The Generation of a Nanobody-Based ELISA for Human Microsomal Epoxide Hydrolase
Abstract
1. Introduction
2. Results and Discussion
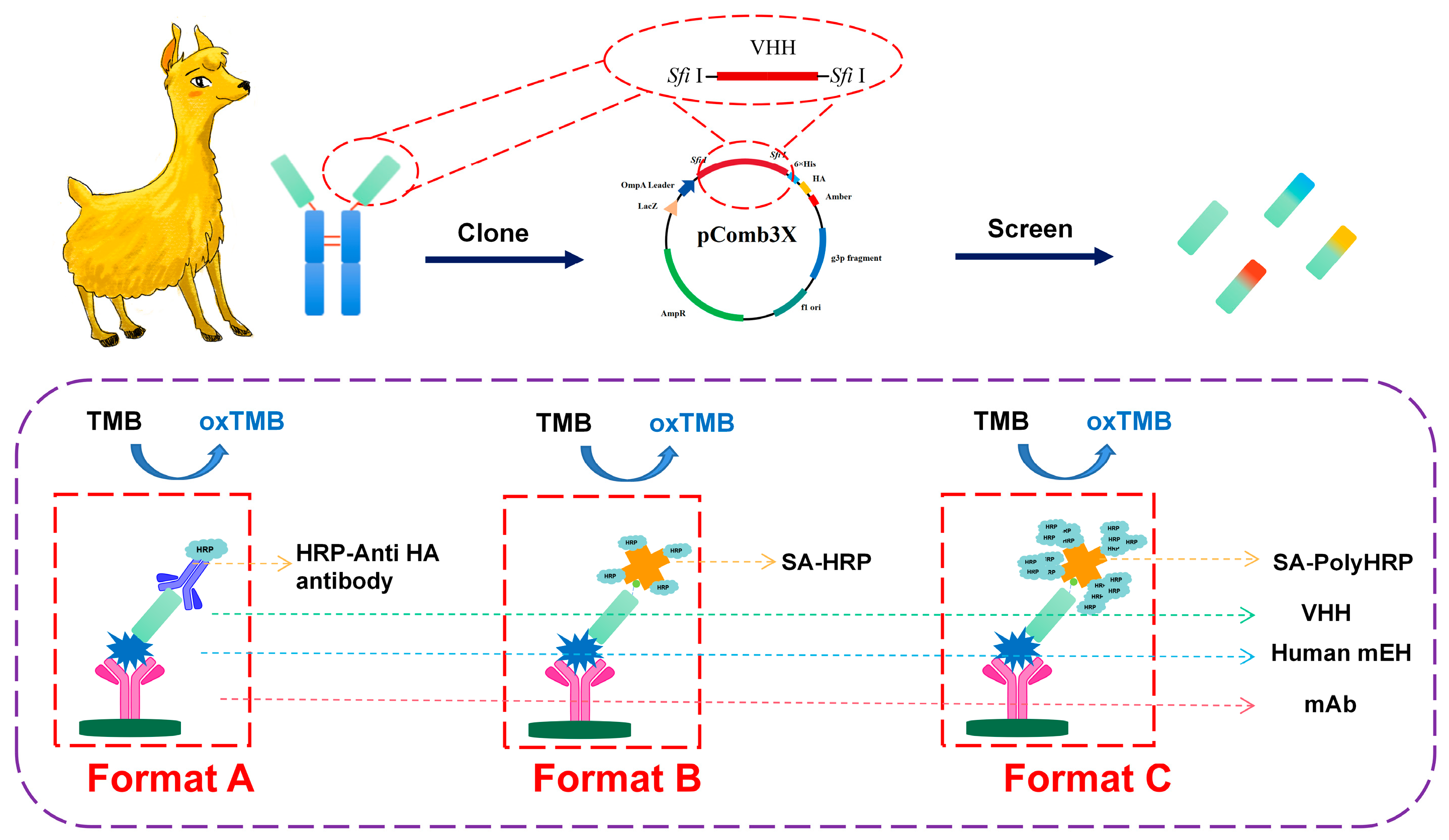
2.1. The Selection of Anti-mEH Nanobodies
2.2. The Performance of Four Unique Nanobodies
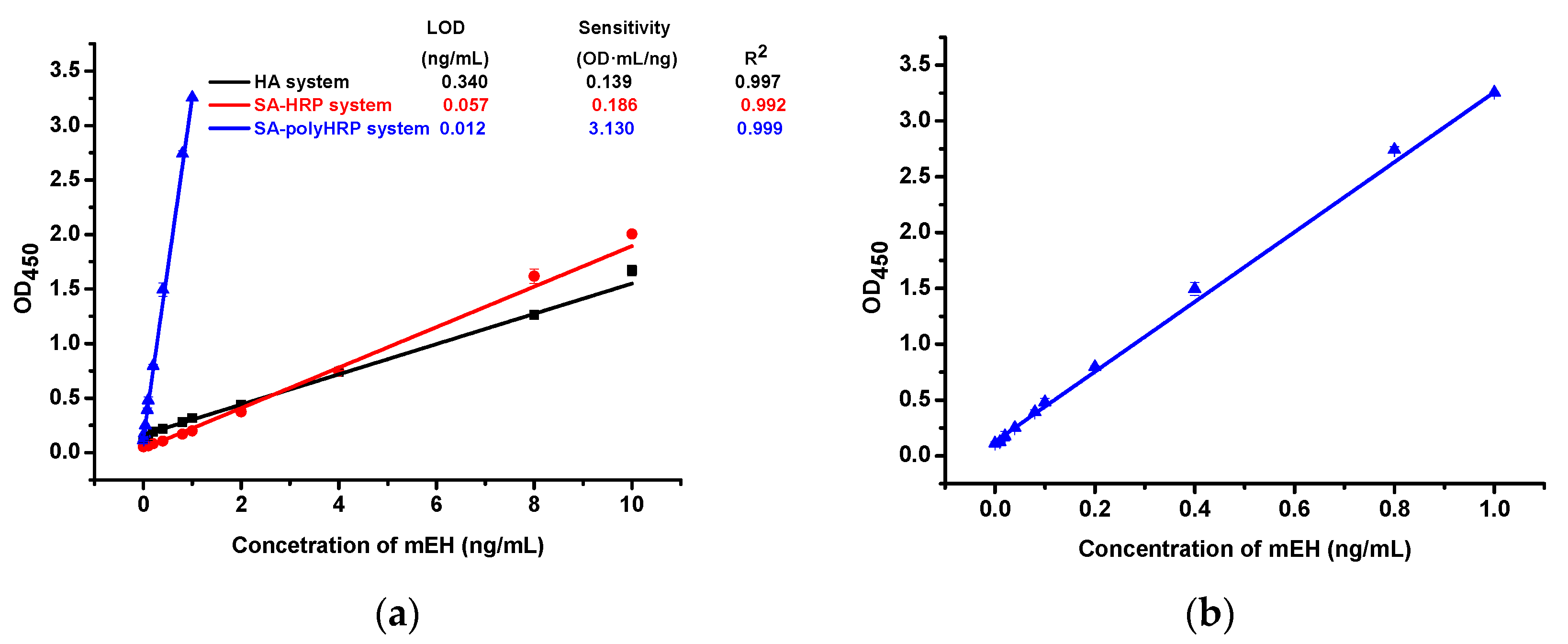
2.3. A Comparison of the Performance of Three ELISA Formats
2.4. Cross-Reactivity
2.5. Matrix Effects
2.6. The Analysis of Human Tissue Samples
3. Materials and Methods
3.1. Materials
3.2. Expression and Purification of Human mEH in Tni Insect Cells
3.3. The Construction of a Nanobody Library and Bio-Panning
3.4. The Expression and Purification of Nanobodies
3.5. The Development of Three Formats of Nanobody-Based Immunoassays for the Detection of Human mEH
3.6. Cross-Reactivity
3.7. Matrix Effects
3.8. Analysis of Clinical Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vaclavikova, R.; Hughes, D.J.; Soucek, P. Microsomal epoxide hydrolase 1 (EPHX1): Gene, structure, function, and role in human disease. Gene 2015, 571, 1–8. [Google Scholar] [CrossRef]
- Omiecinski, C.J.; Aicher, L.; Swenson, L. Developmental expression of human microsomal epoxide hydrolase. J. Pharmacol. Exp. Ther. 1994, 269, 417–423. [Google Scholar] [PubMed]
- Decker, M.; Arand, M.; Cronin, A. Mammalian epoxide hydrolases in xenobiotic metabolism and signalling. Arch. Toxicol. 2009, 83, 297–318. [Google Scholar] [CrossRef] [PubMed]
- Fritz, P.; Mürdter, T.E.; Eichelbaum, M.; Siegle, I.; Weissert, M.; Zanger, U.M. Microsomal epoxide hydrolase expression as a predictor of tamoxifen response in primary breast cancer: A retrospective exploratory study with long-term follow-up. J. Clin. Oncol. 2001, 19, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Brennan, P.; Boffetta, P.; London, S.J.; Benhamou, S.; Rannug, A.; To-Figueras, J.; Ingelman-Sundberg, M.; Shields, P.; Gaspari, L.; et al. Microsomal epoxide hydrolase polymorphisms and lung cancer risk: A quantitative review. Biomarkers 2002, 7, 230–241. [Google Scholar] [CrossRef]
