Salivary Cystatin D Interactome in Patients with Systemic Mastocytosis: An Exploratory Study
Abstract
1. Introduction
2. Results
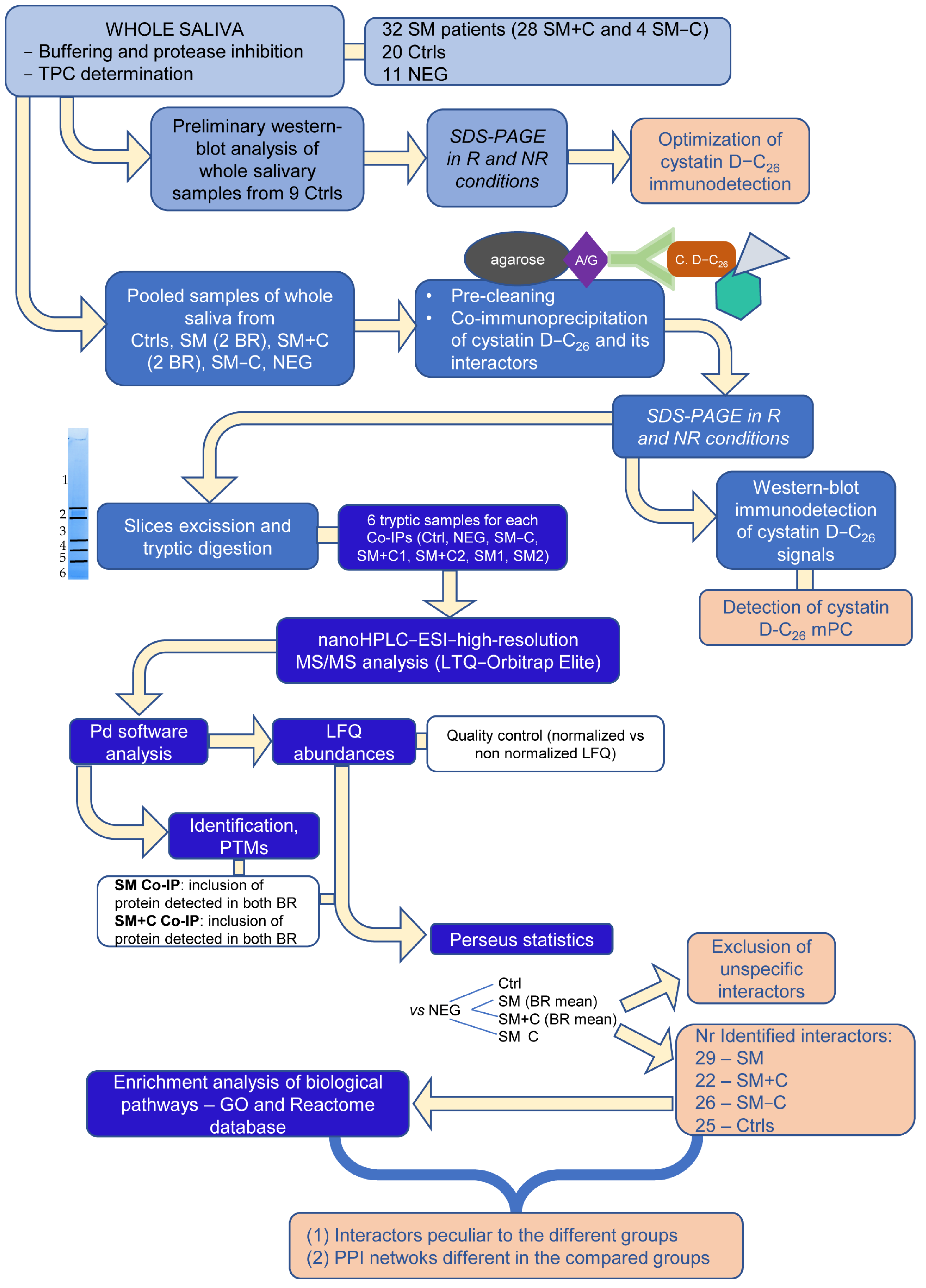
2.1. Explorative Western Blot Analysis of Cystatin D-C26 in Salivary Samples
2.2. Co-Immunoprecipitation of Cystatin D-C26-Bound mPC
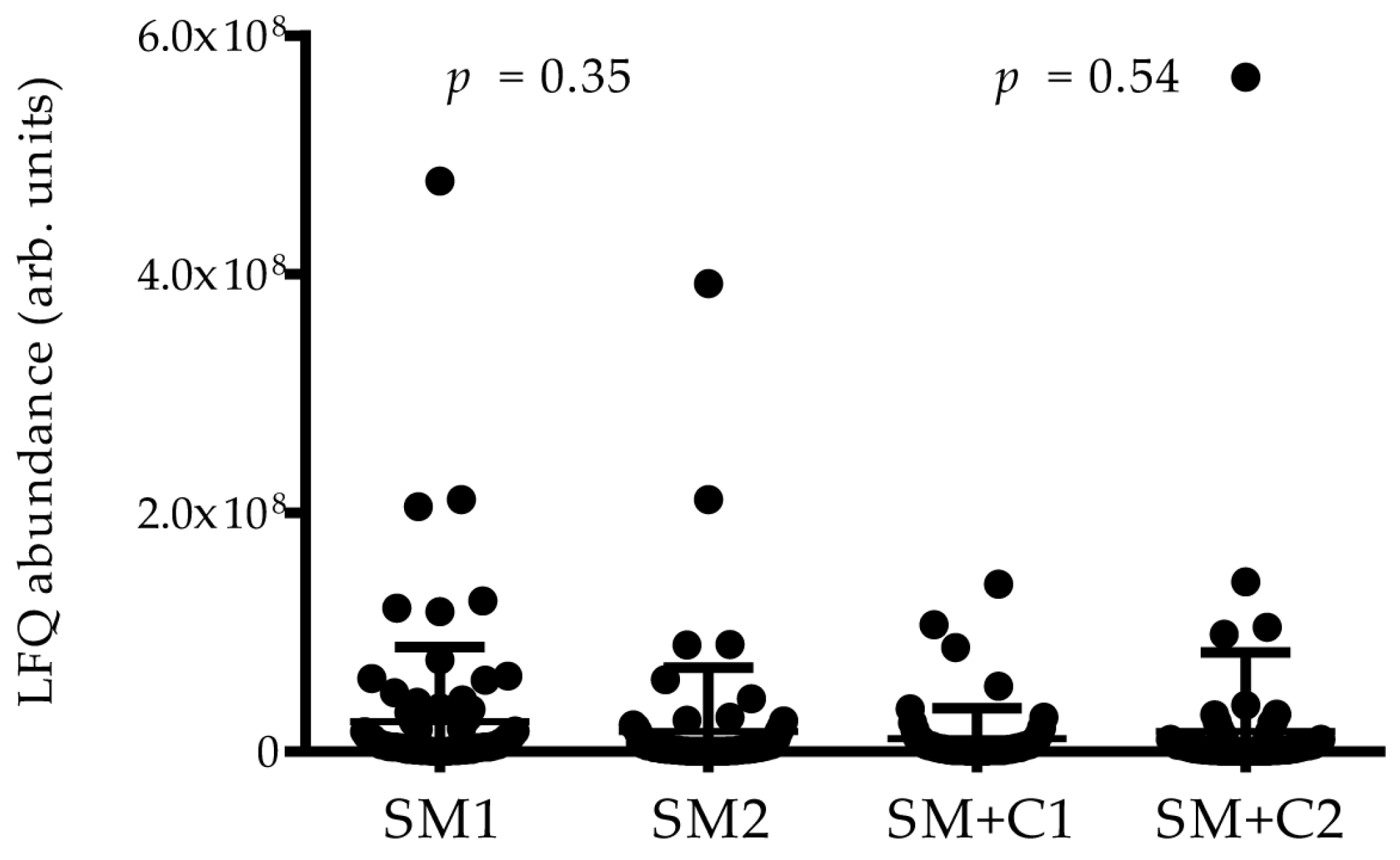
2.3. High-Resolution MS/MS Analysis of Co-IP Samples
2.3.1. Proteins Identified in Co-IPs from Ctrl and Patient Samples
2.3.2. Proteins Identified in Co-IP from Ctrl Samples
2.3.3. Proteins Identified Only in Co-IPs from Patient Samples
2.4. Biological and Functional Analysis
3. Discussion
3.1. Limitations of the Study
3.2. Cystatin D-C26 Interactors and PPI Common to Ctrls and Patients
3.3. Cystatin D-C26 Interactors and PPIs Peculiar to Ctrls
3.4. Cystatin D-C26 Interactors Peculiar to Patients
3.4.1. Cystatin D-C26 Interactors and PPIs Peculiar to SM+C Patients
3.4.2. Cystatin D-C26 Interactors and PPIs Peculiar to SM−C Patients
4. Materials and Methods
4.1. Reagents and Instruments
4.2. Study Subjects and Controls
4.3. Sample Collection
4.4. Immunodetection of Cystatin D-C26
4.5. Co-Immunoprecipitation Experiments and SDS-PAGE
4.6. Tryptic Digestion and High-Resolution MS/MS Analysis
4.7. Statistical Analysis
4.8. Functional Enrichment Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Valent, P.; Akin, C.; Gleixner, K.V.; Sperr, W.R.; Reiter, A.; Arock, M.; Triggiani, M. Multidisciplinary Challenges in Mastocytosis and How to Address with Personalized Medicine Approaches. Int. J. Mol. Sci. 2019, 20, 2976. [Google Scholar] [CrossRef]
- Valent, P.; Akin, C.; Metcalfe, D.D. Mastocytosis: 2016 Updated WHO Classification and Novel Emerging Treatment Concepts. Blood 2017, 129, 1420–1427. [Google Scholar] [CrossRef]
- Pardanani, A. Systemic Mastocytosis in Adults: 2021 Update on Diagnosis, Risk Stratification and Management. Am. J. Hematol. 2021, 96, 508–525. [Google Scholar] [CrossRef]
- Hartmann, K.; Escribano, L.; Grattan, C.; Brockow, K.; Carter, M.C.; Alvarez-Twose, I.; Matito, A.; Broesby-Olsen, S.; Siebenhaar, F.; Lange, M.; et al. Cutaneous Manifestations in Patients with Mastocytosis: Consensus Report of the European Competence Network on Mastocytosis; the American Academy of Allergy, Asthma & Immunology; and the European Academy of Allergology and Clinical Immunology. J. Allergy Clin. Immunol. 2016, 137, 35–45. [Google Scholar] [CrossRef]
- Arock, M.; Sotlar, K.; Akin, C.; Broesby-Olsen, S.; Hoermann, G.; Escribano, L.; Kristensen, T.K.; Kluin-Nelemans, H.C.; Hermine, O.; Dubreuil, P.; et al. KIT Mutation Analysis in Mast Cell Neoplasms: Recommendations of the European Competence Network on Mastocytosis. Leukemia 2015, 29, 1223–1232. [Google Scholar] [CrossRef]
