Spatial Transcriptomic Analysis Reveals Regional Transcript Changes in Early and Late Stages of rd1 Model Mice with Retinitis Pigmentosa
Abstract
:1. Introduction
2. Results
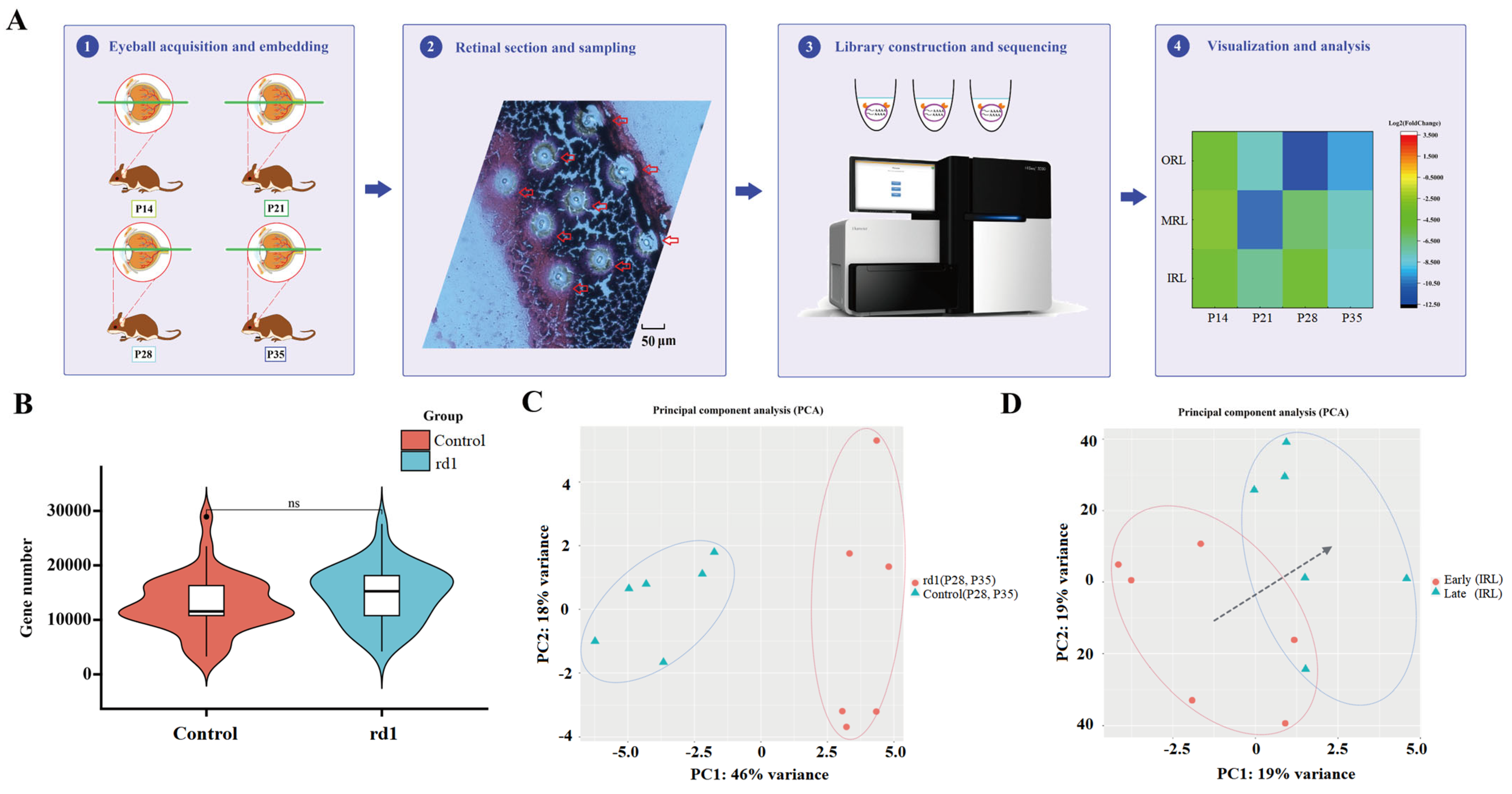
2.1. Spatial Transcriptomic Analysis in rd1 Mice
2.2. The Time Course Is Divided into Early and Late Periods
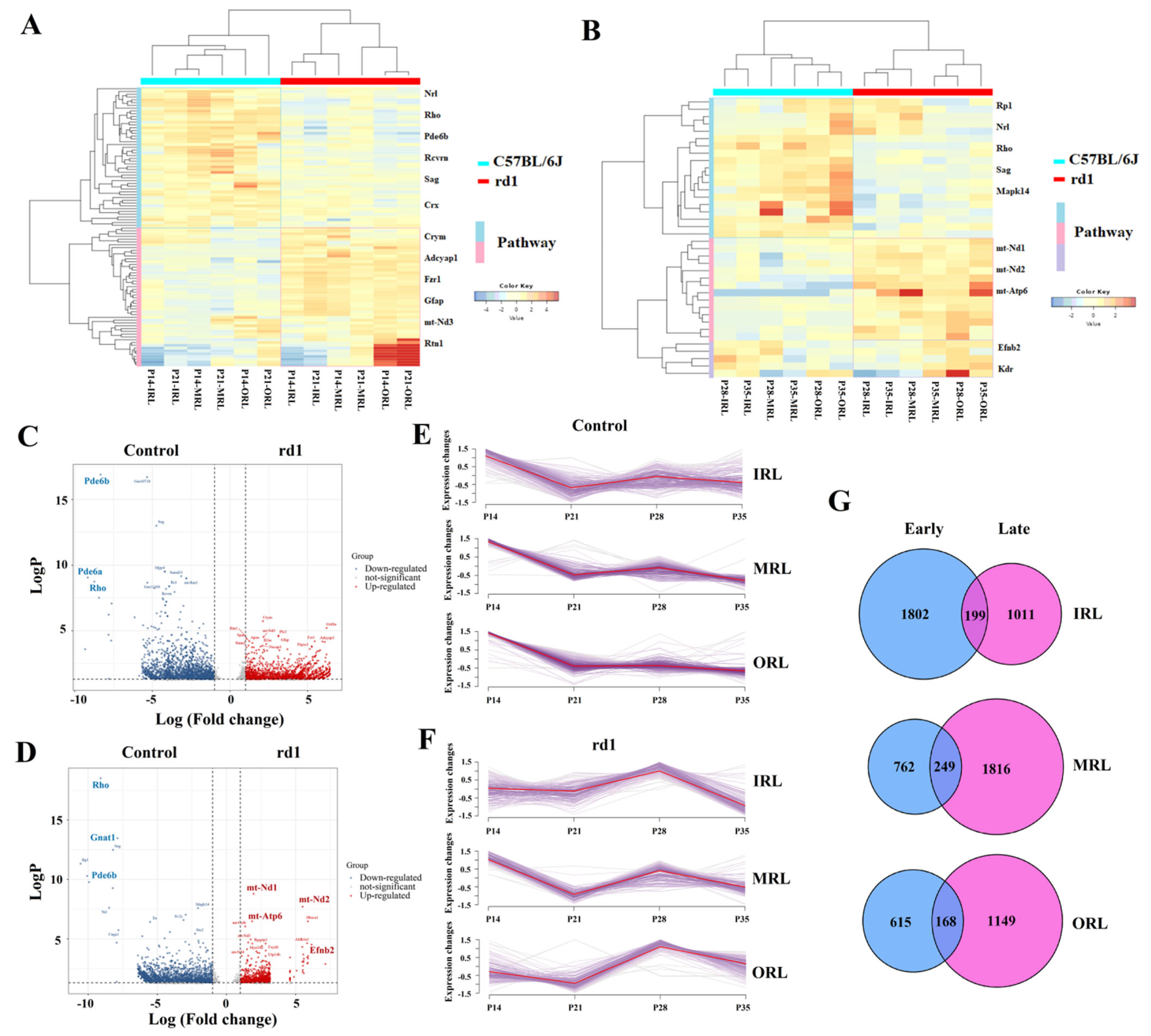
2.3. Identification of DEGs in the Early and Late Stages of RP
2.4. Validation by Quantitative PCR
2.5. Immunofluorescence of VEGF Pathway in RP
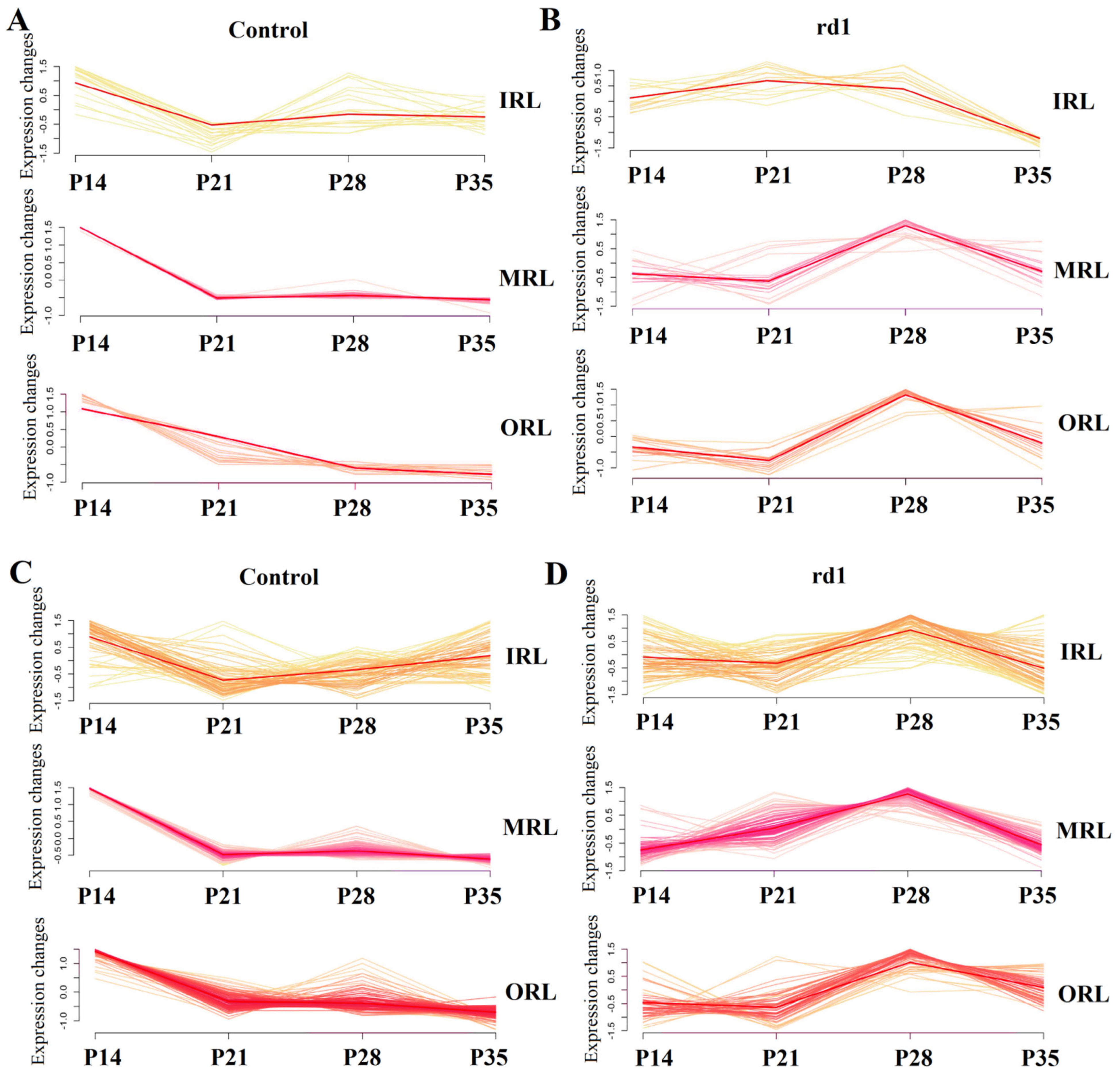
2.6. GO Pathway Trends over Time in the Early and Late Stages of RP
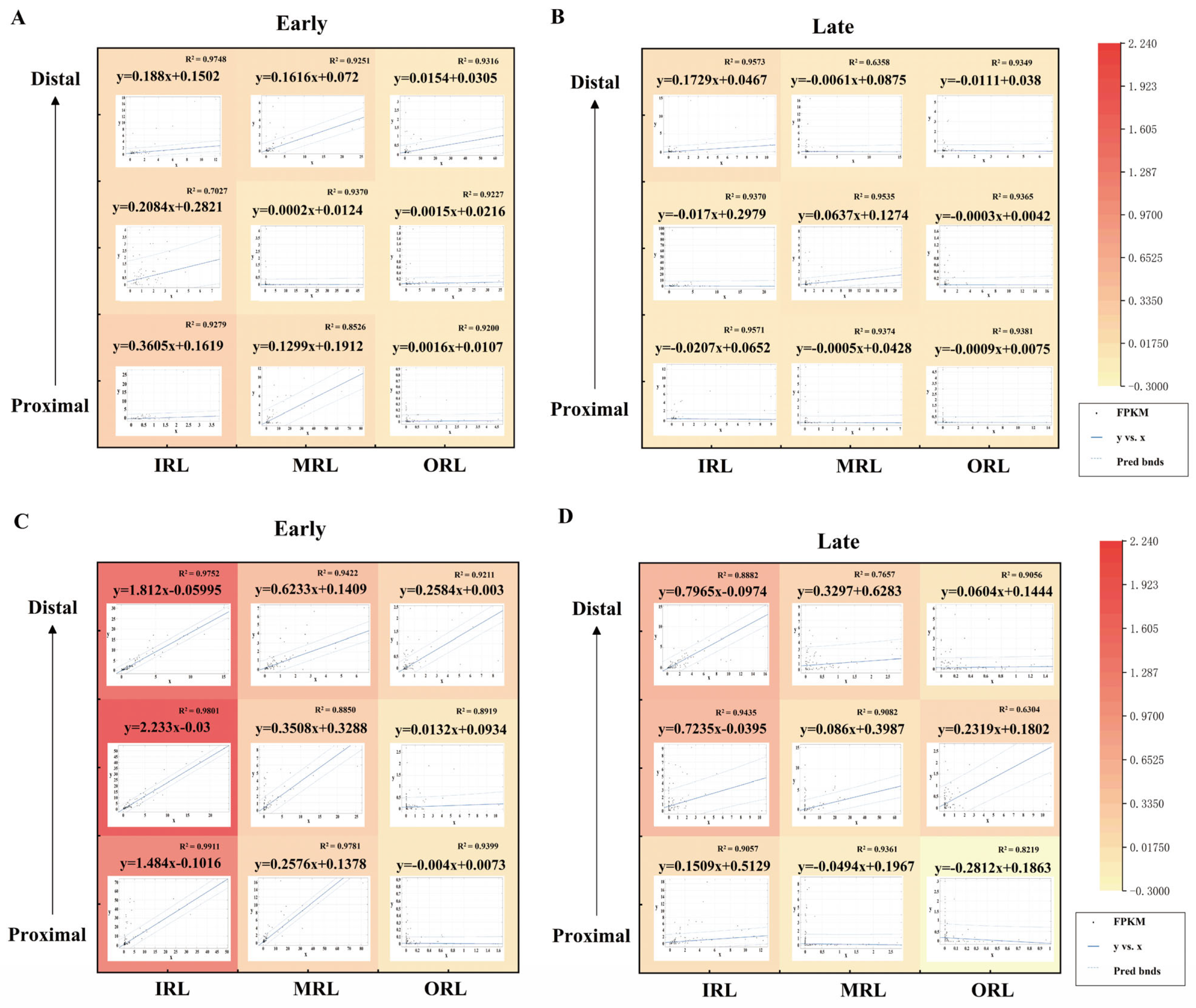
2.7. Fitting of Early and Late Stages at Different Spatial Spots
3. Materials and Methods
3.1. Animals
3.2. Eye Acquisition and Sectioning
3.3. Sampling and Library Construction
3.4. Quantitative PCR
3.5. Immunofluorescence of Frozen Sections
3.6. Data Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sancho-Pelluz, J.; Arango-Gonzalez, B.; Kustermann, S.; Romero, F.J.; van Veen, T.; Zrenner, E.; Ekström, P.; Paquet-Durand, F. Photoreceptor Cell Death Mechanisms in Inherited Retinal Degeneration. Mol. Neurobiol. 2008, 38, 253–269. [Google Scholar] [CrossRef] [PubMed]
- Hartong, D.T.; Berson, E.L.; Dryja, T.P. Retinitis pigmentosa. Lancet 2006, 18, 1795–1809. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Mohan, K.V.; Nagarajan, P.; Iyer, S.; Kesarwani, A.; Nath, M.; Moksha, L.; Bhattacharjee, J.; Das, B.; Jain, K.; et al. Peripheral blood-derived monocytes show neuronal properties and integration in immune-deficient rd1 mouse model upon phenotypic differentiation and induction with retinal growth factors. Stem Cell Res. Ther. 2020, 11, 412. [Google Scholar] [CrossRef] [PubMed]
- Bowes, C.; Li, T.; Danciger, M.; Baxter, L.C.; Applebury, M.L.; Farber, D.B. Retinal degeneration in the rd mouse is caused by a defect in the β subunit of rod cGMP-phosphodiesterase. Nature 1990, 347, 677–680. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.; Hawes, N.L.; Pardue, M.T.; German, A.M.; Hurd, R.E.; Davisson, M.T.; Nusinowitz, S.; Rengarajan, K.; Boyd, A.P.; Sidney, S.S.; et al. Two mouse retinal degenerations caused by missense mutations in the beta-subunit of rod cGMP phosphodiesterase gene. Vision Res. 2007, 47, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Sparrow, J.R. Cellular mechanisms of hereditary photoreceptor degeneration—Focus on cGMP. Prog. Retin. Eye Res. 2021, 74, 100772. [Google Scholar] [CrossRef]
- Pfeiffer, R.L.; Marc, R.E.; Jones, B.W. Persistent remodeling and neurodegeneration in late-stage retinal degeneration. Prog. Retin. Eye Res. 2020, 74, 100771. [Google Scholar] [CrossRef]
- Voisin, A.; Pénaguin, A.; Gaillard, A.; Leveziel, N. Stem cell therapy in retinal diseases. Neural Regen. Res. 2023, 18, 1478. [Google Scholar] [CrossRef]
- Botto, C.; Rucli, M.; Tekinsoy, M.D.; Pulman, J.; Sahel, J.; Dalkara, D. Early and late-stage gene therapy interventions for inherited retinal degenerations. Prog. Retin. Eye Res. 2022, 86, 100975. [Google Scholar] [CrossRef]
