High and Low Levels of ABCB1 Expression Are Associated with Two Distinct Gene Signatures in Lung Tissue of Pulmonary TB Patients with High Inflammation Activity
Abstract
:1. Introduction
2. Results
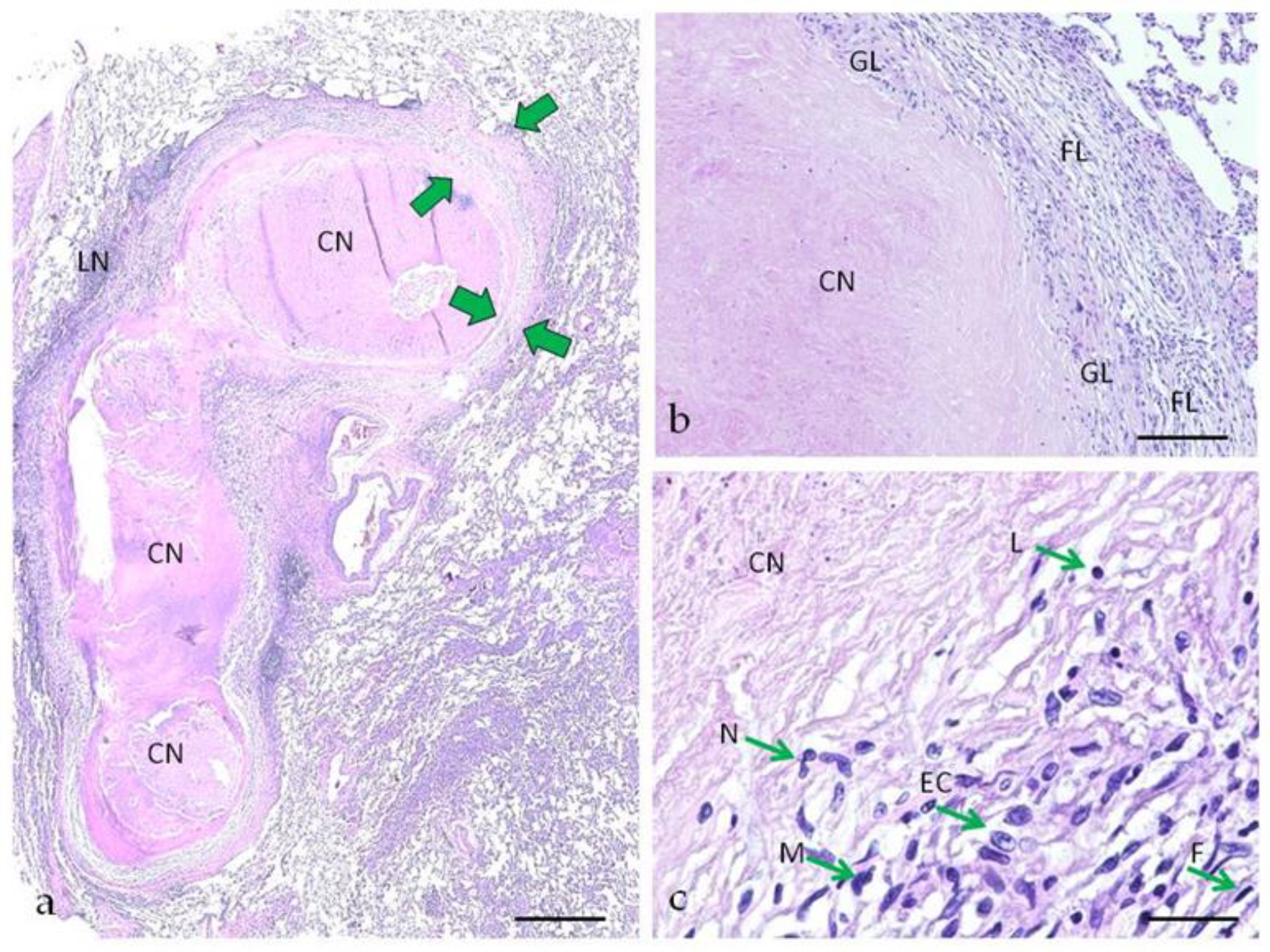
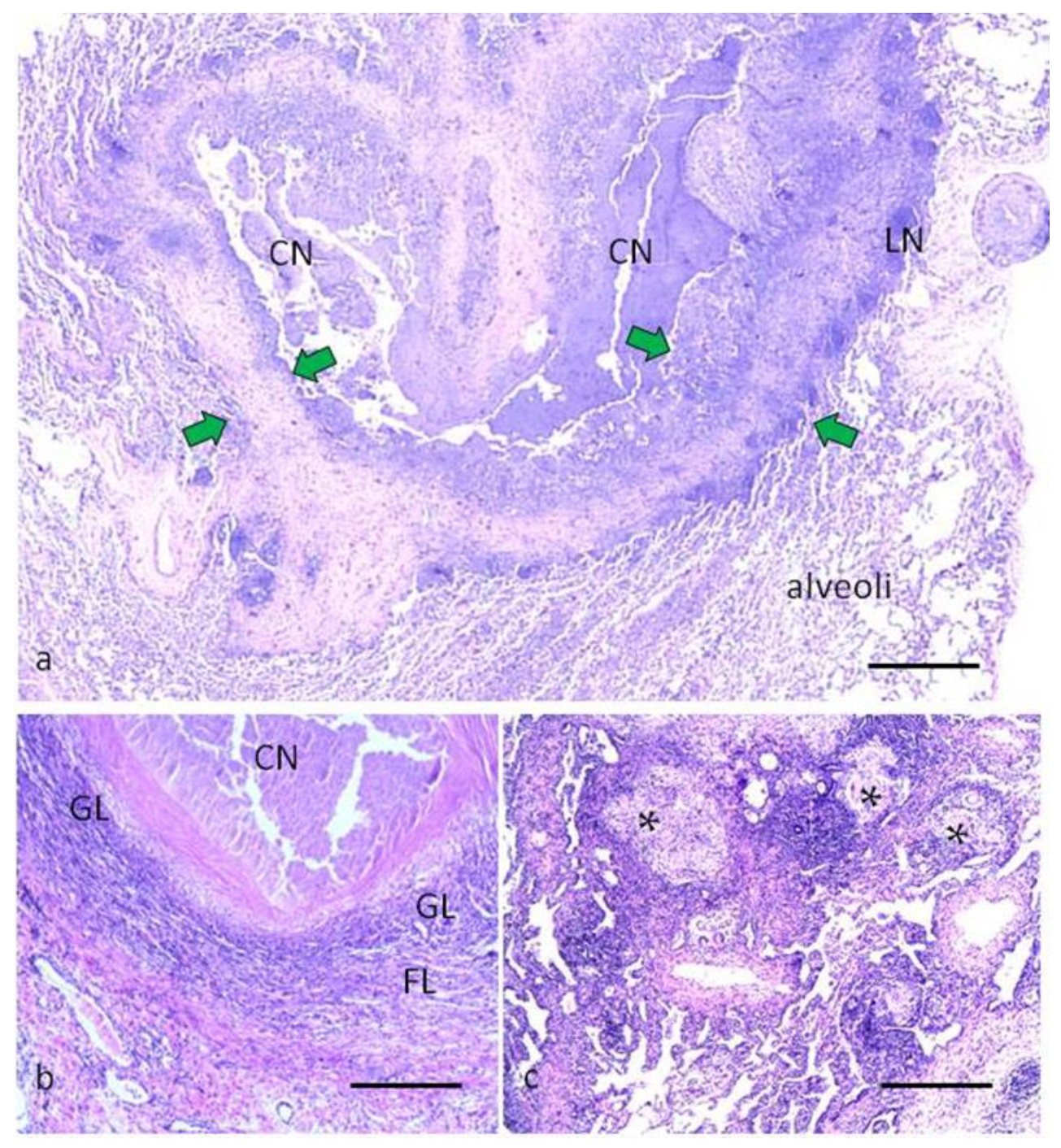
2.1. Morphological Characteristics of Pulmonary Tuberculoma with Moderate and High Activity of Tuberculosis Inflammation
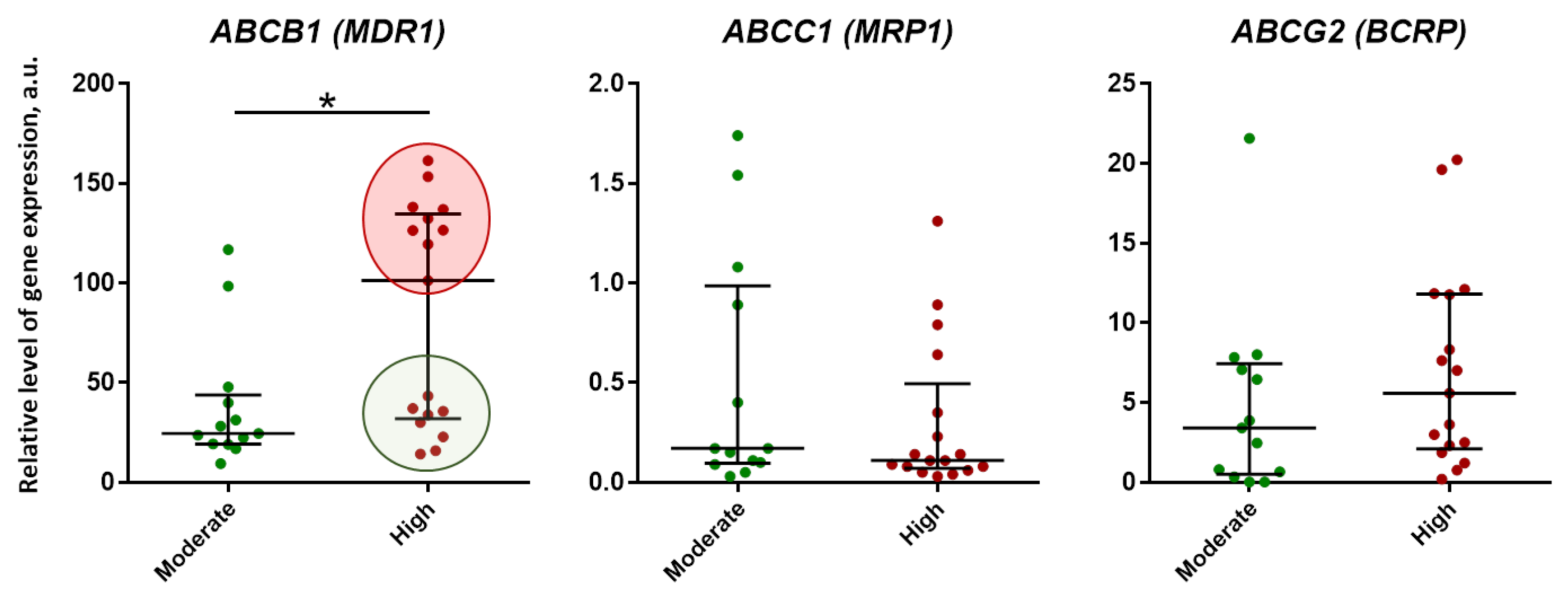
2.2. Expression of ABC-Transporter (P-gp, MRP1, BCRP) Genes in Lung Tissue of TB Patients with Pulmonary Tuberculoma
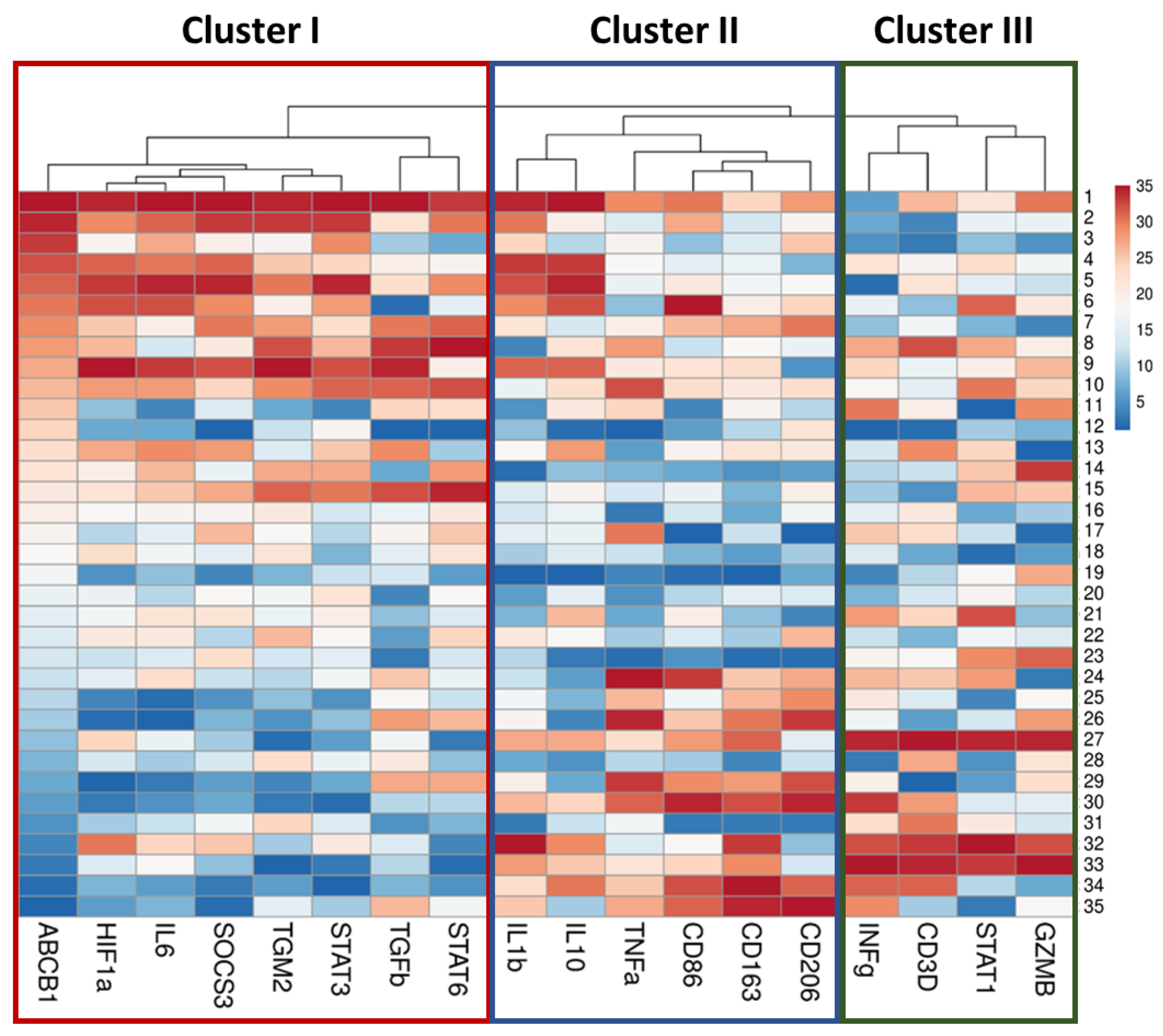
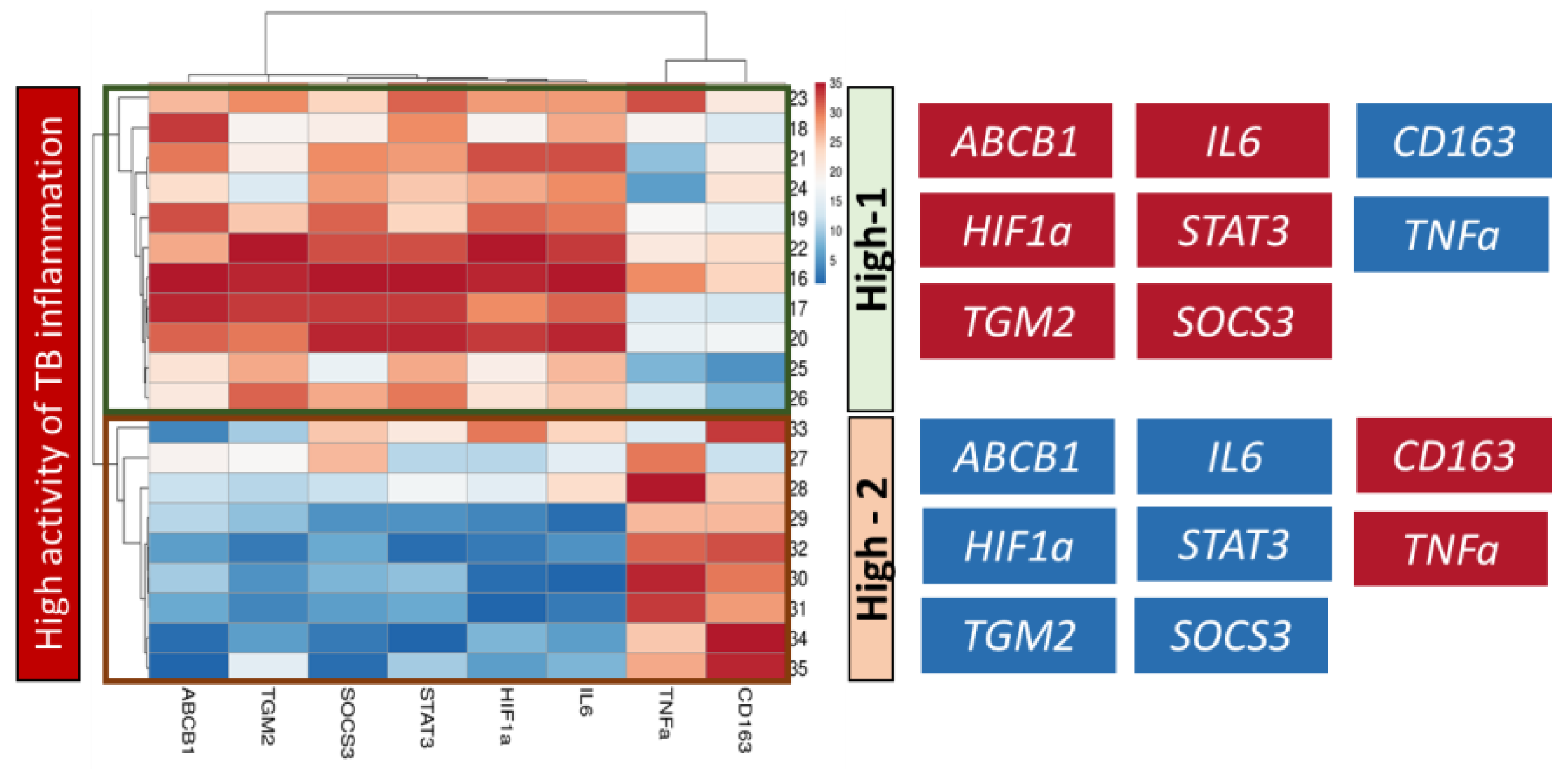
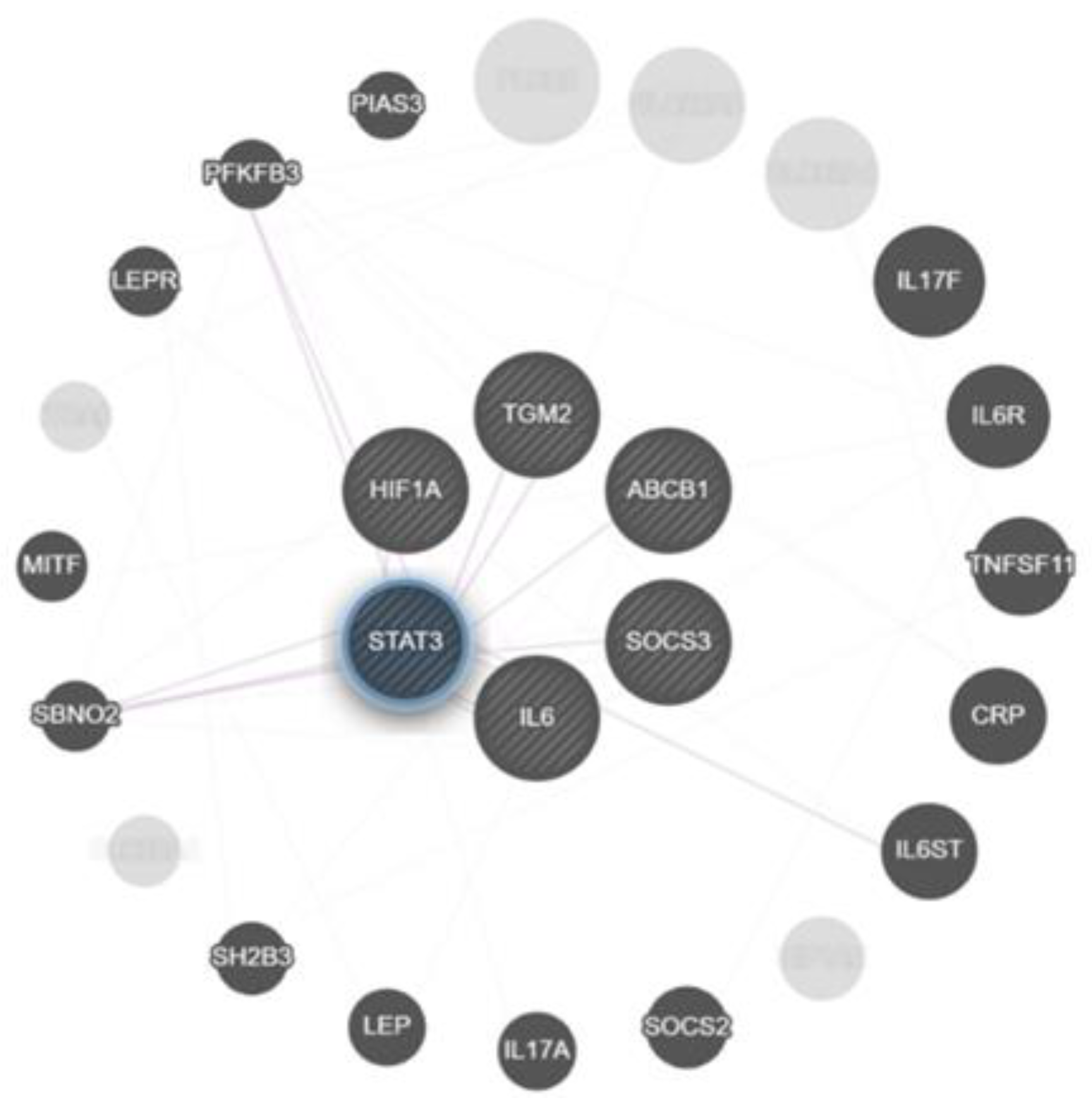
2.3. Correlation Analysis of Genes Expression in Lung Tissue of TB Patients
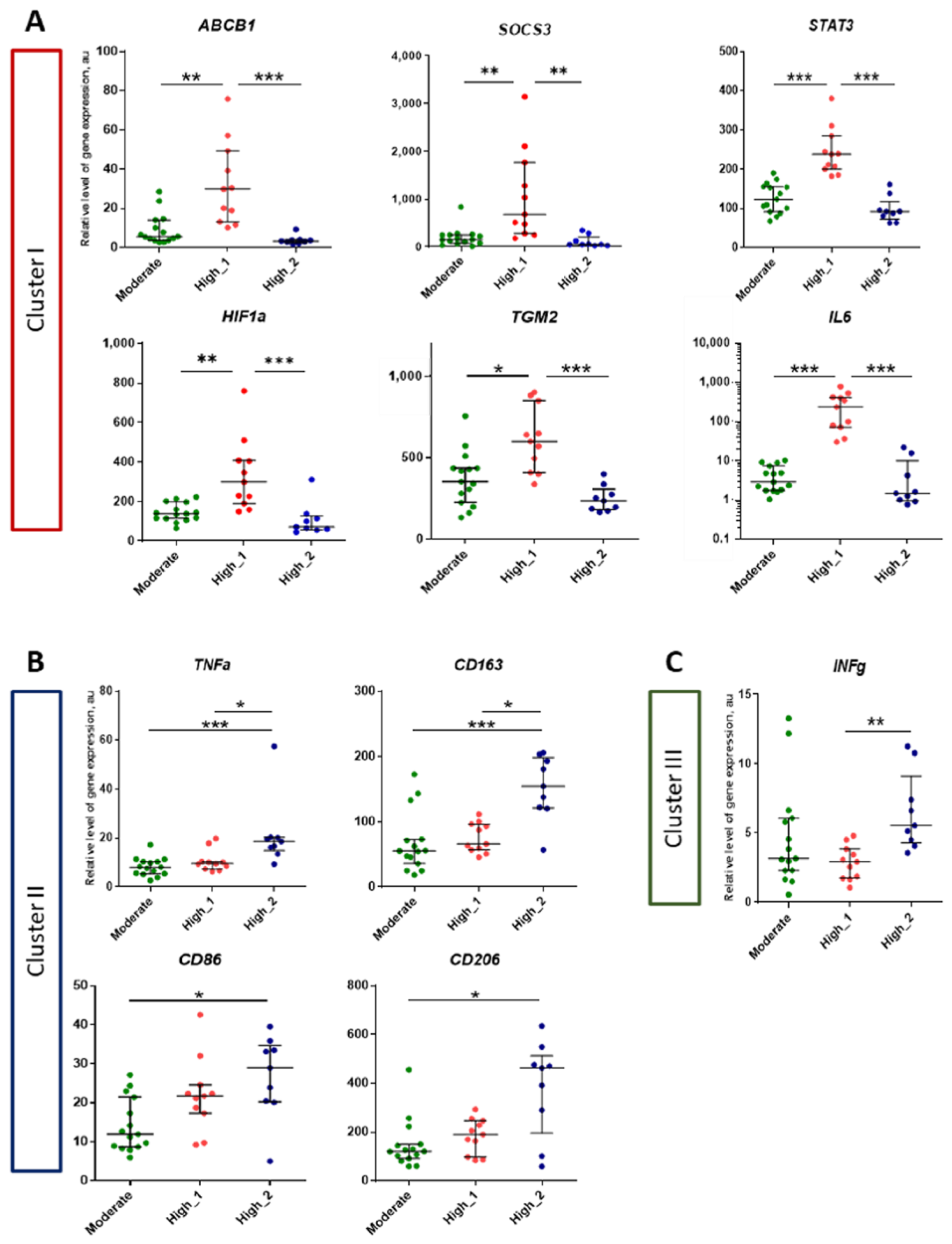
2.4. Analysis of Genes Expression in Lung Tissue of TB Patients Depending on the Activity of Inflammation
3. Discussion
4. Materials and Methods
4.1. Patients and Lung Tissue Samples
4.2. Histology
4.3. Gene Expression Analyses