- Falchi, L.; Verstovsek, S. Kit Mutations: New Insights and Diagnostic Value. Immunol. Allergy Clin. N. Am. 2018, 38, 411–428. [Google Scholar] [CrossRef]
- Laine, E.; Chauvot De Beauchêne, I.; Perahia, D.; Auclair, C.; Tchertanov, L. Mutation D816V Alters the Internal Structure and Dynamics of C-KIT Receptor Cytoplasmic Region: Implications for Dimerization and Activation Mechanisms. PLoS Comput. Biol. 2011, 7, e1002068. [Google Scholar] [CrossRef]
- Parente, R.; Giudice, V.; Cardamone, C.; Serio, B.; Selleri, C.; Triggiani, M. Secretory and Membrane-Associated Biomarkers of Mast Cell Activation and Proliferation. Int. J. Mol. Sci. 2023, 24, 7071. [Google Scholar] [CrossRef]
- Elieh Ali Komi, D.; Wöhrl, S.; Bielory, L. Mast Cell Biology at Molecular Level: A Comprehensive Review. Clin. Rev. Allergy Immunol. 2020, 58, 342–365. [Google Scholar] [CrossRef]
- Pfaffe, T.; Cooper-White, J.; Beyerlein, P.; Kostner, K.; Punyadeera, C. Diagnostic Potential of Saliva: Current State and Future Applications. Clin. Chem. 2011, 57, 675–687. [Google Scholar] [CrossRef]
- Cabras, T.; Iavarone, F.; Manconi, B.; Olianas, A.; Sanna, M.T.; Castagnola, M.; Messana, I. Top-down Analytical Platforms for the Characterization of the Human Salivary Proteome. Bioanalysis 2014, 6, 563–581. [Google Scholar] [CrossRef]
- Bandhakavi, S.; Stone, M.D.; Onsongo, G.; Van Riper, S.K.; Griffin, T.J. A Dynamic Range Compression and Three-Dimensional Peptide Fractionation Analysis Platform Expands Proteome Coverage and the Diagnostic Potential of Whole Saliva. J. Proteome Res. 2009, 8, 5590–5600. [Google Scholar] [CrossRef]
- Serrao, S.; Firinu, D.; Olianas, A.; Deidda, M.; Contini, C.; Iavarone, F.; Sanna, M.T.; Boroumand, M.; Amado, F.; Castagnola, M.; et al. Top-Down Proteomics of Human Saliva Discloses Significant Variations of the Protein Profile in Patients with Mastocytosis. J. Proteome Res. 2020, 19, 3238–3253. [Google Scholar] [CrossRef]
- Soond, S.M.; Kozhevnikova, M.V.; Townsend, P.A.; Zamyatnin, A.A. Cysteine Cathepsin Protease Inhibition: An Update on Its Diagnostic, Prognostic and Therapeutic Potential in Cancer. Pharmaceuticals 2019, 12, 87. [Google Scholar] [CrossRef]
- Magister, Š.; Kos, J. Cystatins in Immune System. J. Cancer 2012, 4, 45–56. [Google Scholar] [CrossRef]
- Fábián, T.K.; Hermann, P.; Beck, A.; Fejérdy, P.; Fábián, G. Salivary Defense Proteins: Their Network and Role in Innate and Acquired Oral Immunity. Int. J. Mol. Sci. 2012, 13, 4295–4320. [Google Scholar] [CrossRef]
- Balbín, M.; Freije, J.P.; Abrahamson, M.; Velasco, G.; Grubb, A.; López-Otín, C. A Sequence Variation in the Human Cystatin D Gene Resulting in an Amino Acid (Cys/Arg) Polymorphism at the Protein Level. Hum. Genet. 1993, 90, 668–669. [Google Scholar] [CrossRef]
