Intervention of AXL in EGFR Signaling via Phosphorylation and Stabilization of MIG6 in Non-Small Cell Lung Cancer
Abstract
1. Introduction
2. Results
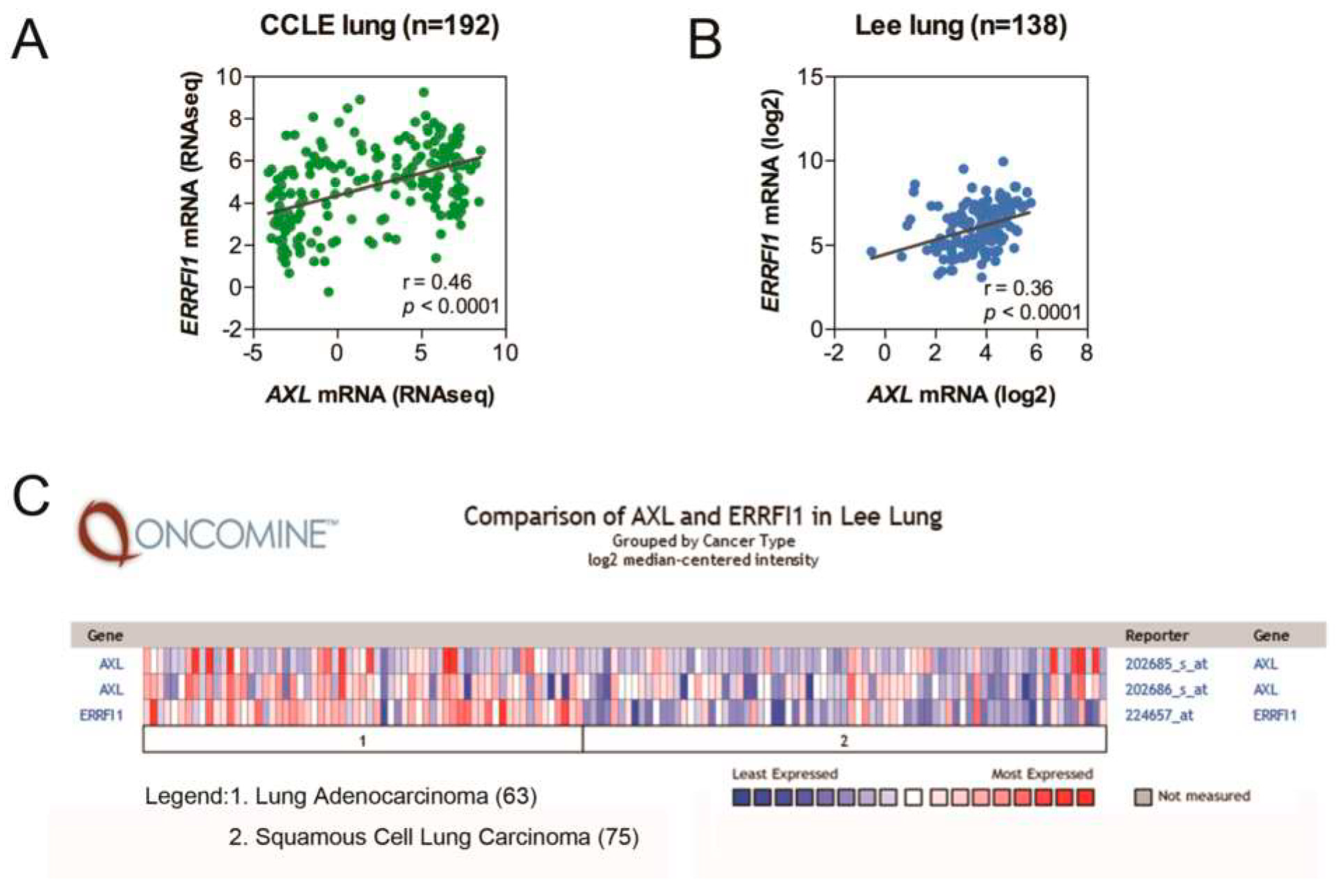
2.1. AXL and MIG6 Are Coexpressed in Lung Cancer
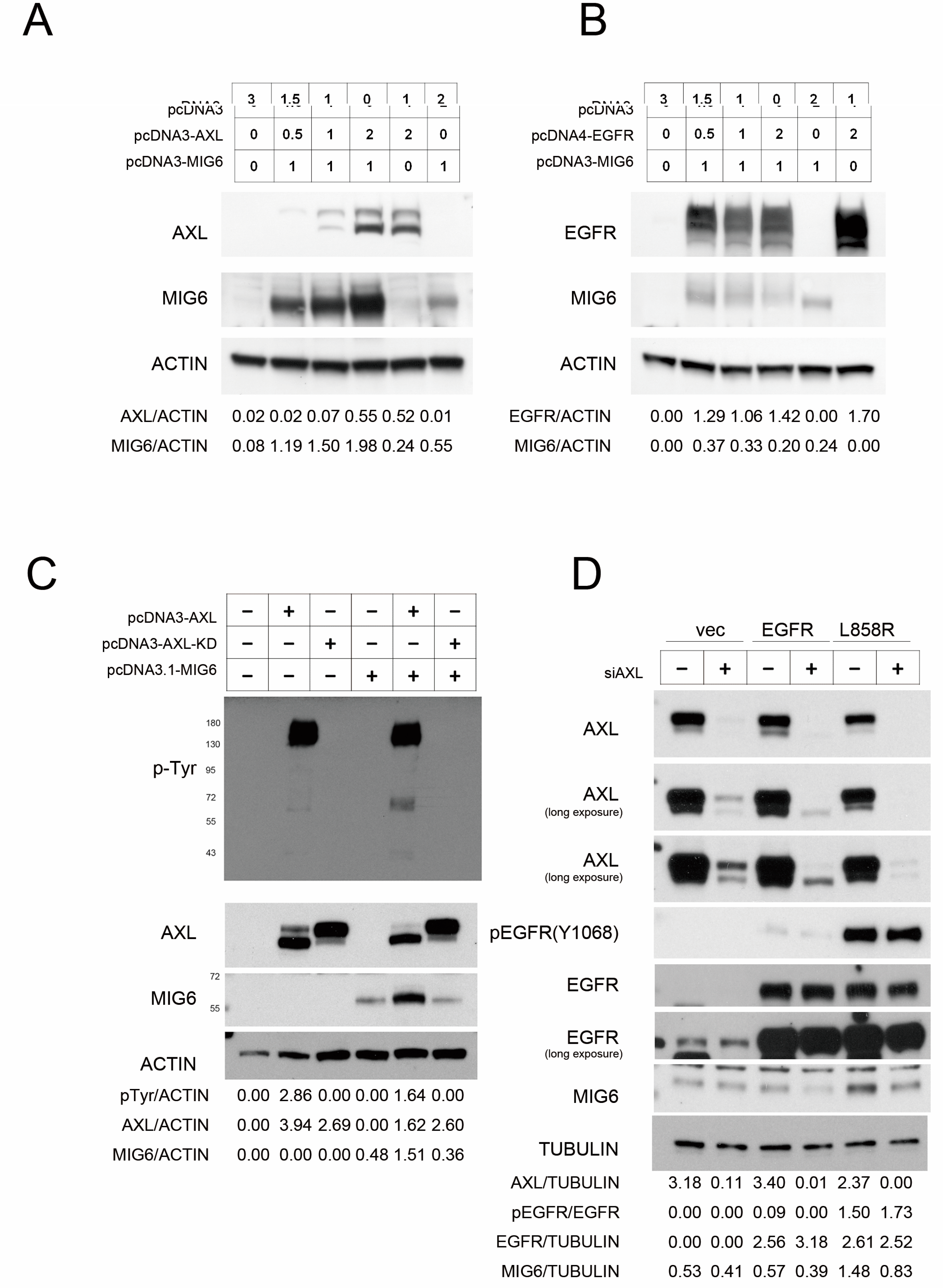
2.2. AXL Overexpression Elevates MIG6 Expression but Downregulates EGFR
2.3. AXL Stabilizes MIG6 Expression
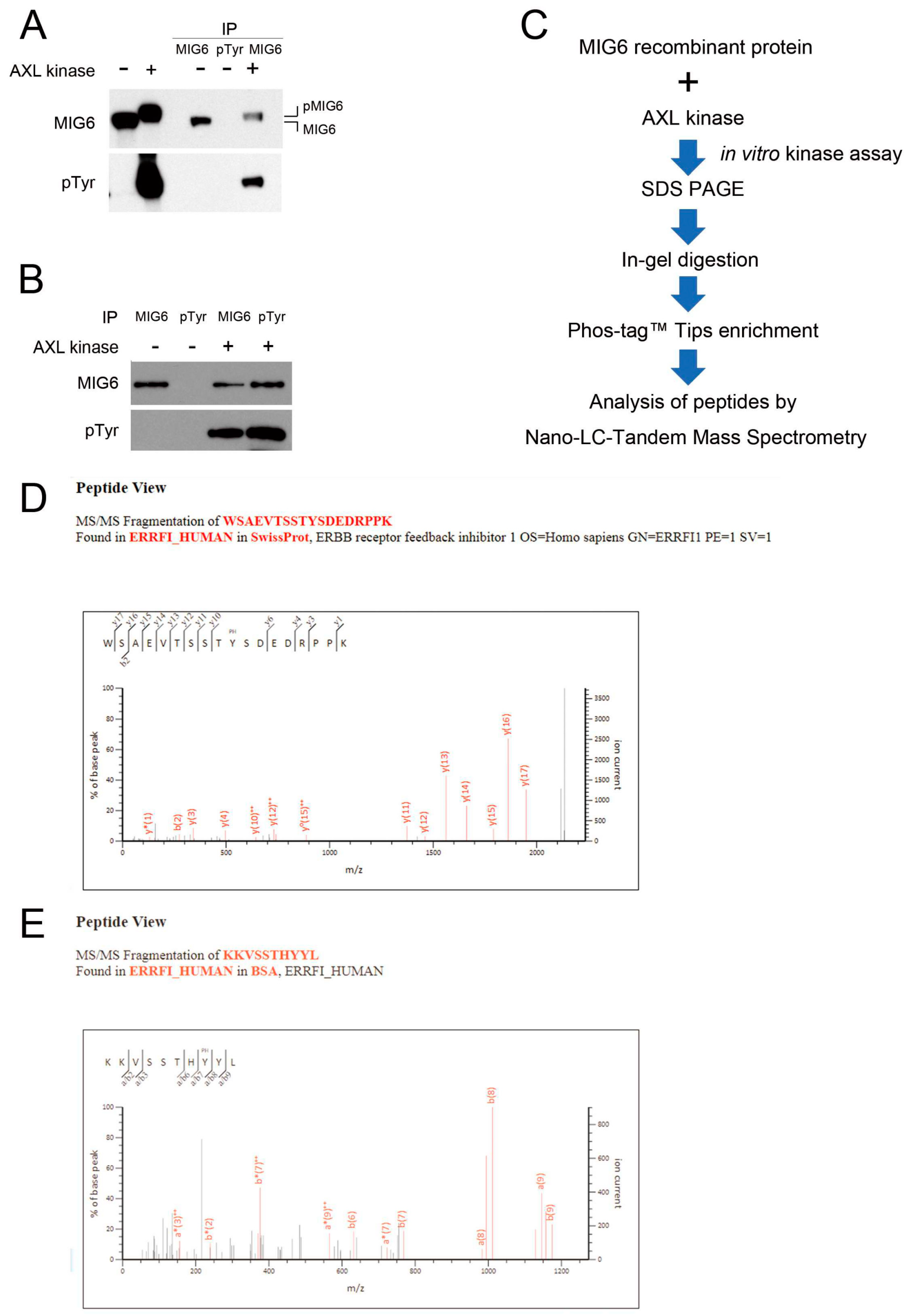
2.4. MIG6 Interacts with AXL and Activated AXL Phosphorylates MIG6 on Residues Y310 and Y394/Y395
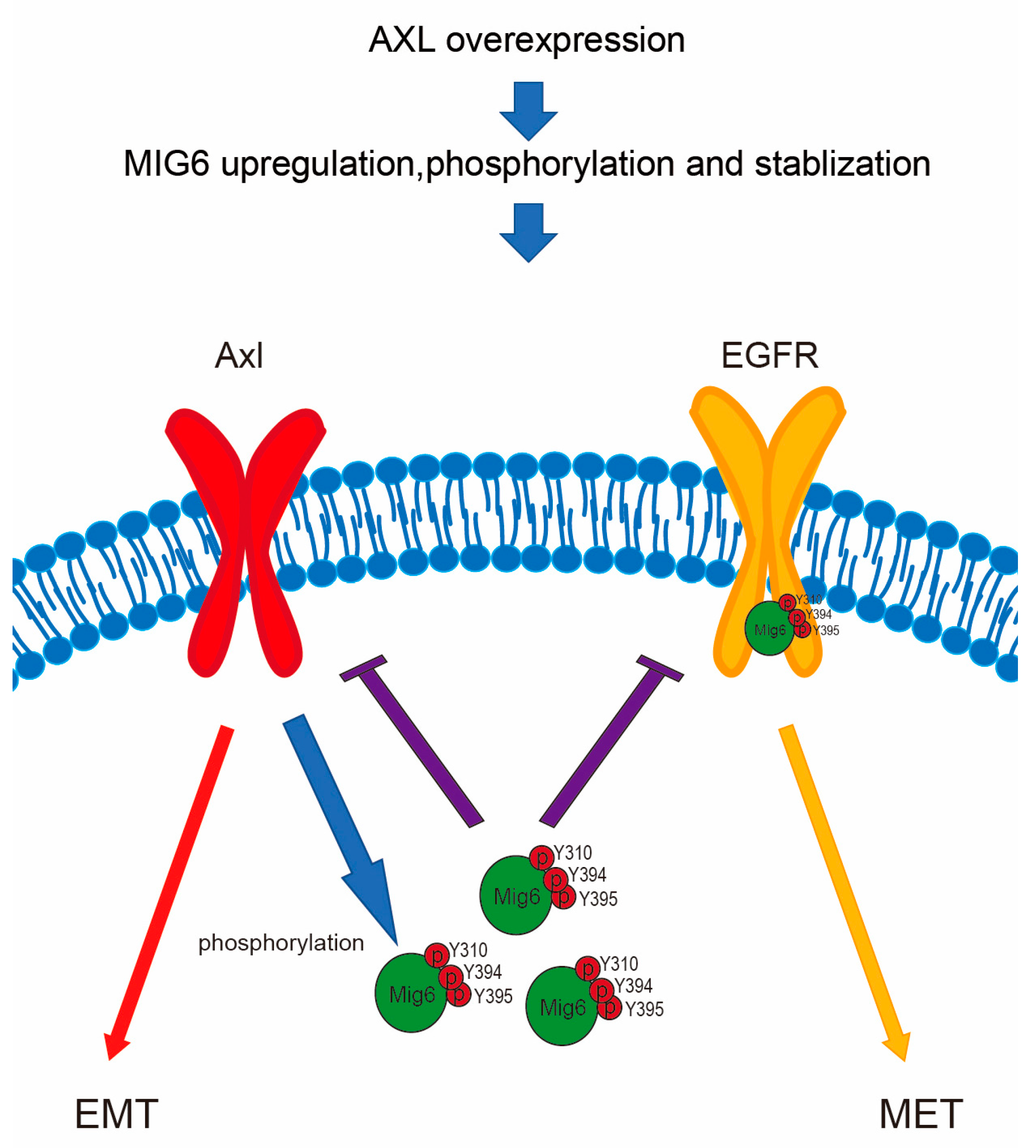
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Public Domain Data Mining and Statistical Analysis
4.3. mRNA Microarray Analysis
4.4. Construction of DNA Vectors
4.5. Reporter Assay
4.6. PLA Assay
4.7. Coimmunoprecipitation (Co–IP) Assay
4.8. Western Blotting
4.9. In Vitro Kinase Assay and Phosphoprotein Enrichment
4.10. Analysis of Peptides with Nano–LC–Tandem Mass Spectrometry
4.11. LC–MS/MS Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paez, J.G.; Janne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Antony, J.; Tan, T.Z.; Kelly, Z.; Low, J.; Choolani, M.; Recchi, C.; Gabra, H.; Thiery, J.P.; Huang, R.Y. The GAS6-AXL signaling network is a mesenchymal (Mes) molecular subtype-specific therapeutic target for ovarian cancer. Sci. Signal. 2016, 9, ra97. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Wang, J.; Shiozawa, Y.; McGee, S.; Kim, J.; Jung, Y.; Joseph, J.; Berry, J.E.; Havens, A.; Pienta, K.J.; et al. Hypoxia stabilizes GAS6/Axl signaling in metastatic prostate cancer. Mol. Cancer Res. 2012, 10, 703–712. [Google Scholar] [CrossRef]