- Zusterzeel, P.L.; Peters, W.H.; Visser, W.; Hermsen, K.J.; Roelofs, H.M.; Steegers, E.A. A polymorphism in the gene for microsomal epoxide hydrolase is associated with pre-eclampsia. J. Med. Genet. 2001, 38, 234–237. [Google Scholar] [CrossRef]
- Liu, M.; Sun, A.; Shin, E.J.; Liu, X.; Kim, S.G.; Runyons, C.R.; Markesbery, W.; Kim, H.C.; Bing, G. Expression of microsomal epoxide hydrolase is elevated in Alzheimer’s hippocampus and induced by exogenous beta-amyloid and trimethyl-tin. Eur. J. Neurosci. 2006, 23, 2027–2034. [Google Scholar] [CrossRef]
- Gautheron, J.; Morisseau, C.; Chung, W.K.; Zammouri, J.; Auclair, M.; Baujat, G.; Capel, E.; Moulin, C.; Wang, Y.; Yang, J.; et al. EPHX1 mutations cause a lipoatrophic diabetes syndrome due to impaired epoxide hydrolysis and increased cellular senescence. eLife 2021, 10, e68445. [Google Scholar] [CrossRef]
- Edin, M.L.; Hamedani, B.G.; Gruzdev, A.; Graves, J.P.; Lih, F.B.; Arbes, S.J., 3rd; Singh, R.; Orjuela Leon, A.C.; Bradbury, J.A.; DeGraff, L.M.; et al. Epoxide hydrolase 1 (EPHX1) hydrolyzes epoxyeicosanoids and impairs cardiac recovery after ischemia. J. Biol. Chem. 2018, 293, 3281–3292. [Google Scholar] [CrossRef]
- Calder, P.C. n–3 Fatty acids and cardiovascular disease: Evidence explained and mechanisms explored. Clin. Sci. 2004, 107, 1–11. [Google Scholar] [CrossRef]
- Morisseau, C.; Kodani, S.D.; Kamita, S.G.; Yang, J.; Lee, K.S.S.; Hammock, B.D. Relative Importance of Soluble and Microsomal Epoxide Hydrolases for the Hydrolysis of Epoxy-Fatty Acids in Human Tissues. Int. J. Mol. Sci. 2021, 22, 4993. [Google Scholar] [CrossRef] [PubMed]
- Edin, M.L.; Zeldin, D.C. Regulation of cardiovascular biology by microsomal epoxide hydrolase. Toxicol. Res. 2021, 37, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Marowsky, A.; Meyer, I.; Erismann-Ebner, K.; Pellegrini, G.; Mule, N.; Arand, M. Beyond detoxification: A role for mouse mEH in the hepatic metabolism of endogenous lipids. Arch. Toxicol. 2017, 91, 3571–3585. [Google Scholar] [CrossRef]
- Lin, J.-C.; Hiasa, Y.; Farber, E. Preneoplastic Antigen as a Marker for Endoplasmic Reticulum of Putative Premalignant Hepatocytes during Liver Carcinogenesis. Cancer Res. 1977, 37, 1972–1981. [Google Scholar] [PubMed]