- Balbín, M.; Hall, A.; Grubb, A.; Mason, R.W.; López-Otín, C.; Abrahamson, M. Structural and Functional Characterization of Two Allelic Variants of Human Cystatin D Sharing a Characteristic Inhibition Spectrum against Mammalian Cysteine Proteinases. J. Biol. Chem. 1994, 269, 23156–23162. [Google Scholar] [CrossRef]
- Manconi, B.; Liori, B.; Cabras, T.; Vincenzoni, F.; Iavarone, F.; Castagnola, M.; Messana, I.; Olianas, A. Salivary Cystatins: Exploring New Post-Translational Modifications and Polymorphisms by Top-Down High-Resolution Mass Spectrometry. J. Proteome Res. 2017, 16, 4196–4207. [Google Scholar] [CrossRef]
- Ryan, C.M.; Souda, P.; Halgand, F.; Wong, D.T.; Loo, J.A.; Faull, K.F.; Whitelegge, J.P. Confident Assignment of Intact Mass Tags to Human Salivary Cystatins Using Top-down Fourier-Transform Ion Cyclotron Resonance Mass Spectrometry. J. Am. Soc. Mass. Spectrom. 2010, 21, 908–917. [Google Scholar] [CrossRef][Green Version]
- Liori, B.; Olianas, A.; Cabras, T.; Vincenzoni, F.; Serrao, S.; Messana, I.; Castagnola, M.; Manconi, B. Interactors of Human Salivary Cystatin D Explored by Immunoprecipitation Coupled to Mass Spectrometry. In Proceedings of the XII ItPA National Congress, Lecce, Italy, 12–15 June 2017. [Google Scholar]
- Richards, A.L.; Eckhardt, M.; Krogan, N.J. Mass Spectrometry-based Protein–Protein Interaction Networks for the Study of Human Diseases. Mol. Syst. Biol. 2021, 17, e8792. [Google Scholar] [CrossRef] [PubMed]
- Guard, S.E.; Ebmeier, C.C.; Old, W.M. Label-Free Immunoprecipitation Mass Spectrometry Workflow for Large-Scale Nuclear Interactome Profiling. JoVE 2019, 153, e60432. [Google Scholar] [CrossRef]
- Mellacheruvu, D.; Wright, Z.; Couzens, A.L.; Lambert, J.-P.; St-Denis, N.; Li, T.; Miteva, Y.V.; Hauri, S.; Sardiu, M.E.; Low, T.Y.; et al. The CRAPome: A Contaminant Repository for Affinity Purification Mass Spectrometry Data. Nat. Methods 2013, 10, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Padiglia, A.; Orrù, R.; Boroumand, M.; Olianas, A.; Manconi, B.; Sanna, M.T.; Desiderio, C.; Iavarone, F.; Liori, B.; Messana, I.; et al. Extensive Characterization of the Human Salivary Basic Proline-Rich Protein Family by Top-Down Mass Spectrometry. J. Proteome Res. 2018, 17, 3292–3307. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.J.; McMullen, A.A.; Moore, G.; Dempsey-Hibbert, N.C.; Myers, B.; Graham, C. SWATH-MS Identification of CXCL7, LBP, TGFβ1 and PDGFRβ as Novel Biomarkers in Human Systemic Mastocytosis. Sci. Rep. 2022, 12, 5087. [Google Scholar] [CrossRef]