- Bae, C.A.; Ham, I.H.; Oh, H.J.; Lee, D.; Woo, J.; Son, S.Y.; Yoon, J.H.; Lorens, J.B.; Brekken, R.A.; Kim, T.M.; et al. Inhibiting the GAS6/AXL axis suppresses tumor progression by blocking the interaction between cancer-associated fibroblasts and cancer cells in gastric carcinoma. Gastric Cancer 2020, 23, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Wei, Y.; Wei, X. AXL receptor tyrosine kinase as a promising anti-cancer approach: Functions, molecular mechanisms and clinical applications. Mol. Cancer 2019, 18, 153. [Google Scholar]
- Goyette, M.A.; Duhamel, S.; Aubert, L.; Pelletier, A.; Savage, P.; Thibault, M.P.; Johnson, R.M.; Carmeliet, P.; Basik, M.; Gaboury, L.; et al. The Receptor Tyrosine Kinase AXL Is Required at Multiple Steps of the Metastatic Cascade during HER2-Positive Breast Cancer Progression. Cell. Rep. 2018, 23, 1476–1490. [Google Scholar] [CrossRef]
- Zhang, Z.; Lee, J.C.; Lin, L.; Olivas, V.; Au, V.; LaFramboise, T.; Abdel-Rahman, M.; Wang, X.; Levine, A.D.; Rho, J.K.; et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet. 2012, 44, 852–860. [Google Scholar] [CrossRef]
- Kim, D.; Bach, D.H.; Fan, Y.H.; Luu, T.T.; Hong, J.Y.; Park, H.J.; Lee, S.K. AXL degradation in combination with EGFR-TKI can delay and overcome acquired resistance in human non-small cell lung cancer cells. Cell. Death Dis. 2019, 10, 361. [Google Scholar] [CrossRef]
- Wilson, C.; Ye, X.; Pham, T.; Lin, E.; Chan, S.; McNamara, E.; Neve, R.M.; Belmont, L.; Koeppen, H.; Yauch, R.L.; et al. AXL inhibition sensitizes mesenchymal cancer cells to antimitotic drugs. Cancer Res. 2014, 74, 5878–5890. [Google Scholar] [CrossRef]