4.4. Data Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Global Tuberculosis Report. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2022 (accessed on 25 June 2023).
- Dean, A.S.; Auguet, O.T.; Glaziou, P.; Zignol, M.; Ismail, N.; Kasaeva, T.; Floyd, K. 25 Years of Surveillance of Drug-Resistant Tuberculosis: Achievements, Challenges, and Way Forward. Lancet Infect. Dis. 2022, 22, e191–e196. [Google Scholar] [CrossRef]
- Andres, S.; Merker, M.; Heyckendorf, J.; Kalsdorf, B.; Rumetshofer, R.; Indra, A.; Hofmann-Thiel, S.; Hoffmann, H.; Lange, C.; Niemann, S.; et al. Bedaquiline-Resistant Tuberculosis: Dark Clouds on the Horizon. Am. J. Respir. Crit. Care Med. 2020, 201, 1564–1568. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.A.; Omar, S.V.; Moultrie, H.; Bhyat, Z.; Conradie, F.; Enwerem, M.; Ferreira, H.; Hughes, J.; Joseph, L.; Kock, Y.; et al. Assessment of Epidemiological and Genetic Characteristics and Clinical Outcomes of Resistance to Bedaquiline in Patients Treated for Rifampicin-Resistant Tuberculosis: A Cross-Sectional and Longitudinal Study. Lancet Infect. Dis. 2022, 22, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Tobin, D.M. Host-Directed Therapies for Tuberculosis. Cold Spring Harb. Perspect. Med. 2015, 5, a021196. [Google Scholar] [CrossRef] [PubMed]
- Kolloli, A.; Subbian, S. Host-Directed Therapeutic Strategies for Tuberculosis. Front. Med. 2017, 4, 171. [Google Scholar] [CrossRef]
- Ahmed, S.; Raqib, R.; Guðmundsson, G.H.; Bergman, P.; Agerberth, B.; Rekha, R.S. Host-Directed Therapy as a Novel Treatment Strategy to Overcome Tuberculosis: Targeting Immune Modulation. Antibiotics 2020, 9, 21. [Google Scholar] [CrossRef]
- Tiwari, D.; Martineau, A.R. Inflammation-Mediated Tissue Damage in Pulmonary Tuberculosis and Host-Directed Therapeutic Strategies. Semin. Immunol. 2023, 65, 101672. [Google Scholar] [CrossRef]
- Young, C.; Walzl, G.; du Plessis, N. Therapeutic Host-Directed Strategies to Improve Outcome in Tuberculosis. Mucosal Immunol. 2020, 13, 190–204. [Google Scholar] [CrossRef]