- Hermans, M.A.W.; Heeringa, J.J.; Swagemakers, S.G.A.; Schrijver, B.; Van Daele, P.L.A.; Van Der Spek, P.J.; Van Hagen, P.M.; Van Zelm, M.C.; Dik, W.A. Altered Leukocyte Subsets and Immune Proteome Indicate Proinflammatory Mechanisms in Mastocytosis. J. Allergy Clin. Immunol. 2022, 150, 146–156.e10. [Google Scholar] [CrossRef]
- Söderlund, S.; Boey, D.; Van Midden, W.; Kjellander, M.; Ax, K.; Qian, H.; Dahlin, J.S.; Ungerstedt, J. Proteomic and Transcriptomic Screening Demonstrates Increased Mast Cell–Derived CCL23 in Systemic Mastocytosis. J. Allergy Clin. Immunol. 2023, 152, 205–213. [Google Scholar] [CrossRef]
- Yuasa, K.; Ota, R.; Matsuda, S.; Isshiki, K.; Inoue, M.; Tsuji, A. Suppression of Death-Associated Protein Kinase 2 by Interaction with 14-3-3 Proteins. Biochem. Biophys. Res. Commun. 2015, 464, 70–75. [Google Scholar] [CrossRef]
- Ferrer-Mayorga, G.; Alvarez-Díaz, S.; Valle, N.; De Las Rivas, J.; Mendes, M.; Barderas, R.; Canals, F.; Tapia, O.; Casal, J.I.; Lafarga, M.; et al. Cystatin D Locates in the Nucleus at Sites of Active Transcription and Modulates Gene and Protein Expression. J. Biol. Chem. 2015, 290, 26533–26548. [Google Scholar] [CrossRef]
- Turato, C.; Pontisso, P. SERPINB3 (serpin peptidase inhibitor, clade B (ovalbumin), member 3). Atlas Genet. Cytogenet. Oncol. Haematol. 2015, 19, 202–209. [Google Scholar] [CrossRef]
- Zamolodchikova, T.S.; Tolpygo, S.M.; Svirshchevskaya, E.V. Cathepsin G-Not Only Inflammation: The Immune Protease Can Regulate Normal Physiological Processes. Front. Immunol. 2020, 11, 411. [Google Scholar] [CrossRef] [PubMed]
- Ligtenberg, A.J.M.; Karlsson, N.G.; Veerman, E.C.I. Deleted in Malignant Brain Tumors-1 Protein (DMBT1): A Pattern Recognition Receptor with Multiple Binding Sites. Int. J. Mol. Sci. 2010, 11, 5212–5233. [Google Scholar] [CrossRef] [PubMed]
- Tur-Gracia, S.; Martinez-Quiles, N. Emerging functions of cytoskeletal proteins in immune diseases. J. Cell Sci. 2021, 134, jcs253534. [Google Scholar] [CrossRef]
- Contini, C.; Serrao, S.; Manconi, B.; Olianas, A.; Iavarone, F.; Guadalupi, G.; Messana, I.; Castagnola, M.; Masullo, C.; Bizzarro, A.; et al. Characterization of Cystatin B Interactome in Saliva from Healthy Elderly and Alzheimer’s Disease Patients. Life 2023, 13, 748. [Google Scholar] [CrossRef] [PubMed]
- Messana, I.; Cabras, T.; Pisano, E.; Sanna, M.T.; Olianas, A.; Manconi, B.; Pellegrini, M.; Paludetti, G.; Scarano, E.; Fiorita, A.; et al. Trafficking and Postsecretory Events Responsible for the Formation of Secreted Human Salivary Peptides. Mol. Cell. Proteom. 2008, 7, 911–926. [Google Scholar] [CrossRef]
- Manconi, B.; Castagnola, M.; Cabras, T.; Olianas, A.; Vitali, A.; Desiderio, C.; Sanna, M.T.; Messana, I. The Intriguing Heterogeneity of Human Salivary Proline-Rich Proteins. J. Proteom. 2016, 134, 47–56. [Google Scholar] [CrossRef]