- Wu, F.; Li, J.; Jang, C.; Wang, J.; Xiong, J. The role of Axl in drug resistance and epithelial-to-mesenchymal transition of non-small cell lung carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 6653–6661. [Google Scholar]
- Ghiso, E.; Migliore, C.; Ciciriello, V.; Morando, E.; Petrelli, A.; Corso, S.; De Luca, E.; Gatti, G.; Volante, M.; Giordano, S. YAP-Dependent AXL Overexpression Mediates Resistance to EGFR Inhibitors in NSCLC. Neoplasia 2017, 19, 1012–1021. [Google Scholar] [CrossRef]
- Brand, T.M.; Iida, M.; Stein, A.P.; Corrigan, K.L.; Braverman, C.M.; Luthar, N.; Toulany, M.; Gill, P.S.; Salgia, R.; Kimple, R.J.; et al. AXL mediates resistance to cetuximab therapy. Cancer Res. 2014, 74, 5152–5164. [Google Scholar] [CrossRef]
- Gjerdrum, C.; Tiron, C.; Hoiby, T.; Stefansson, I.; Haugen, H.; Sandal, T.; Collett, K.; Li, S.; McCormack, E.; Gjertsen, B.T.; et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc. Natl. Acad. Sci. USA 2010, 107, 1124–1129. [Google Scholar] [CrossRef]
- Antony, J.; Huang, R.Y. AXL-Driven EMT State as a Targetable Conduit in Cancer. Cancer Res. 2017, 77, 3725–3732. [Google Scholar] [CrossRef]
- Vouri, M.; Croucher, D.R.; Kennedy, S.P.; An, Q.; Pilkington, G.J.; Hafizi, S. Axl-EGFR receptor tyrosine kinase hetero-interaction provides EGFR with access to pro-invasive signalling in cancer cells. Oncogenesis 2016, 5, e266. [Google Scholar] [CrossRef]
- Li, N.; Wei, M. Conversion of MIG6 peptide from the nonbinder to binder of lung cancer-related EGFR by phosphorylation and cyclization. Artif. Cells Nanomed. Biotechnol. 2017, 45, 1023–1028. [Google Scholar] [CrossRef]