- Wallis, R.S.; Ginindza, S.; Beattie, T.; Arjun, N.; Likoti, M.; Sebe, M.; Edward, V.A.; Rassool, M.; Ahmed, K.; Fielding, K.; et al. Lung and Blood Early Biomarkers for Host-Directed Tuberculosis Therapies: Secondary Outcome Measures from a Randomized Controlled Trial. PLoS ONE 2022, 17, e0252097. [Google Scholar] [CrossRef]
- Te Brake, L.H.M.; de Knegt, G.J.; de Steenwinkel, J.E.; van Dam, T.J.P.; Burger, D.M.; Russel, F.G.M.; van Crevel, R.; Koenderink, J.B.; Aarnoutse, R.E. The Role of Efflux Pumps in Tuberculosis Treatment and Their Promise as a Target in Drug Development: Unraveling the Black Box. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 271–291. [Google Scholar] [CrossRef]
- Juliano, R.L.; Ling, V. A Surface Glycoprotein Modulating Drug Permeability in Chinese Hamster Ovary Cell Mutants. Biochim. Biophys. Acta 1976, 455, 152–162. [Google Scholar] [CrossRef] [PubMed]
- P-Glycoprotein Substrates|DrugBank Online. Available online: https://go.drugbank.com/categories/DBCAT002668 (accessed on 25 June 2023).