- Boroumand, M.; Olianas, A.; Manconi, B.; Serrao, S.; Iavarone, F.; Desiderio, C.; Pieroni, L.; Faa, G.; Messana, I.; Castagnola, M.; et al. Mapping of Transglutaminase-2 Sites of Human Salivary Small Basic Proline-Rich Proteins by HPLC–High-Resolution ESI–MS/MS. J. Proteome Res. 2020, 19, 300–313. [Google Scholar] [CrossRef]
- Choi, Y.J.; Kim, S.; Choi, Y.; Nielsen, T.B.; Yan, J.; Lu, A.; Ruan, J.; Lee, H.-R.; Wu, H.; Spellberg, B.; et al. SERPINB1-Mediated Checkpoint of Inflammatory Caspase Activation. Nat. Immunol. 2019, 20, 276–287. [Google Scholar] [CrossRef]
- Cooley, J.; Takayama, T.K.; Shapiro, S.D.; Schechter, N.M.; Remold-O’Donnell, E. The Serpin MNEI Inhibits Elastase-like and Chymotrypsin-like Serine Proteases through Efficient Reactions at Two Active Sites. Biochemistry 2001, 40, 15762–15770. [Google Scholar] [CrossRef]
- Taipale, J.; Lohi, J.; Saarinen, J.; Kovanen, P.T.; Keski-Oja, J. Human Mast Cell Chymase and Leukocyte Elastase Release Latent Transforming Growth Factor-Β1 from the Extracellular Matrix of Cultured Human Epithelial and Endothelial Cells. J. Biol. Chem. 1995, 270, 4689–4696. [Google Scholar] [CrossRef]
- Caughey, G.H. Mast Cell Proteases as Pharmacological Targets. Eur. J. Pharmacol. 2016, 778, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Campanelli, D.; Detmers, P.A.; Nathan, C.F.; Gabay, J.E. Azurocidin and a Homologous Serine Protease from Neutrophils. Differential Antimicrobial and Proteolytic Properties. J. Clin. Investig. 1990, 85, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 Proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.Y.; Raftery, M.; Cai, H.; Hsu, K.; Yan, W.X.; Hseih, H.-L.; Watts, R.N.; Richardson, D.; Thomas, S.; Perry, M.; et al. S-Nitrosylated S100A8: Novel Anti-Inflammatory Properties. J. Immunol. 2008, 181, 5627–5636. [Google Scholar] [CrossRef] [PubMed]
- Castegna, A.; Menga, A. Glutamine Synthetase: Localization Dictates Outcome. Genes 2018, 9, 108. [Google Scholar] [CrossRef]
- Palmieri, E.M.; Menga, A.; Lebrun, A.; Hooper, D.C.; Butterfield, D.A.; Mazzone, M.; Castegna, A. Blockade of Glutamine Synthetase Enhances Inflammatory Response in Microglial Cells. Antioxid. Redox Signal. 2016, 26, 351–363. [Google Scholar] [CrossRef]
- Gangemi, S.; Minciullo, P.L.; Magliacane, D.; Saitta, S.; Loffredo, S.; Saija, A.; Cristani, M.; Marone, G.; Triggiani, M. Oxidative Stress Markers Are Increased in Patients with Mastocytosis. Allergy 2015, 70, 436–442. [Google Scholar] [CrossRef]
- Yamamoto, N.; Kaneko, I.; Motohashi, K.; Sakagami, H.; Adachi, Y.; Tokuda, N.; Sawada, T.; Furukawa, H.; Ueyama, Y.; Fukunaga, K.; et al. Fatty Acid-Binding Protein Regulates LPS-Induced TNF-α Production in Mast Cells. Prostaglandins Leukot. Essent. Fat. Acids 2008, 79, 21–26. [Google Scholar] [CrossRef]