- Megaw, R.; Hurd, T.W. Photoreceptor actin dysregulation in syndromic and non-syndromic retinitis pigmentosa. Biochem. Soc. Trans. 2018, 46, 1463–1473. [Google Scholar] [CrossRef]
- Yang, M.; So, K.F.; Lam, W.C.; Lo, A. Cell Ferroptosis: New Mechanism and New Hope for Retinitis Pigmentosa. Cells 2021, 10, 2153. [Google Scholar] [CrossRef]
- Jiang, K.; Mondal, A.K.; Adlakha, Y.K.; Gumerson, J.; Aponte, A.; Gieser, L.; Kim, J.; Boleda, A.; Brooks, M.J.; Nellissery, J.; et al. Multiomics analyses reveal early metabolic imbalance and mitochondrial stress in neonatal photoreceptors leading to cell death in Pde6brd1/rd1 mouse model of retinal degeneration. Hum. Mol. Genet. 2022, 31, 2137–2154. [Google Scholar] [CrossRef]
- Cavusoglu, N.; Thierse, D.; Mohand-Saïd, S.; Chalmel, F.; Poch, O.; Van-Dorsselaer, A.; Sahel, J.; Léveillard, T. Differential proteomic analysis of the mouse retina: The induction of crystallin proteins by retinal degeneration in the rd1 mouse. Mol. Cell Proteom. 2003, 2, 494–505. [Google Scholar] [CrossRef] [PubMed]
- Gimeno-Hernández, R.; Cantó, A.; Fernández-Carbonell, A.; Olivar, T.; Hernández-Rabaza, V.; Almansa, I.; Miranda, M. Thioredoxin Delays Photoreceptor Degeneration, Oxidative and Inflammation Alterations in Retinitis Pigmentosa. Front. Pharmacol. 2020, 11, 590572. [Google Scholar] [CrossRef] [PubMed]