- Seelig, A. P-Glycoprotein: One Mechanism, Many Tasks and the Consequences for Pharmacotherapy of Cancers. Front. Oncol. 2020, 10, 576559. [Google Scholar] [CrossRef] [PubMed]
- Campbell, L.; Abulrob, A.-N.G.; Kandalaft, L.E.; Plummer, S.; Hollins, A.J.; Gibbs, A.; Gumbleton, M. Constitutive Expression of P-Glycoprotein in Normal Lung Alveolar Epithelium and Functionality in Primary Alveolar Epithelial Cultures. J. Pharmacol. Exp. Ther. 2003, 304, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Bossennec, M.; di Roio, A.; Caux, C.; Ménétrier-Caux, C. MDR1 in Immunity: Friend or Foe? Oncoimmunology 2018, 7, e1499388. [Google Scholar] [CrossRef] [PubMed]
- Kooij, G.; Backer, R.; Koning, J.J.; Reijerkerk, A.; van Horssen, J.; van der Pol, S.M.A.; Drexhage, J.; Schinkel, A.; Dijkstra, C.D.; den Haan, J.M.M.; et al. P-Glycoprotein Acts as an Immunomodulator during Neuroinflammation. PLoS ONE 2009, 4, e8212. [Google Scholar] [CrossRef]
- van der Deen, M.; Timens, W.; Timmer-Bosscha, H.; van der Strate, B.W.; Scheper, R.J.; Postma, D.S.; de Vries, E.G.; Kerstjens, H.A. Reduced Inflammatory Response in Cigarette Smoke Exposed Mrp1/Mdr1a/1b Deficient Mice. Respir. Res. 2007, 8, 49. [Google Scholar] [CrossRef]
- Hartkoorn, R.C.; Chandler, B.; Owen, A.; Ward, S.A.; Bertel Squire, S.; Back, D.J.; Khoo, S.H. Differential Drug Susceptibility of Intracellular and Extracellular Tuberculosis, and the Impact of P-Glycoprotein. Tuberculosis 2007, 87, 248–255. [Google Scholar] [CrossRef]
- Gollapudi, S.; Reddy, M.; Gangadharam, P.; Tsuruo, T.; Gupta, S. Mycobacterium Tuberculosis Induces Expression of P-Glycoprotein in Promonocytic U1 Cells Chronically Infected with HIV Type 1. Biochem. Biophys. Res. Commun. 1994, 199, 1181–1187. [Google Scholar] [CrossRef]
- Ding, Z.; Yang, L.; Xie, X.; Xie, F.; Pan, F.; Li, J.; He, J.; Liang, H. Expression and Significance of Hypoxia-Inducible Factor-1 Alpha and MDR1/P-Glycoprotein in Human Colon Carcinoma Tissue and Cells. J. Cancer Res. Clin. Oncol. 2010, 136, 1697–1707. [Google Scholar] [CrossRef]
- Xie, A.; Robles, R.J.; Mukherjee, S.; Zhang, H.; Feldbrügge, L.; Csizmadia, E.; Wu, Y.; Enjyoji, K.; Moss, A.C.; Otterbein, L.E.; et al. HIF-1α-Induced Xenobiotic Transporters Promote Th17 Responses in Crohn’s Disease. J. Autoimmun. 2018, 94, 122–133. [Google Scholar] [CrossRef]
- Ho, E.A.; Piquette-Miller, M. Regulation of Multidrug Resistance by Pro-Inflammatory Cytokines. Curr. Cancer Drug Targets 2006, 6, 295–311. [Google Scholar] [CrossRef]
- Liptrott, N.J.; Penny, M.; Bray, P.G.; Sathish, J.; Khoo, S.H.; Back, D.J.; Owen, A. The Impact of Cytokines on the Expression of Drug Transporters, Cytochrome P450 Enzymes and Chemokine Receptors in Human PBMC. Br. J. Pharmacol. 2009, 156, 497–508. [Google Scholar] [CrossRef]
- Bhagyaraj, E.; Tiwari, D.; Ahuja, N.; Nanduri, R.; Saini, A.; Kalra, R.; Kumar, S.; Janmeja, A.K.; Gupta, P. A Human Xenobiotic Nuclear Receptor Contributes to Nonresponsiveness of Mycobacterium Tuberculosis to the Antituberculosis Drug Rifampicin. J. Biol. Chem. 2018, 293, 3747–3757. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, E.N.; Erokhina, M.V.; Rybalkina, E.Y.; Potashnikova, D.M.; Masyutin, A.G.; Lepekha, L.N.; Ergeshov, A.E. The Effect of Rifampicin on the Induction of MDR1/P-gp Activity in Proinflammatory Human Macrophages. Antibiot. Chemother. 2022, 67, 16–22. [Google Scholar] [CrossRef]
- Kholodok, O.A.; Grigorenko, A.A.; Cheryemkin, M.I. Pulmonary tuberculoma as a form of tuberculous process. Bull. Physiol. Pathol. Respir. 2014, 53, 126–131. [Google Scholar]
- Giller, D.B.; Giller, B.D.; Giller, G.V.; Shcherbakova, G.V.; Bizhanov, A.B.; Enilenis, I.I.; Glotov, A.A. Treatment of Pulmonary Tuberculosis: Past and Present. Eur. J. Cardiothorac. Surg. 2018, 53, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Krasnikova, E.; Lepeha, L.; Tarasov, R.; Erokhina, M.; Aliev, V.; Sadovnikova, S.; Bagirov, M. Indicaions to Surgery for Lung Tuberculomas Today. In Proceedings of the Tuberculosis, European Respiratory Society, Madrid, Spain, 28 September 2019; p. PA2960. [Google Scholar]
- Rottenberg, M.E.; Carow, B. SOCS3 and STAT3, Major Controllers of the Outcome of Infection with Mycobacterium Tuberculosis. Semin. Immunol. 2014, 26, 518–532. [Google Scholar] [CrossRef]
- Yi, X.; Zhang, B.; Fu, Y.; Yi, Z. STAT1 and Its Related Molecules as Potential Biomarkers in Mycobacterium Tuberculosis Infection. J. Cell. Mol. Med. 2020, 24, 2866–2878. [Google Scholar] [CrossRef]
- Cronan, M.R.; Hughes, E.J.; Brewer, W.J.; Viswanathan, G.; Hunt, E.G.; Singh, B.; Mehra, S.; Oehlers, S.H.; Gregory, S.G.; Kaushal, D.; et al. A Non-Canonical Type 2 Immune Response Coordinates Tuberculous Granuloma Formation and Epithelialization. Cell 2021, 184, 1757–1774.e14. [Google Scholar] [CrossRef]
- Akter, S.; Chauhan, K.S.; Dunlap, M.D.; Choreño-Parra, J.A.; Lu, L.; Esaulova, E.; Zúñiga, J.; Artyomov, M.N.; Kaushal, D.; Khader, S.A. Mycobacterium Tuberculosis Infection Drives a Type I IFN Signature in Lung Lymphocytes. Cell Rep. 2022, 39, 110983. [Google Scholar] [CrossRef]
- Lungu, P.; Mushota, K.; Njelesani, E.; Sukwa, T.; Lakhi, S.; Mwaba, P. Expression of mycobacterium tuberculosis induced SOCS3 and STAT3 and the implications on innate immunity in TB patients vs healthy contacts in high TB/HIV endemic setting: A cross-sectional analytical study. PLoS ONE 2022, 17, e0263624. [Google Scholar] [CrossRef]
- Braverman, J.; Sogi, K.M.; Benjamin, D.; Nomura, D.K.; Stanley, S.A. HIF-1α Is an Essential Mediator of IFN-γ-Dependent Immunity to Mycobacterium Tuberculosis. J. Immunol. 2016, 197, 1287–1297. [Google Scholar] [CrossRef]
- Belton, M.; Brilha, S.; Manavaki, R.; Mauri, F.; Nijran, K.; Hong, Y.T.; Patel, N.H.; Dembek, M.; Tezera, L.; Green, J.; et al. Hypoxia and Tissue Destruction in Pulmonary TB. Thorax 2016, 71, 1145–1153. [Google Scholar] [CrossRef]
- McGettrick, A.F.; O’Neill, L.A.J. The Role of HIF in Immunity and Inflammation. Cell Metab. 2020, 32, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Silva Miranda, M.; Breiman, A.; Allain, S.; Deknuydt, F.; Altare, F. The Tuberculous Granuloma: An Unsuccessful Host Defence Mechanism Providing a Safety Shelter for the Bacteria? Clin. Dev. Immunol. 2012, 2012, 139127. [Google Scholar] [CrossRef]
- Domingo-Gonzalez, R.; Prince, O.; Cooper, A.; Khader, S.A. Cytokines and Chemokines in Mycobacterium Tuberculosis Infection. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Flynn, J.; Chan, J.; Lin, P. Macrophages and Control of Granulomatous Inflammation in Tuberculosis. Mucosal Immunol. 2011, 4, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Marino, S.; Cilfone, N.A.; Mattila, J.T.; Linderman, J.J.; Flynn, J.L.; Kirschner, D.E. Macrophage Polarization Drives Granuloma Outcome during Mycobacterium Tuberculosis Infection. Infect. Immun. 2015, 83, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Lim, Y.-J.; Kim, J.; Koh, W.-J.; Song, C.-H.; Kang, M.-W. Different Macrophage Polarization between Drug-Susceptible and Multidrug-Resistant Pulmonary Tuberculosis. BMC Infect. Dis. 2020, 20, 81. [Google Scholar] [CrossRef]
- Cory, T.J.; He, H.; Winchester, L.C.; Kumar, S.; Fletcher, C.V. Alterations in P-Glycoprotein Expression and Function Between Macrophage Subsets. Pharm. Res. 2016, 33, 2713–2721. [Google Scholar] [CrossRef]
- Immune Cell—ABCB1—The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000085563-ABCB1/immune+cell (accessed on 25 June 2023).