- Frosi, Y.; Anastasi, S.; Ballaro, C.; Varsano, G.; Castellani, L.; Maspero, E.; Polo, S.; Alema, S.; Segatto, O. A two-tiered mechanism of EGFR inhibition by RALT/MIG6 via kinase suppression and receptor degradation. J. Cell. Biol. 2010, 189, 557–571. [Google Scholar] [CrossRef]
- Ferby, I.; Reschke, M.; Kudlacek, O.; Knyazev, P.; Pante, G.; Amann, K.; Sommergruber, W.; Kraut, N.; Ullrich, A.; Fassler, R.; et al. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat. Med. 2006, 12, 568–573. [Google Scholar] [CrossRef]
- Wang, Z.; Raines, L.L.; Hooy, R.M.; Roberson, H.; Leahy, D.J.; Cole, P.A. Tyrosine phosphorylation of mig6 reduces its inhibition of the epidermal growth factor receptor. ACS Chem. Biol. 2013, 8, 2372–2376. [Google Scholar] [CrossRef]
- Bose, R.; Zhang, X. The ErbB kinase domain: Structural perspectives into kinase activation and inhibition. Exp. Cell. Res. 2009, 315, 649–658. [Google Scholar] [CrossRef]
- Anastasi, S.; Baietti, M.F.; Frosi, Y.; Alema, S.; Segatto, O. The evolutionarily conserved EBR module of RALT/MIG6 mediates suppression of the EGFR catalytic activity. Oncogene 2007, 26, 7833–7846. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Kim, N.; Ficarro, S.B.; Zhang, Y.; Lee, B.I.; Cho, A.; Kim, K.; Park, A.K.J.; Park, W.Y.; Murray, B.; et al. and mechanism of activity-based inhibition of the EGF receptor by Mig6. Nat. Struct. Mol. Biol. 2015, 22, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Okami, J.; Okuyama, H.; Nishizawa, Y.; Imamura, F.; Inoue, M. The induction of MIG6 under hypoxic conditions is critical for dormancy in primary cultured lung cancer cells with activating EGFR mutations. Oncogene 2017, 36, 2824–2834. [Google Scholar] [CrossRef] [PubMed]