- Griffin, M.J.; Noda, K. Quantitation of Epoxide Hydrolase Released from Hyperplastic Nodule and Hepatoma Microsomes. Cancer Res. 1980, 40, 2768–2773. [Google Scholar]
- Hammock, B.D.; Loury, D.N.; Moody, D.E.; Ruebner, B.; Baselt, R.; Milam, K.M.; Volberding, P.; Ketterman, A.; Talcott, R. A methodology for the analysis of the preneoplastic antigen. Carcinogenesis 1984, 5, 1467–1473. [Google Scholar] [CrossRef]
- Akatsuka, T.; Kobayashi, N.; Ishikawa, T.; Saito, T.; Shindo, M.; Yamauchi, M.; Kurokohchi, K.; Miyazawa, H.; Duan, H.; Matsunaga, T.; et al. Autoantibody response to microsomal epoxide hydrolase in hepatitis C and A. J. Autoimmun. 2007, 28, 7–18. [Google Scholar] [CrossRef][Green Version]
- Wu, T.; Xi, X.; Chen, Y.; Jiang, C.; Zhang, Q.; Dai, G.; Bai, Y.; Zhang, W.; Ni, T.; Zou, J.; et al. Absolute protein assay for the simultaneous quantification of two epoxide hydrolases in rats by mass spectrometry-based targeted proteomics. J. Sep. Sci. 2021, 44, 2754–2763. [Google Scholar] [CrossRef]
- Morisseau, C.; Bernay, M.; Escaich, A.; Sanborn, J.R.; Lango, J.; Hammock, B.D. Development of fluorescent substrates for microsomal epoxide hydrolase and application to inhibition studies. Anal. Biochem. 2011, 414, 154–162. [Google Scholar] [CrossRef]
- Duan, H.; Yoshimura, K.; Kobayashi, N.; Sugiyama, K.; Sawada, J.; Saito, Y.; Morisseau, C.; Hammock, B.D.; Akatsuka, T. Development of monoclonal antibodies to human microsomal epoxide hydrolase and analysis of “preneoplastic antigen”-like molecules. Toxicol. Appl. Pharmacol. 2012, 260, 17–26. [Google Scholar] [CrossRef][Green Version]
- Backman, J.T.; Siegle, I.; Zanger, U.M.; Fritz, P. Immunohistochemical detection of microsomal epoxide hydrolase in human synovial tissue. Histochem. J. 1999, 31, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Wie, S.I.; Guenthner, T.M.; Oesch, F.; Hammock, B.D. Rapid and sensitive enzyme-linked immunosorbent assay for the microsomal epoxide hydrolase. Carcinogenesis 1982, 3, 1307–1310. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, L.; Jin, D.; Liu, Y. Nanobody-A versatile tool for cancer diagnosis and therapeutics. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 13, e1697. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Li, D.; Morisseau, C.; Dong, J.X.; Yang, J.; Wan, D.; Rossotti, M.A.; Gee, S.J.; González-Sapienza, G.G.; Hammock, B.D. Heavy chain single-domain antibodies to detect native human soluble epoxide hydrolase. Anal. Bioanal. Chem. 2015, 407, 7275–7283. [Google Scholar] [CrossRef]
- Skoda, R.C.; Demierre, A.; McBride, O.W.; Gonzalez, F.J.; Meyer, U.A. Human microsomal xenobiotic epoxide hydrolase. Complementary DNA sequence, complementary DNA-directed expression in COS-1 cells, and chromosomal localization. J. Biol. Chem. 1988, 263, 1549–1554. [Google Scholar] [CrossRef]
- Decker, M.; Adamska, M.; Cronin, A.; Di Giallonardo, F.; Burgener, J.; Marowsky, A.; Falck, J.R.; Morisseau, C.; Hammock, B.D.; Gruzdev, A.; et al. EH3 (ABHD9): The first member of a new epoxide hydrolase family with high activity for fatty acid epoxides. J. Lipid Res. 2012, 53, 2038–2045. [Google Scholar] [CrossRef]
- Barbas, C.F., III; Burton, D.R.; Scott, J.K.; Silverman, G.J. Phage Display: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Los Angeles, CA, USA, 2001. [Google Scholar]
- Li, D.; Cui, Y.; Morisseau, C.; Gee, S.J.; Bever, C.S.; Liu, X.; Wu, J.; Hammock, B.D.; Ying, Y. Nanobody Based Immunoassay for Human Soluble Epoxide Hydrolase Detection Using Polymeric Horseradish Peroxidase (PolyHRP) for Signal Enhancement: The Rediscovery of PolyHRP? Anal. Chem. 2017, 89, 6248–6256. [Google Scholar] [CrossRef]

| Epoxide Hydrolases | Spiked Concentration (ng mL−1) | Measured Concentration Mean ± SD (ng mL−1) | Cross-Reactivity (%) |
|---|---|---|---|
| Denatured Meh a | 1000 | 0.21 ± 0.06 | 0.02 |
| Rat mEH | 1000 | 1.05 ± 0.28 | 0.11 |
| Human sEH | 1000 | N.D. | <0.01 |
| Human EH-3 | 1000 | N.D. | <0.01 |
| Human EH-4 | 1000 | N.D. | <0.01 |
| Format A | ||||||||||||
| Intra-Assay | Inter-Assay | |||||||||||
| Spiked (ng mL−1) | 1:10 Dilution | 1:100 Dilution | 1:1000 Dilution | 1:10 Dilution | 1:100 Dilution | 1:1000 Dilution | ||||||
| Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | |
| 8.00 | 6.48 ± 0.81 | 81 | 8.21 ± 0.25 | 103 | 7.65 ± 0.38 | 96 | 6.14 ± 0.17 | 77 | 7.86 ± 0.82 | 98 | 6.53 ± 0.63 | 82 |
| 4.00 | 4.53 ± 0.31 | 113 | 4.59 ± 0.23 | 115 | 4.53 ± 0.08 | 113 | 4.65 ± 0.12 | 116 | 4.66 ± 0.15 | 117 | 4.50 ± 0.12 | 112 |
| 2.00 | 2.02 ± 0.11 | 101 | 2.38 ± 0.06 | 119 | 2.35 ± 0.04 | 118 | 2.08 ± 0.08 | 104 | 2.20 ± 0.13 | 110 | 2.12 ± 0.18 | 106 |
| 1.00 | 0.86 ± 0.07 | 86 | 0.96 ± 0.09 | 96 | 1.28 ± 0.10 | 105 | 0.87 ± 0.06 | 87 | 1.15 ± 0.01 | 115 | 1.20 ± 0.07 | 120 |
| 0.50 | 0.57 ± 0.01 | 115 | 0.59 ± 0.13 | 118 | 0.46 ± 0.01 | 93 | 0.65 ± 0.04 | 130 | 0.58 ± 0.09 | 115 | 0.55 ± 0.16 | 110 |
| Format B | ||||||||||||
| Intra-Assay | Inter-Assay | |||||||||||
| Spiked (ng mL−1) | 1:10 Dilution | 1:100 Dilution | 1:1000 Dilution | 1:10 Dilution | 1:100 Dilution | 1:1000 Dilution | ||||||
| Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | |
| 8.00 | 6.90 ± 0.46 | 86 | 8.23 ± 0.30 | 103 | 8.30 ± 0.61 | 104 | 6.07 ± 0.23 | 76 | 7.17 ± 0.17 | 90 | 7.18 ± 0.28 | 90 |
| 4.00 | 4.95 ± 0.01 | 124 | 4.81 ± 0.03 | 120 | 4.81 ± 0.19 | 120 | 4.79 ± 0.32 | 120 | 4.73 ± 0.14 | 118 | 4.83 ± 0.13 | 121 |
| 2.00 | 2.30 ± 0.13 | 115 | 2.23 ± 0.09 | 112 | 2.37 ± 0.10 | 118 | 1.71 ± 0.09 | 85 | 2.29 ± 0.10 | 114 | 2.35 ± 0.22 | 117 |
| 1.00 | 0.99 ± 0.05 | 99 | 1.05 ± 0.10 | 105 | 1.22 ± 0.04 | 122 | 0.74 ± 0.05 | 74 | 0.81 ± 0.03 | 81 | 1.20 ± 0.15 | 120 |
| 0.50 | 0.45 ± 0.13 | 90 | 0.42 ± 0.07 | 83 | 0.49 ± 0.05 | 98 | 0.37 ± 0.04 | 75 | 0.41 ± 0.08 | 82 | 0.40 ± 0.06 | 80 |
| Format C | ||||||||||||
| Intra-Assay | Inter-Assay | |||||||||||
| Spiked (ng mL−1) | 1:10 Dilution | 1:100 Dilution | 1:1000 Dilution | 1:10 Dilution | 1:100 Dilution | 1:1000 Dilution | ||||||
| Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | Found (ng mL−1) | Recovery (%) | |
| 1.00 | 0.75 ± 0.02 | 75 | 0.84 ± 0.06 | 84 | 0.75 ± 0.02 | 75 | 0.72 ± 0.02 | 72 | 0.76 ± 0.03 | 76 | 0.77 ± 0.05 | 77 |
| 0.80 | 0.60 ± 0.03 | 75 | 0.75 ± 0.02 | 94 | 0.63 ± 0.01 | 79 | 0.58 ± 0.01 | 73 | 0.60 ± 0.01 | 75 | 0.59 ± 0.05 | 73 |