- Henry, J.; Hsu, C.-Y.; Haftek, M.; Nachat, R.; de Koning, H.D.; Gardinal-Galera, I.; Hitomi, K.; Balica, S.; Jean-Decoster, C.; Schmitt, A.-M.; et al. Hornerin Is a Component of the Epidermal Cornified Cell Envelopes. FASEB J. 2011, 25, 1567–1576. [Google Scholar] [CrossRef]
- Sehra, S.; Serezani, A.P.M.; Ocaña, J.A.; Travers, J.B.; Kaplan, M.H. Mast Cells Regulate Epidermal Barrier Function and the Development of Allergic Skin Inflammation. J. Investig. Dermatol. 2016, 136, 1429–1437. [Google Scholar] [CrossRef]
- Markiewicz, A.; Sigorski, D.; Markiewicz, M.; Owczarczyk-Saczonek, A.; Placek, W. Caspase-14-From Biomolecular Basics to Clinical Approach. A Review of Available Data. Int. J. Mol. Sci. 2021, 22, 5575. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.; Hatzfeld, M.; Keil, R. Desmosomes as Signaling Hubs in the Regulation of Cell Behavior. Front. Cell Dev. Biol. 2021, 9, 745670. [Google Scholar] [CrossRef] [PubMed]
- Rosenstiel, P.; Sina, C.; End, C.; Renner, M.; Lyer, S.; Till, A.; Hellmig, S.; Nikolaus, S.; Fölsch, U.R.; Helmke, B.; et al. Regulation of DMBT1 via NOD2 and TLR4 in Intestinal Epithelial Cells Modulates Bacterial Recognition and Invasion1. J. Immunol. 2007, 178, 8203–8211. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, X.; Shen, P.; Ni, Y.; Han, X. The basic functions of phosphoglycerate kinase 1 and its roles in cancer and other diseases. Eur. J. Pharmacol. 2022, 920, 174835. [Google Scholar] [CrossRef] [PubMed]
- Sinniah, A.; Yazid, S.; Bena, S.; Oliani, S.M.; Perretti, M.; Flower, R.J. Endogenous Annexin-A1 Negatively Regulates Mast Cell-Mediated Allergic Reactions. Front. Pharmacol. 2019, 10, 1313. [Google Scholar] [CrossRef]
- Oshi, M.; Tokumaru, Y.; Mukhopadhyay, S.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. Annexin A1 Expression Is Associated with Epithelial–Mesenchymal Transition (EMT), Cell Proliferation, Prognosis, and Drug Response in Pancreatic Cancer. Cells 2021, 10, 653. [Google Scholar] [CrossRef]
- Krajewski, D.; Polukort, S.H.; Gelzinis, J.; Rovatti, J.; Kaczenski, E.; Galinski, C.; Pantos, M.; Shah, N.N.; Schneider, S.S.; Kennedy, D.R.; et al. Protein Disulfide Isomerases Regulate IgE-Mediated Mast Cell Responses and Their Inhibition Confers Protective Effects During Food Allergy. Front. Immunol. 2020, 11, 606837. [Google Scholar] [CrossRef]
- Dráber, P.; Sulimenko, V.; Dráberová, E. Cytoskeleton in Mast Cell Signaling. Front. Immunol. 2012, 3, 130. [Google Scholar] [CrossRef]
- Valent, P.; Akin, C.; Sperr, W.R.; Horny, H.-P.; Arock, M.; Lechner, K.; Bennett, J.M.; Metcalfe, D.D. Diagnosis and Treatment of Systemic Mastocytosis: State of the Art. Br. J. Haematol. 2003, 122, 695–717. [Google Scholar] [CrossRef]