- Hackam, A.S.; Strom, R.; Liu, D.; Qian, J.; Wang, C.; Otteson, D.; Gunatilaka, T.; Farkas, R.H.; Chowers, I.; Kageyama, M.; et al. Identification of gene expression changes associated with the progression of retinal degeneration in the rd1 mouse. Investig. Ophthalmol. Vis. Sci. 2004, 45, 2929. [Google Scholar] [CrossRef]
- Hwang, Y.H.; Kim, S.; Kim, Y.Y.; Na, J.H.; Kim, H.K.; Sohn, Y.H. Optic Nerve Head, Retinal Nerve Fiber Layer, and Macular Thickness Measurements in Young Patients with Retinitis Pigmentosa. Curr. Eye Res. 2012, 37, 914–920. [Google Scholar] [CrossRef]
- Gong, Y.; Xia, H.; Zhang, A.; Chen, L.J.; Chen, H. Optical coherence tomography biomarkers of photoreceptor degeneration in retinitis pigmentosa. Int. Ophthalmol. 2021, 41, 3949–3959. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Humayun, M.S.; de Juan, E.J.; Greenburg, R.J.; Marsh, M.J.; Klock, I.B.; Milam, A.H. Preservation of the inner retina in retinitis pigmentosa. A morphometric analysis. Arch. Ophthalmol. 1997, 115, 511–515. [Google Scholar] [CrossRef]
- Jang, M.J.; Coughlin, G.M.; Jackson, C.R.; Chen, X.; Chuapoco, M.R.; Vendemiatti, J.L.; Wang, A.Z.; Gradinaru, V. Spatial transcriptomics for profiling the tropism of viral vectors in tissues. Nat. Biotechnol. 2023, 41, 1272–1286. [Google Scholar] [CrossRef]
- Picelli, S.; Faridani, O.R.; Bjorklund, A.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171–181. [Google Scholar] [CrossRef]
- Nichterwitz, S.; Chen, G.; Aguila Benitez, J.; Yilmaz, M.; Storvall, H.; Cao, M.; Sandberg, R.; Deng, Q.; Hedlund, E. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat. Commun. 2016, 7, 12139. [Google Scholar] [CrossRef] [PubMed]