- Bond, T. Expression of Mig6 linked to resistance to EGFR kinase inhibitors. Pharmacogenomics 2013, 14, 1946. [Google Scholar]
- Chang, X.F.; Izumchenko, E.; Solis, L.M.; Kim, M.S.; Chatterjee, A.; Ling, S.Z.; Monitto, C.L.; Harari, P.M.; Hidalgo, M.; Goodman, S.N.; et al. The Relative Expression of Mig6 and EGFR Is Associated with Resistance to EGFR Kinase Inhibitors. PLoS ONE 2013, 8, e68966. [Google Scholar] [CrossRef]
- Chang, X.F.; Chatterjee, A.; Huang, S.M.; Kim, M.S.; Shao, C.B.; Monitto, C.L.; Ha, P.K.; Hidalgo, M.; Harari, P.M.; Berman, D.M.; et al. Loss of dependence on EGFR signaling by upregulation of Mig6 confers drug resistance to erlotinib. Cancer Res. 2010, 70, 2703. [Google Scholar] [CrossRef]
- Kang, D.H.; Jung, S.S.; Yeo, M.K.; Lee, D.H.; Yoo, G.; Cho, S.Y.; Oh, I.J.; Kim, J.O.; Park, H.S.; Chung, C.; et al. Suppression of Mig-6 overcomes the acquired EGFR-TKI resistance of lung adenocarcinoma. BMC Cancer 2020, 20, 571. [Google Scholar] [CrossRef]
- Liu, N.; Matsumoto, M.; Kitagawa, K.; Kotake, Y.; Suzuki, S.; Shirasawa, S.; Nakayama, K.I.; Nakanishi, M.; Niida, H.; Kitagawa, M. Chk1 phosphorylates the tumour suppressor Mig-6, regulating the activation of EGF signalling. EMBO J. 2012, 31, 2365–2377. [Google Scholar] [CrossRef]