- Iwamoto, A.; Inoue, Y.; Tachibana, H.; Kawahara, H. Immunomodulatory Effect of Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) in Allergic Conditions in Vitro and in Vivo. Cytotechnology 2021, 73, 333–342. [Google Scholar] [CrossRef]
- Dyer, R.R.; Ford, K.I.; Robinson, R.A.S. The roles of S-nitrosylation and S-glutathionylation in Alzheimer’s disease. Methods Enzymol. 2019, 626, 499–538. [Google Scholar] [PubMed]
- Schittek, B.; Hipfel, R.; Sauer, B.; Bauer, J.; Kalbacher, H.; Stevanovic, S.; Schirle, M.; Schroeder, K.; Blin, N.; Meier, F.; et al. Dermcidin: A Novel Human Antibiotic Peptide Secreted by Sweat Glands. Nat. Immunol. 2001, 2, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Toufiq, M.; Roelands, J.; Alfaki, M.; Syed Ahamed Kabeer, B.; Saadaoui, M.; Lakshmanan, A.P.; Bangarusamy, D.K.; Murugesan, S.; Bedognetti, D.; Hendrickx, W.; et al. Annexin A3 in Sepsis: Novel Perspectives from an Exploration of Public Transcriptome Data. Immunology 2020, 161, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Du, X.; Zhang, L.; Zheng, Y.; Jia, T.; Song, X.; Che, D.; Geng, S. Suprabasin-Derived Polypeptides: SBSN(50-63) Induces Inflammatory Response via TLR4-Mediated Mast Cell Activation. Inflammopharmacology 2023, 31, 1329–1339. [Google Scholar] [CrossRef]
- Gundry, R.L.; White, M.Y.; Murray, C.I.; Kane, L.A.; Fu, Q.; Stanley, B.A.; Van Eyk, J.E. Preparation of Proteins and Peptides for Mass Spectrometry Analysis in a Bottom-Up Proteomics Workflow. Curr. Protoc. Mol. Biol. 2009, 90, 10.25.1–10.25.23. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Bandeira, N.; Sharma, V.; Perez-Riverol, Y.; Carver, J.J.; Kundu, D.J.; García-Seisdedos, D.; Jarnuczak, A.F.; Hewapathirana, S.; Pullman, B.S.; et al. The ProteomeXchange Consortium in 2020: Enabling ‘Big Data’ Approaches in Proteomics. Nucleic Acids Res. 2020, 48, D1145–D1152. [Google Scholar] [CrossRef]
- Tyanova, S.; Cox, J. Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research. Methods Mol. Biol. 2018, 1711, 133–148. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]

| Uniprot-KB Code | Description | PTMs | Ctrl | SM+C | SM−C | SM |
|---|---|---|---|---|---|---|
| P28325 | Cystatin D-C26 | • | • | • | • | |
| Q9UGM3 | Deleted in malignant brain tumors 1 protein (DMBT1) | • | • | • | • | |
| P15924 | Desmoplakin | • | • | • | • | |
| P04075 | Fructose-bisphosphate aldolase A | All: M1-loss | • | • | • | • |
| Q9HC84 | Mucin 5B | • | • | • | • | |
| P01833 | Polymeric immunoglobulin receptor (PIgR) | • | • | • | • | |
| P06702 | S100A9 | • | • | • | • | |
| P29508 | Serpin B3 | • | • | • | • | |
| P31947 | 14-3-3 protein sigma | • | • | |||