- Frost, A.R.; Eltoum, I.E.; Siegal, G.P.; Emmert Buck, M.R.; Tangrea, M.A. Laser Microdissection. Curr. Protoc. Mol. Biol. 2015, 112, 25A-1. [Google Scholar] [CrossRef] [PubMed]
- Pembroke, W.G.; Babbs, A.; Davies, K.E.; Ponting, C.P.; Oliver, P.L. Temporal transcriptomics suggest that twin-peaking genes reset the clock. eLife 2015, 4, e10518. [Google Scholar] [CrossRef]
- Zechel, S.; Zajac, P.; Lonnerberg, P.; Ibanez, C.F.; Linnarsson, S. Topographical transcriptome mapping of the mouse medial ganglionic eminence by spatially resolved RNA-seq. Genome Biol. 2014, 15, 486. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Vingolo, E.M.; Casillo, L.; Contento, L.; Toja, F.; Florido, A. Retinitis Pigmentosa (RP): The Role of Oxidative Stress in the Degenerative Process Progression. Biomedicines 2022, 10, 582. [Google Scholar] [CrossRef]
- Malik, A.; Sood, S.; Narang, S. Successful treatment of choroidal neovascular membrane in retinitis pigmentosa with intravitreal bevacizumab. Int. Ophthalmol. 2010, 30, 425–428. [Google Scholar] [CrossRef]
- Miyata, M.; Oishi, A.; Oishi, M.; Hasegawa, T.; Ikeda, H.O.; Tsujikawa, A. Long-term efficacy and safety of anti-VEGF therapy in retinitis pigmentosa: A case report. BMC Ophthalmol. 2018, 18, 248. [Google Scholar] [CrossRef]
- Hong, M.; Wen, J.; Lou, J.; Xu, J.; Xu, T. Outer Macular Microvascular Supply in Retinitis Pigmentosa Examined using Optical Coherence Tomography Angiography. J. Ophthalmol. 2021, 2021, 5575851. [Google Scholar] [CrossRef]