- Martinez, F.O.; Gordon, S. The M1 and M2 Paradigm of Macrophage Activation: Time for Reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef]
- Cho, S.-Y.; Oh, Y.; Jeong, E.M.; Park, S.; Lee, D.; Wang, X.; Zeng, Q.; Qin, H.; Hu, F.; Gong, H.; et al. Amplification of Transglutaminase 2 Enhances Tumor-Promoting Inflammation in Gastric Cancers. Exp. Mol. Med. 2020, 52, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Agnihotri, N.; Kumar, S.; Mehta, K. Tissue Transglutaminase as a Central Mediator in Inflammation-Induced Progression of Breast Cancer. Breast Cancer Res. 2013, 15, 202. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.-M.; Jang, S.-J.; Park, S.-Y.; Choi, Y.-B.; Jeong, J.-H.; Kim, D.-S.; Kim, H.-K.; Park, K.-S.; Nam, B.-H.; Kim, H.-R.; et al. Transglutaminase 2 as an Independent Prognostic Marker for Survival of Patients with Non-Adenocarcinoma Subtype of Non-Small Cell Lung Cancer. Mol. Cancer 2011, 10, 119. [Google Scholar] [CrossRef]
- Palucci, I.; Matic, I.; Falasca, L.; Minerva, M.; Maulucci, G.; de Spirito, M.; Petruccioli, E.; Goletti, D.; Rossin, F.; Piacentini, M.; et al. Transglutaminase Type 2 Plays a Key Role in the Pathogenesis of Mycobacterium Tuberculosis Infection. J. Intern. Med. 2018, 283, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Nwabufo, C.K.; Hoque, M.T.; Yip, L.; Khara, M.; Mubareka, S.; Pollanen, M.S.; Bendayan, R. SARS-CoV-2 Infection Dysregulates the Expression of Clinically Relevant Drug Metabolizing Enzymes in Vero E6 Cells and Membrane Transporters in Human Lung Tissues. Front. Pharmacol. 2023, 14, 1124693. [Google Scholar] [CrossRef]
- Rodríguez-Castillo, J.A.; Arce-Mendoza, A.Y.; Quintanilla-Siller, A.; Rendon, A.; Salinas-Carmona, M.C.; Rosas-Taraco, A.G. Possible Association of Rare Polymorphism in the ABCB1 Gene with Rifampin and Ethambutol Drug-Resistant Tuberculosis. Tuberculosis 2015, 95, 532–537. [Google Scholar] [CrossRef]
- Pontual, Y.; Pacheco, V.S.S.; Monteiro, S.P.; Quintana, M.S.B.; Costa, M.J.M.; Rolla, V.C.; de Castro, L. ABCB1 Gene Polymorphism Associated with Clinical Factors Can Predict Drug-Resistant Tuberculosis. Clin. Sci. 2017, 131, 1831–1840. [Google Scholar] [CrossRef]
- Zhang, X.; Xiao, W.; Wang, L.; Tian, Z.; Zhang, J. Deactivation of Signal Transducer and Activator of Transcription 3 Reverses Chemotherapeutics Resistance of Leukemia Cells via Down-Regulating P-Gp. PLoS ONE 2011, 6, e20965. [Google Scholar] [CrossRef]
- Chen, Q.; Bian, Y.; Zeng, S. Involvement of AP-1 and NF-κB in the Up-Regulation of P-Gp in Vinblastine Resistant Caco-2 Cells. Drug Metab. Pharmacokinet. 2014, 29, 223–226. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, F.; Chen, Q.; Kang, A.; Lu, M.; Liu, W.; Zang, X.; Wang, G.; Zhang, J. Chronic Inflammation Up-Regulates P-Gp in Peripheral Mononuclear Blood Cells via the STAT3/Nf-Κb Pathway in 2,4,6-Trinitrobenzene Sulfonic Acid-Induced Colitis Mice. Sci. Rep. 2015, 5, 13558. [Google Scholar] [CrossRef] [PubMed]
- Comerford, K.M.; Wallace, T.J.; Karhausen, J.; Louis, N.A.; Montalto, M.C.; Colgan, S.P. Hypoxia-Inducible Factor-1-Dependent Regulation of the Multidrug Resistance (MDR1) Gene. Cancer Res. 2002, 62, 3387–3394. [Google Scholar] [PubMed]
- Subbian, S.; Tsenova, L.; Kim, M.-J.; Wainwright, H.C.; Visser, A.; Bandyopadhyay, N.; Bader, J.S.; Karakousis, P.C.; Murrmann, G.B.; Bekker, L.-G.; et al. Lesion-Specific Immune Response in Granulomas of Patients with Pulmonary Tuberculosis: A Pilot Study. PLoS ONE 2015, 10, e0132249. [Google Scholar] [CrossRef] [PubMed]
- Abengozar-Muela, M.; Esparza, M.V.; Garcia-Ros, D.; Vásquez, C.E.; Echeveste, J.I.; Idoate, M.A.; Lozano, M.D.; Melero, I.; de Andrea, C.E. Diverse Immune Environments in Human Lung Tuberculosis Granulomas Assessed by Quantitative Multiplexed Immunofluorescence. Mod. Pathol. 2020, 33, 2507–2519. [Google Scholar] [CrossRef]
- McCaffrey, E.F.; Donato, M.; Keren, L.; Chen, Z.; Delmastro, A.; Fitzpatrick, M.B.; Gupta, S.; Greenwald, N.F.; Baranski, A.; Graf, W.; et al. The Immunoregulatory Landscape of Human Tuberculosis Granulomas. Nat. Immunol. 2022, 23, 318–329. [Google Scholar] [CrossRef]
- Sawyer, A.J.; Patrick, E.; Edwards, J.; Wilmott, J.S.; Fielder, T.; Yang, Q.; Barber, D.L.; Ernst, J.D.; Britton, W.J.; Palendira, U.; et al. Spatial Mapping Reveals Granuloma Diversity and Histopathological Superstructure in Human Tuberculosis. J. Exp. Med. 2023, 220, e20221392. [Google Scholar] [CrossRef]
- Lenaerts, A.; Barry, C.E.; Dartois, V. Heterogeneity in Tuberculosis Pathology, Microenvironments and Therapeutic Responses. Immunol. Rev. 2015, 264, 288–307. [Google Scholar] [CrossRef]
- Hunter, R.L. The Pathogenesis of Tuberculosis–The Koch Phenomenon Reinstated. Pathogens 2020, 9, 813. [Google Scholar] [CrossRef]
- Erokhina, M.V.; Lepekha, L.N.; Voronezhskaya, E.E.; Nezlin, L.P.; Avdienko, V.G.; Ergeshov, A.E. Application of Laser Scanning Confocal Microscopy for the Visualization of M. Tuberculosis in Lung Tissue Samples with Weak Ziehl–Neelsen Staining. J. Clin. Med. 2019, 8, 1185. [Google Scholar] [CrossRef]
- Suzuki, Y.; Shirai, M.; Asada, K.; Miwa, S.; Karayama, M.; Nakamura, Y.; Inui, N.; Shirai, T.; Hayakawa, H.; Baba, S.; et al. Utility of Macrophage-Activated Marker CD163 for Diagnosis and Prognosis in Pulmonary Tuberculosis. Ann. Am. Thorac. Soc. 2017, 14, 57–64. [Google Scholar] [CrossRef]
- Suzuki, Y.; Shirai, M.; Asada, K.; Yasui, H.; Karayama, M.; Hozumi, H.; Furuhashi, K.; Enomoto, N.; Fujisawa, T.; Nakamura, Y.; et al. Macrophage Mannose Receptor, CD206, Predict Prognosis in Patients with Pulmonary Tuberculosis. Sci. Rep. 2018, 8, 13129. [Google Scholar] [CrossRef]
- Liu, Q.; Ou, Q.; Chen, H.; Gao, Y.; Liu, L.; Xu, Y.; Ruan, Q.; Zhang, W.; Zhang, L. Differential Expression and Predictive Value of Monocyte Scavenger Receptor CD163 in Populations with Different Tuberculosis Infection Statuses. BMC Infect. Dis. 2019, 19, 1006. [Google Scholar] [CrossRef]