- Boopathy, G.T.K.; Lynn, J.L.S.; Wee, S.; Gunaratne, J.; Hong, W. Phosphorylation of Mig6 negatively regulates the ubiquitination and degradation of EGFR mutants in lung adenocarcinoma cell lines. Cell. Signal. 2018, 43, 21–31. [Google Scholar] [CrossRef]
- Park, S.Y.; Choi, H.K.; Seo, J.S.; Yoo, J.Y.; Jeong, J.W.; Choi, Y.; Choi, K.C.; Yoon, H.G. DNAJB1 negatively regulates MIG6 to promote epidermal growth factor receptor signaling. Biochim. Biophys. Acta 2015, 1853, 2722–2730. [Google Scholar] [CrossRef]
- Lay, J.D.; Hong, C.C.; Huang, J.S.; Yang, Y.Y.; Pao, C.Y.; Liu, C.H.; Lai, Y.P.; Lai, G.M.; Cheng, A.L.; Su, I.J.; et al. Sulfasalazine suppresses drug resistance and invasiveness of lung adenocarcinoma cells expressing AXL. Cancer Res. 2007, 67, 3878–3887. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.S.; Cho, C.Y.; Hong, C.C.; Yan, M.D.; Hsieh, M.C.; Lay, J.D.; Lai, G.M.; Cheng, A.L.; Chuang, S.E. Oxidative stress enhances Axl-mediated cell migration through an Akt1/Rac1-dependent mechanism. Free. Radic. Biol. Med. 2013, 65, 1246–1256. [Google Scholar] [CrossRef] [PubMed]
- Labots, M.; Gotink, K.J.; Dekker, H.; Azijli, K.; van der Mijn, J.C.; Huijts, C.M.; Piersma, S.R.; Jiménez, C.R.; Verheul, H.M. Evaluation of a tyrosine kinase peptide microarray for tyrosine kinase inhibitor therapy selection in cancer. Exp. Mol. Med. 2016, 48, e279. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hopkins, S.; Linderoth, E.; Hantschel, O.; Suarez-Henriques, P.; Pilia, G.; Kendrick, H.; Smalley, M.J.; Superti-Furga, G.; Ferby, I. Mig6 is a sensor of EGF receptor inactivation that directly activates c-Abl to induce apoptosis during epithelial homeostasis. Dev. Cell. 2012, 23, 547–559. [Google Scholar] [CrossRef] [PubMed]