- Lahdenranta, J.; Pasqualini, R.; Schlingemann, R.O.; Hagedorn, M.; Stallcup, W.B.; Bucana, C.D.; Sidman, R.L.; Arap, W. An anti-angiogenic state in mice and humans with retinal photoreceptor cell degeneration. Proc. Natl. Acad. Sci. USA 2001, 98, 10368–10373. [Google Scholar] [CrossRef]
- Broggini, T.; Stange, L.; Lucia, K.E.; Vajkoczy, P.; Czabanka, M. Endothelial EphrinB2 Regulates Sunitinib Therapy Response in Murine Glioma. Life 2022, 12, 691. [Google Scholar] [CrossRef]
- Li, Y.; Chen, D.; Sun, L.; Wu, Y.; Zou, Y.; Liang, C.; Bao, Y.; Yi, J.; Zhang, Y.; Hou, J.; et al. Induced Expression of VEGFC, ANGPT, and EFNB2 and Their Receptors Characterizes Neovascularization in Proliferative Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2019, 60, 4084–4096. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, C.; Fantin, A.; Lampropoulou, A.; Denti, L.; Chikh, A.; Ruhrberg, C. Imatinib inhibits VEGF-independent angiogenesis by targeting neuropilin 1-dependent ABL1 activation in endothelial cells. J. Exp. Med. 2014, 211, 1167–1183. [Google Scholar] [CrossRef]
- Petros, T.J.; Bryson, J.B.; Mason, C. Ephrin-B2 elicits differential growth cone collapse and axon retraction in retinal ganglion cells from distinct retinal regions. Dev. Neurobiol. 2010, 70, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Sawamiphak, S.; Seidel, S.; Essmann, C.L.; Wilkinson, G.A.; Pitulescu, M.E.; Acker, T.; Acker-Palmer, A. Ephrin-B2 regulates VEGFR2 function in developmental and tumour angiogenesis. Nature 2010, 465, 487–491. [Google Scholar] [CrossRef]