- Chowdhury, I.H.; Ahmed, A.M.; Choudhuri, S.; Sen, A.; Hazra, A.; Pal, N.K.; Bhattacharya, B.; Bahar, B. Alteration of Serum Inflammatory Cytokines in Active Pulmonary Tuberculosis Following Anti-Tuberculosis Drug Therapy. Mol. Immunol. 2014, 62, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Fiorenza, G.; Rateni, L.; Farroni, M.A.; Bogué, C.; Dlugovitzky, D.G. TNF-α, TGF-β and NO Relationship in Sera from Tuberculosis (TB) Patients of Different Severity. Immunol. Lett. 2005, 98, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.P.; Moideen, K.; Banurekha, V.V.; Nair, D.; Babu, S. Plasma Proinflammatory Cytokines Are Markers of Disease Severity and Bacterial Burden in Pulmonary Tuberculosis. Open Forum Infect. Dis. 2019, 6, ofz257. [Google Scholar] [CrossRef] [PubMed]
- De Andrade Júnior, D.R.; dos Santos, S.A.; de Castro, I.; de Andrade, D.R. Correlation between Serum Tumor Necrosis Factor Alpha Levels and Clinical Severity of Tuberculosis. Braz. J. Infect. Dis. 2008, 12, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Beig, T.Y.; Khan, U.H.; Ganie, B.A.; Tahir, S.; Shah, S.; Dhobi, G.N. Correlation Between Serum Tumor Necrosis Factor-Alpha (TNF-α) and Clinical Severity of Tuberculosis: A Hospital-Based Study. Cureus 2023, 15, e35626. [Google Scholar] [CrossRef]
- Ghanavi, J.; Farnia, P.; Farnia, P.; Velayati, A.A. The Role of Interferon-Gamma and Interferon-Gamma Receptor in Tuberculosis and Nontuberculous Mycobacterial Infections. Int. J. Mycobacteriology 2021, 10, 349. [Google Scholar] [CrossRef]
- Dorhoi, A.; Kaufmann, S.H.E. Tumor Necrosis Factor Alpha in Mycobacterial Infection. Semin. Immunol. 2014, 26, 203–209. [Google Scholar] [CrossRef]
- van Loo, G.; Bertrand, M.J.M. Death by TNF: A Road to Inflammation. Nat. Rev. Immunol. 2023, 23, 289–303. [Google Scholar] [CrossRef]
- Tang, S.; Cui, H.; Yao, L.; Hao, X.; Shen, Y.; Fan, L.; Sun, H.; Zhang, Z.; Huang, J.A. Increased Cytokines Response in Patients with Tuberculosis Complicated with Chronic Obstructive Pulmonary Disease. PLoS ONE 2013, 8, e62385. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Beltrán, S.; Carreto-Binaghi, L.E.; Carranza, C.; Torres, M.; Gonzalez, Y.; Muñoz-Torrico, M.; Juárez, E. Oxidative Stress and Inflammatory Mediators in Exhaled Breath Condensate of Patients with Pulmonary Tuberculosis. A Pilot Study with a Biomarker Perspective. Antioxidants 2021, 10, 1572. [Google Scholar] [CrossRef]
- Heinrich, P.C.; Behrmann, I.; Haan, S.; Hermanns, H.M.; Müller-Newen, G.; Schaper, F. Principles of Interleukin (IL)-6-Type Cytokine Signalling and Its Regulation. Biochem. J. 2003, 374, 1–20. [Google Scholar] [CrossRef]
- Boni, F.G.; Hamdi, I.; Koundi, L.M.; Shrestha, K.; Xie, J. Cytokine Storm in Tuberculosis and IL-6 Involvement. Infect. Genet. Evol. 2022, 97, 105166. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Huang, G.; Shen, L.; Peng, Y.; Sha, W.; Chen, Z.W.; Shen, H. Genetics and Functional Mechanisms of STAT3 Polymorphisms in Human Tuberculosis. Front. Cell. Infect. Microbiol. 2021, 11, 669394. [Google Scholar] [CrossRef]
- Zhong, Z.; Wen, Z.; Darnell, J.E. Stat3: A STAT Family Member Activated by Tyrosine Phosphorylation in Response to Epidermal Growth Factor and Interleukin-6. Science 1994, 264, 95–98. [Google Scholar] [CrossRef]
- Yang, J.; Liao, X.; Agarwal, M.K.; Barnes, L.; Auron, P.E.; Stark, G.R. Unphosphorylated STAT3 Accumulates in Response to IL-6 and Activates Transcription by Binding to NFkappaB. Genes Dev. 2007, 21, 1396–1408. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhao, H.; Wang, P.; Wang, J.; Zou, L. The Roles of SOCS3 and STAT3 in Bacterial Infection and Inflammatory Diseases. Scand. J. Immunol. 2018, 88, e12727. [Google Scholar] [CrossRef] [PubMed]
- Adankwah, E.; Seyfarth, J.; Phillips, R.; Jacobsen, M. Aberrant Cytokine Milieu and Signaling Affect Immune Cell Phenotypes and Functions in Tuberculosis Pathology: What Can We Learn from This Phenomenon for Application to Inflammatory Syndromes? Cell. Mol. Immunol. 2021, 18, 2062–2064. [Google Scholar] [CrossRef] [PubMed]
- Harling, K.; Adankwah, E.; Güler, A.; Afum-Adjei Awuah, A.; Adu-Amoah, L.; Mayatepek, E.; Owusu-Dabo, E.; Nausch, N.; Jacobsen, M. Constitutive STAT3 Phosphorylation and IL-6/IL-10 Co-Expression Are Associated with Impaired T-Cell Function in Tuberculosis Patients. Cell. Mol. Immunol. 2019, 16, 275–287. [Google Scholar] [CrossRef]
- Piacentini, M.; D’Eletto, M.; Farrace, M.G.; Rodolfo, C.; del Nonno, F.; Ippolito, G.; Falasca, L. Characterization of Distinct Sub-Cellular Location of Transglutaminase Type II: Changes in Intracellular Distribution in Physiological and Pathological States. Cell Tissue Res. 2014, 358, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, N.; Sun, T.; Zhao, H.; Chen, Y.; Liu, C. Role of TGM2 in T-cell Lymphoblastic Lymphoma via Regulation of IL-6/JAK/STAT3 Signalling. Mol. Med. Rep. 2022, 25, 76. [Google Scholar] [CrossRef] [PubMed]
- Palucci, I.; Maulucci, G.; de Maio, F.; Sali, M.; Romagnoli, A.; Petrone, L.; Fimia, G.M.; Sanguinetti, M.; Goletti, D.; de Spirito, M.; et al. Inhibition of Transglutaminase 2 as a Potential Host-Directed Therapy Against Mycobacterium Tuberculosis. Front. Immunol. 2019, 10, 3042. [Google Scholar] [CrossRef]
- Kumar, S.; Mehta, K. Tissue Transglutaminase Constitutively Activates HIF-1α Promoter and Nuclear Factor-κB via a Non-Canonical Pathway. PLoS ONE 2012, 7, e49321. [Google Scholar] [CrossRef]
- O’Donnell, J.L.; Joyce, M.R.; Shannon, A.M.; Harmey, J.; Geraghty, J.; Bouchier-Hayes, D. Oncological Implications of Hypoxia Inducible Factor-1alpha (HIF-1alpha) Expression. Cancer Treat. Rev. 2006, 32, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Briggs, J.; Park, S.; Niu, G.; Kortylewski, M.; Zhang, S.; Gritsko, T.; Turkson, J.; Kay, H.; Semenza, G.L.; et al. Targeting Stat3 Blocks Both HIF-1 and VEGF Expression Induced by Multiple Oncogenic Growth Signaling Pathways. Oncogene 2005, 24, 5552–5560. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Muliadi, V.; Mou, W.; Li, H.; Yuan, J.; Holmberg, J.; Chambers, B.J.; Ullah, N.; Wurth, J.; Alzrigat, M.; et al. HIF-1 Stabilization in T Cells Hampers the Control of Mycobacterium Tuberculosis Infection. Nat. Commun. 2022, 13, 5093. [Google Scholar] [CrossRef]