- Pao-Chun, L.; Chan, P.M.; Chan, W.; Manser, E. Cytoplasmic ACK1 interaction with multiple receptor tyrosine kinases is mediated by Grb2: An analysis of ACK1 effects on Axl signaling. J. Biol. Chem. 2009, 284, 34954–34963. [Google Scholar] [CrossRef]
- Byers, L.A.; Diao, L.; Wang, J.; Saintigny, P.; Girard, L.; Peyton, M.; Shen, L.; Fan, Y.; Giri, U.; Tumula, P.K.; et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer Res. 2013, 19, 279–290. [Google Scholar] [CrossRef]
- Izumchenko, E.; Chang, X.F.; Michailidi, C.; Kagohara, L.; Ravi, R.; Paz, K.; Brait, M.; Hoque, M.; Ling, S.Z.; Bedi, A.; et al. The TGF beta-miR200-MIG6 Pathway Orchestrates the EMT-Associated Kinase Switch That Induces Resistance to EGFR Inhibitors. Cancer Res. 2014, 74, 3995–4005. [Google Scholar] [CrossRef]
- Maity, T.K.; Venugopalan, A.; Linnoila, I.; Cultraro, C.M.; Giannakou, A.; Nemati, R.; Zhang, X.; Webster, J.D.; Ritt, D.; Ghosal, S.; et al. Loss of MIG6 Accelerates Initiation and Progression of Mutant Epidermal Growth Factor Receptor-Driven Lung Adenocarcinoma. Cancer Discov. 2015, 5, 534–549. [Google Scholar] [CrossRef]
- Taniguchi, H.; Yamada, T.; Wang, R.; Tanimura, K.; Adachi, Y.; Nishiyama, A.; Tanimoto, A.; Takeuchi, S.; Araujo, L.H.; Boroni, M.; et al. AXL confers intrinsic resistance to osimertinib and advances the emergence of tolerant cells. Nat. Commun. 2019, 10, 259. [Google Scholar] [CrossRef]
- Chu, Y.W.; Yang, P.C.; Yang, S.C.; Shyu, Y.C.; Hendrix, M.J.; Wu, R.; Wu, C.W. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am. J. Respir. Cell. Mol. Biol. 1997, 17, 353–360. [Google Scholar] [CrossRef]