- Wang, Y.; Nakayama, M.; Pitulescu, M.E.; Schmidt, T.S.; Bochenek, M.L.; Sakakibara, A.; Adams, S.; Davy, A.; Deutsch, U.; Luthi, U.; et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 2010, 465, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Steinle, J.J.; Meininger, C.J.; Chowdhury, U.; Wu, G.; Granger, H.J. Role of ephrin B2 in human retinal endothelial cell proliferation and migration. Cell. Signal. 2003, 15, 1011–1017. [Google Scholar] [CrossRef]
- Palmer, A.; Zimmer, M.; Erdmann, K.S.; Eulenburg, V.; Porthin, A.; Heumann, R.; Deutsch, U.; Klein, R. EphrinB phosphorylation and reverse signaling: Regulation by Src kinases and PTP-BL phosphatase. Mol. Cell 2002, 9, 725–737. [Google Scholar] [CrossRef]
- Abengozar, M.A.; de Frutos, S.; Ferreiro, S.; Soriano, J.; Perez-Martinez, M.; Olmeda, D.; Marenchino, M.; Canamero, M.; Ortega, S.; Megias, D.; et al. Blocking ephrinB2 with highly specific antibodies inhibits angiogenesis, lymphangiogenesis, and tumor growth. Blood 2012, 119, 4565–4576. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, F.; Ma, Y.; Xue, T.; Shen, Y. Identification of early-onset photoreceptor degeneration in transgenic mouse models of Alzheimer’s disease. Iscience 2021, 24, 103327. [Google Scholar] [CrossRef]