- Bandarra, D.; Biddlestone, J.; Mudie, S.; Müller, H.-A.J.; Rocha, S. HIF-1α Restricts NF-κB-Dependent Gene Expression to Control Innate Immunity Signals. Dis. Models Mech. 2015, 8, 169–181. [Google Scholar] [CrossRef]
- Suresh, M.V.; Balijepalli, S.; Solanki, S.; Aktay, S.; Choudhary, K.; Shah, Y.M.; Raghavendran, K. Hypoxia-Inducible Factor 1α and Its Role in Lung Injury: Adaptive or Maladaptive. Inflammation 2023, 46, 491–508. [Google Scholar] [CrossRef]
- Nocera, A.L.; Meurer, A.T.; Miyake, M.M.; Sadow, P.M.; Han, X.; Bleier, B.S. Secreted P-Glycoprotein Is a Noninvasive Biomarker of Chronic Rhinosinusitis. Laryngoscope 2017, 127, E1–E4. [Google Scholar] [CrossRef]
- Perez-Guerrero, E.E.; Gonzalez-Lopez, L.; Muñoz-Valle, J.F.; Vasquez-Jimenez, J.C.; Ramirez-Villafaña, M.; Sanchez-Rodriguez, E.N.; Gutierrez-Ureña, S.R.; Cerpa-Cruz, S.; Aguilar-Chavez, E.A.; Cardona-Muñoz, E.G.; et al. Serum P-Glycoprotein Level: A Potential Biomarker of DMARD Failure in Patients with Rheumatoid Arthritis. Inflammopharmacology 2018. [Google Scholar] [CrossRef] [PubMed]
- Perez-Guerrero, E.E.; Gamez-Nava, J.I.; Muñoz-Valle, J.F.; Cardona-Muñoz, E.G.; Bonilla-Lara, D.; Fajardo-Robledo, N.S.; Nava-Zavala, A.H.; Garcia-Cobian, T.A.; Rincón-Sánchez, A.R.; Murillo-Vazquez, J.D.; et al. Serum Levels of P-Glycoprotein and Persistence of Disease Activity despite Treatment in Patients with Systemic Lupus Erythematosus. Clin. Exp. Med. 2018, 18, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, N.; Arumugam, M. Meta-Analysis of Active Tuberculosis Gene Expression Ascertains Host Directed Drug Targets. Front. Cell. Infect. Microbiol. 2022, 12, 1010771. [Google Scholar] [CrossRef] [PubMed]
- Burel, J.G.; Lindestam Arlehamn, C.S.; Khan, N.; Seumois, G.; Greenbaum, J.A.; Taplitz, R.; Gilman, R.H.; Saito, M.; Vijayanand, P.; Sette, A.; et al. Transcriptomic Analysis of CD4+ T Cells Reveals Novel Immune Signatures of Latent Tuberculosis. J. Immunol. 2018, 200, 3283–3290. [Google Scholar] [CrossRef]
- Perelman, M.I. (Ed.) Phthisiology. National Guidelines; GEOTAR-Media: Moscow, Russia, 2007; pp. 288–295. (In Russian) [Google Scholar]
- Metsalu, T.; Vilo, J. ClustVis: A Web Tool for Visualizing Clustering of Multivariate Data Using Principal Component Analysis and Heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]



| Gene | Forward Primer | Reverse Primer | Product Length, bp |
|---|---|---|---|
| ABCB1 | GGGATGGTCAGTGTTGATGGA | GCTATCGTGGTGGCAAACAATA | 110 |
| ABCC1 | GTGAATCGTGGCATCGACATA | GCTTGGGACGGAAGGGAATC | 184 |
| ABCG2 | TGAGCCTACAACTGGCTTAGA | CCCTGCTTAGACATCCTTTTCAG | 75 |
| RPL27 | ACCGCTACCCCCGCAAAGTG | CCCGTCGGGCCTTGCGTTTA | 198 |
| Gene | Forward Primer | Reverse Primer | Product Length, bp |
|---|---|---|---|
| B2M | GGGTTTCATCCATCCGACATTG | ACACGGCAGGCATACTC | 161 |
| RPL27 | ACCGCTACCCCCGCAAAGTG | CCCGTCGGGCCTTGCGTTTA | 198 |
| ABCB1 | TTGCTGCTTACATTCAGGTTTCA | AGCCTATCTCCTGTCGCATTA | 105 |
| HIF1a | CTGAACGTCGAAAAGAAAAGTC | AAATCACCAGCATCCAGAAGT | 190 |
| IL1b | TTACAGTGGCAATGAGGATGAC | TGTAGTGGTGGTCGGAGATTC | 131 |
| IL6 | ACTCACCTCTTCAGAACGAATTG | CCATCTTTGGAAGGTTCAGGTTG | 149 |
| IL10 | CGCTGTCATCGATTTCTTCCC | AGAGTCGCCACCCTGATGTC | 185 |
| TNFa | TCAGCAAGGACAGCAGAGGA | GTCAGTATGTGAGAGGAAGAGAACC | 128 |
| INFg | TCGGTAACTGACTTGAATGTCCA | TCGCTTCCCTGTTTTAGCTGC | 93 |
| TGFb1 | AACAATTCCTGGCGATACCTCA | AAGCCCTCAATTTCCCCTCC | 125 |
| STAT1 | TGGTTCACTATAGTTGCGGAGAG | TGAATGAGCTGCTGGAAAAGA | 158 |
| STAT3 | CATATGCGGCCAGCAAAGAA | ATACCTGCTCTGAAGAAACT | 152 |
| STAT6 | AGGATGGCAGGGGTTATGT | GGGGGATGGAGTGAGAGTG | 198 |
| SOCS3 | AAGATCCCCCTGGTGTTGA | TTAAAGCGGGGCATCGTA | 162 |
| CD3D | GAGGGAACGGTGGGAACA | CAAAGCAAGGAGCAGAGTGG | 228 |
| GZMB | CCCTGGGAAAACACTCACAC | CCCCACGCACAACTCAAT | 116 |
| CD86 | CAAAACCAAAGCCTGAGTGA | GGCTTTTTGTGATGGATGATAC | 215 |
| CD163 | ATCCCGTCAGTCATCCTTTATT | CTGTCTCTGTCTTCGCTTTTTAGT | 103 |
| CD206 | CTACAAGGGATCGGGTTTATGGA | TTGGCATTGCCTAGTAGCGTA | 105 |
| TGM2 | CGTGACCAACTACAACTCGG | CATCCACGACTCCACCCAG | 136 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlova, E.N.; Lepekha, L.N.; Rybalkina, E.Y.; Tarasov, R.V.; Sychevskaya, K.A.; Voronezhskaya, E.E.; Masyutin, A.G.; Ergeshov, A.E.; Erokhina, M.V. High and Low Levels of ABCB1 Expression Are Associated with Two Distinct Gene Signatures in Lung Tissue of Pulmonary TB Patients with High Inflammation Activity. Int. J. Mol. Sci. 2023, 24, 14839. https://doi.org/10.3390/ijms241914839
Pavlova EN, Lepekha LN, Rybalkina EY, Tarasov RV, Sychevskaya KA, Voronezhskaya EE, Masyutin AG, Ergeshov AE, Erokhina MV. High and Low Levels of ABCB1 Expression Are Associated with Two Distinct Gene Signatures in Lung Tissue of Pulmonary TB Patients with High Inflammation Activity. International Journal of Molecular Sciences. 2023; 24(19):14839. https://doi.org/10.3390/ijms241914839
Chicago/Turabian StylePavlova, Ekaterina N., Larisa N. Lepekha, Ekaterina Yu. Rybalkina, Ruslan V. Tarasov, Ksenia A. Sychevskaya, Elena E. Voronezhskaya, Alexander G. Masyutin, Atadzhan E. Ergeshov, and Maria V. Erokhina. 2023. "High and Low Levels of ABCB1 Expression Are Associated with Two Distinct Gene Signatures in Lung Tissue of Pulmonary TB Patients with High Inflammation Activity" International Journal of Molecular Sciences 24, no. 19: 14839. https://doi.org/10.3390/ijms241914839
APA StylePavlova, E. N., Lepekha, L. N., Rybalkina, E. Y., Tarasov, R. V., Sychevskaya, K. A., Voronezhskaya, E. E., Masyutin, A. G., Ergeshov, A. E., & Erokhina, M. V. (2023). High and Low Levels of ABCB1 Expression Are Associated with Two Distinct Gene Signatures in Lung Tissue of Pulmonary TB Patients with High Inflammation Activity. International Journal of Molecular Sciences, 24(19), 14839. https://doi.org/10.3390/ijms241914839