| 0.40 | 0.33 ± 0.02 | 83 | 0.39 ± 0.03 | 98 | 0.35 ± 0.02 | 88 | 0.28 ± 0.01 | 70 | 0.39 ± 0.02 | 98 | 0.39 ± 0.03 | 96 |
| 0.20 | 0.19 ± 0.01 | 93 | 0.20 ± 0.01 | 101 | 0.18 ± 0.03 | 90 | 0.20 ± 0.02 | 98 | 0.20 ± 0.02 | 118 | 0.21 ± 0.02 | 103 |
| 0.10 | 0.08 ± 0.01 | 83 | 0.09 ± 0.01 | 86 | 0.10 ± 0.02 | 104 | 0.11 ± 0.01 | 113 | 0.13 ± 0.01 | 126 | 0.10 ± 0.01 | 99 |
| Human Tissues | mEH Concentration (nM) a | |||
|---|---|---|---|---|
| Enzyme Activity b | HRP-Anti-HA Antibody ELISA | SA-HRP ELISA | SA-Poly HRP ELISA | |
| Adrenal | 290 ± 50 | 159 ± 11 | 172 ± 10 | 136 ± 8 |
| Esophagus | 12 ± 1 | N.D. c | 3 ± 0.4 | 2 ± 0.03 |
| Liver | 850 ± 50 | 292 ± 9 | 322 ± 8 | 261 ± 24 |
| Pancreas | 67 ± 4 | 17 ± 0.6 | 20 ± 1.0 | 21 ± 2 |
| Ovary | 6 ± 1 | N.D. | N.D. | 0.1 ± 0.05 |
| Heart | 10 ± 4 | N.D. | N.D. | 0.2 ± 0.1 |
| Hippocampus | 17 ± 2 | 8 ± 0.2 | 7 ± 0.1 | 6 ± 0.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Q.; McCoy, M.R.; Qi, M.; Morisseau, C.; Yang, H.; Xu, C.; Shey, R.; Goodman, M.C.; Zhao, S.; Hammock, B.D. The Generation of a Nanobody-Based ELISA for Human Microsomal Epoxide Hydrolase. Int. J. Mol. Sci. 2023, 24, 14698. https://doi.org/10.3390/ijms241914698
He Q, McCoy MR, Qi M, Morisseau C, Yang H, Xu C, Shey R, Goodman MC, Zhao S, Hammock BD. The Generation of a Nanobody-Based ELISA for Human Microsomal Epoxide Hydrolase. International Journal of Molecular Sciences. 2023; 24(19):14698. https://doi.org/10.3390/ijms241914698
Chicago/Turabian StyleHe, Qiyi, Mark R. McCoy, Meng Qi, Christophe Morisseau, Huiyi Yang, Chengpeng Xu, Rachel Shey, Michael C. Goodman, Suqing Zhao, and Bruce D. Hammock. 2023. "The Generation of a Nanobody-Based ELISA for Human Microsomal Epoxide Hydrolase" International Journal of Molecular Sciences 24, no. 19: 14698. https://doi.org/10.3390/ijms241914698
APA StyleHe, Q., McCoy, M. R., Qi, M., Morisseau, C., Yang, H., Xu, C., Shey, R., Goodman, M. C., Zhao, S., & Hammock, B. D. (2023). The Generation of a Nanobody-Based ELISA for Human Microsomal Epoxide Hydrolase. International Journal of Molecular Sciences, 24(19), 14698. https://doi.org/10.3390/ijms241914698







