_003971235.1 | 2497456 | type | TRUE | 85.8461 | 1326 | 1760 |
| Chryseobacterium gleum | strain=NCTC11432 | GCA_900636535.1 | 250 | type | TRUE | 85.8060 | 1312 | 1760 |
| Chryseobacterium gleum | strain=FDAARGOS_1103 | GCA_016766875.1 | 250 | type | TRUE | 85.7937 | 1318 | 1760 |
| Chryseobacterium culicis | strain=DSM 23031 | GCA_900108365.1 | 680127 | type | TRUE | 85.5231 | 1323 | 1760 |
| Chryseobacterium jejuense | strain=DSM 19299 | GCA_900100075.1 | 445960 | type | TRUE | 83.2892 | 1147 | 1760 |
| Chryseobacterium ureilyticum | strain=DSM 18017 | GCA_900156735.1 | 373668 | type | TRUE | 83.1204 | 1126 | 1760 |
| Chryseobacterium nakagawai | strain=NCTC13529 | GCA_900637665.1 | 1241982 | type | TRUE | 82.9748 | 1147 | 1760 |
| 11-11 MM | ||||||||
| Streptomyces vinaceus | strain=JCM 4849 | GCA_014656155.1 | 1960 | type | TRUE | 86.7781 | 1870 | 3057 |
| Streptomyces xanthophaeus | strain=NBRC 12829 | GCA_016755895.1 | 67385 | type | TRUE | 86.7459 | 1974 | 3057 |
| Streptomyces xanthophaeus | strain=NRRL B-5414 | GCA_000725805.1 | 67385 | type | TRUE | 86.7181 | 1950 | 3057 |
| Streptomyces goshikiensis | strain=JCM 4640 | GCA_014650555.1 | 1942 | type | TRUE | 86.6907 | 1786 | 3057 |
| Streptomyces nojiriensis | strain=JCM 3382 | GCA_014648615.1 | 66374 | type | TRUE | 86.6852 | 1933 | 3057 |
| Streptomyces spororaveus | strain=NBRC 15456 | GCA_016755875.1 | 284039 | type | TRUE | 86.6749 | 1886 | 3057 |
| Streptomyces nojiriensis | strain=NBRC 13794 | GCA_016755855.1 | 66374 | type | TRUE | 86.6735 | 1924 | 3057 |
| Streptomyces avidinii | strain=JCM 4726 | GCA_014650695.1 | 1895 | type | TRUE | 86.5841 | 1893 | 3057 |
| Streptomyces subrutilus | strain=ATCC 27467 | GCA_008704535.1 | 36818 | type | TRUE | 86.4921 | 1785 | 3057 |
| Streptomyces subrutilus | strain=JCM 4834 | GCA_014650935.1 | 36818 | type | TRUE | 86.4834 | 1766 | 3057 |
| Streptomyces cinnamonensis | strain=JCM 4019 | GCA_014648795.1 | 1900 | type | TRUE | 86.4664 | 1876 | 3057 |
| Streptomyces cinnamonensis | strain=NBRC 15873 | GCA_016755835.1 | 1900 | type | TRUE | 86.4132 | 1914 | 3057 |
| Streptomyces virginiae | strain=NRRL ISP-5094 | GCA_000720455.1 | 1961 | type | TRUE | 86.4089 | 1896 | 3057 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dimkić, I.; Ćopić, M.; Petrović, M.; Stupar, M.; Savković, Ž.; Knežević, A.; Subakov Simić, G.; Ljaljević Grbić, M.; Unković, N. Bacteriobiota of the Cave Church of Sts. Peter and Paul in Serbia—Culturable and Non-Culturable Communities’ Assessment in the Bioconservation Potential of a Peculiar Fresco Painting. Int. J. Mol. Sci. 2023, 24, 1016. https://doi.org/10.3390/ijms24021016
Dimkić I, Ćopić M, Petrović M, Stupar M, Savković Ž, Knežević A, Subakov Simić G, Ljaljević Grbić M, Unković N. Bacteriobiota of the Cave Church of Sts. Peter and Paul in Serbia—Culturable and Non-Culturable Communities’ Assessment in the Bioconservation Potential of a Peculiar Fresco Painting. International Journal of Molecular Sciences. 2023; 24(2):1016. https://doi.org/10.3390/ijms24021016
Chicago/Turabian StyleDimkić, Ivica, Milica Ćopić, Marija Petrović, Miloš Stupar, Željko Savković, Aleksandar Knežević, Gordana Subakov Simić, Milica Ljaljević Grbić, and Nikola Unković. 2023. "Bacteriobiota of the Cave Church of Sts. Peter and Paul in Serbia—Culturable and Non-Culturable Communities’ Assessment in the Bioconservation Potential of a Peculiar Fresco Painting" International Journal of Molecular Sciences 24, no. 2: 1016. https://doi.org/10.3390/ijms24021016
APA StyleDimkić, I., Ćopić, M., Petrović, M., Stupar, M., Savković, Ž., Knežević, A., Subakov Simić, G., Ljaljević Grbić, M., & Unković, N. (2023). Bacteriobiota of the Cave Church of Sts. Peter and Paul in Serbia—Culturable and Non-Culturable Communities’ Assessment in the Bioconservation Potential of a Peculiar Fresco Painting. International Journal of Molecular Sciences, 24(2), 1016. https://doi.org/10.3390/ijms24021016