- Móvio, M.I.; de Lima-Vasconcellos, T.H.; Dos Santos, G.B.; Echeverry, M.B.; Colombo, E.; Mattos, L.S.; Resende, R.R.; Kihara, A.H. Retinal organoids from human-induced pluripotent stem cells: From studying retinal dystrophies to early diagnosis of Alzheimer’s and Parkinson’s disease. Semin. Cell Dev. Biol. 2023, 144, 77–86. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Sheng, Y.; Pan, M.; Tu, J.; Zhao, X.; Ge, Q.; Lu, Z. Spatial Transcriptomic Analysis Reveals Regional Transcript Changes in Early and Late Stages of rd1 Model Mice with Retinitis Pigmentosa. Int. J. Mol. Sci. 2023, 24, 14869. https://doi.org/10.3390/ijms241914869
Zhou Y, Sheng Y, Pan M, Tu J, Zhao X, Ge Q, Lu Z. Spatial Transcriptomic Analysis Reveals Regional Transcript Changes in Early and Late Stages of rd1 Model Mice with Retinitis Pigmentosa. International Journal of Molecular Sciences. 2023; 24(19):14869. https://doi.org/10.3390/ijms241914869
Chicago/Turabian StyleZhou, Ying, Yuqi Sheng, Min Pan, Jing Tu, Xiangwei Zhao, Qinyu Ge, and Zuhong Lu. 2023. "Spatial Transcriptomic Analysis Reveals Regional Transcript Changes in Early and Late Stages of rd1 Model Mice with Retinitis Pigmentosa" International Journal of Molecular Sciences 24, no. 19: 14869. https://doi.org/10.3390/ijms241914869
APA StyleZhou, Y., Sheng, Y., Pan, M., Tu, J., Zhao, X., Ge, Q., & Lu, Z. (2023). Spatial Transcriptomic Analysis Reveals Regional Transcript Changes in Early and Late Stages of rd1 Model Mice with Retinitis Pigmentosa. International Journal of Molecular Sciences, 24(19), 14869. https://doi.org/10.3390/ijms241914869