- Chen, Y.R.; Fu, Y.N.; Lin, C.H.; Yang, S.T.; Hu, S.F.; Chen, Y.T.; Tsai, S.F.; Huang, S.F. Distinctive activation patterns in constitutively active and gefitinib-sensitive EGFR mutants. Oncogene 2006, 25, 1205–1215. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.-Y.; Lin, S.-C.; Lay, J.-D.; Cho, C.-Y.; Jang, T.-H.; Ku, H.-Y.; Yao, C.-J.; Chuang, S.-E. Intervention of AXL in EGFR Signaling via Phosphorylation and Stabilization of MIG6 in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2023, 24, 14879. https://doi.org/10.3390/ijms241914879
Yang Y-Y, Lin S-C, Lay J-D, Cho C-Y, Jang T-H, Ku H-Y, Yao C-J, Chuang S-E. Intervention of AXL in EGFR Signaling via Phosphorylation and Stabilization of MIG6 in Non-Small Cell Lung Cancer. International Journal of Molecular Sciences. 2023; 24(19):14879. https://doi.org/10.3390/ijms241914879
Chicago/Turabian StyleYang, Ya-Yu, Sheng-Chieh Lin, Jong-Ding Lay, Chun-Yu Cho, Te-Hsuan Jang, Hsiu-Ying Ku, Chih-Jung Yao, and Shuang-En Chuang. 2023. "Intervention of AXL in EGFR Signaling via Phosphorylation and Stabilization of MIG6 in Non-Small Cell Lung Cancer" International Journal of Molecular Sciences 24, no. 19: 14879. https://doi.org/10.3390/ijms241914879
APA StyleYang, Y.-Y., Lin, S.-C., Lay, J.-D., Cho, C.-Y., Jang, T.-H., Ku, H.-Y., Yao, C.-J., & Chuang, S.-E. (2023). Intervention of AXL in EGFR Signaling via Phosphorylation and Stabilization of MIG6 in Non-Small Cell Lung Cancer. International Journal of Molecular Sciences, 24(19), 14879. https://doi.org/10.3390/ijms241914879