| P05141 | ADP/ATP translocase 2 | Ctrl/SM: M1-loss +N-term.-Acetyl. | • | • | ||
| P04080 | Cystatin B | SM: N-term.-Acetyl. | • | • | ||
| P02788 | Lactotransferrin | • | • | |||
| Q9BYD5 | Cornifelin | • | • | • | ||
| P01037 | Cystatin SN | • | • | • | ||
| P01036 | Cystatin S | • | • | |||
| P12273 | Prolactin-inducible protein (PIP) | • | • | • | ||
| P63104 | 14-3-3 protein zeta/delta | Ctrl: N-term.-Acetyl. | • | |||
| P59998 | Actin-related protein 2/3 complex subunit 4 (ARPC4) | • | ||||
| P08311 | Cathepsin G | • | ||||
| O95833 | Chloride intracellular channel protein 3 | • | ||||
| P23528 | Cofilin-1 | Ctrl: M1-loss +N-term.-Acetyl. | • | |||
| O43240 | Kallikrein-10 | • | ||||
| P61626 | Lysozyme C | • | ||||
| P35232 | Prohibitin | • | ||||
| P13489 | Ribonuclease inhibitor | • | ||||
| P30740 | Leukocyte elastase inhibitor (Serpin B1) | • | • | • | ||
| P00558 | Phosphoglycerate kinase 1 | • | • | • | ||
| P02812 | Basic salivary proline-rich protein 2 (bPRP-2L) | • | • | |||
| P02814 | Submaxillary gland androgen-regulated protein 3B (P-B peptide) | • | • | |||
| P20160 | Azurocidin | • | ||||
| Q01469 | Fatty acid-binding protein 5 (FABP-5) | SM: M1-loss +N-term.-Acetyl. | • | |||
| P15104 | Glutamine synthetase | • | ||||
| P14923 | Junction plakoglobin | • | ||||
| P59665 | Neutrophil defensin 1 | • | ||||
| Q13835 | Plakophilin-1 | • | ||||
| P05109 | S100A8 | • | ||||
| P62987 | Ubiquitin-60S ribosomal protein L40 | • | ||||
| P21796 | Voltage-dependent anion-selective channel protein 1 | • | ||||
| P04083 | Annexin A1 | SM: M1-loss +N-term.-Acetyl. Phospho-S37 | • | • | ||
| P31944 | Caspase-14 | • | ||||
| P20930 | Filaggrin | • | ||||
| Q86YZ3 | Hornerin | • | ||||
| P07237 | Protein disulfide-isomerase (PDI) | • | ||||
| P68371 | Tubulin beta-4B chain | • | ||||
| P25311 | Zinc-alpha-2-glycoprotein | • | ||||
| P12429 | Annexin A3 | • | • | |||
| P52907 | F-actin-capping protein subunit alpha-1 | SM: M1-loss +N-term.-Acetyl. | • | • | ||
| P00338 | L-lactate dehydrogenase A chain (LDH) | SM/SM−C: M1-loss +N-term- Acetyl | • | • | ||
| P29401 | Transketolase | • | • | |||
| P81605 | Dermcidin | • | ||||
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | • | ||||
| P15515 | Histatin-1 (Hst-1) | • | ||||
| Q8TAX7 | Mucin 7 | • | ||||
| Q6UWP8 | Suprabasin | • | ||||
| P07951 | Tropomyosin beta chain | • | ||||
| P08670 | Vimentin | • |
| Term ID | Term | FDR * | Associated Genes | Uniprot KB Code |
|---|---|---|---|---|
| Gene Ontology (GO) Biological Process | ||||
| GO:0001895 | Retina homeostasis | 0.00028 | CST4, LTF, LYZ, PIP, PIGR | P01036, P02788, P61626, P12273, P01833 |
| GO:0060249 | Anatomical structure homeostasis | 0.0019 | CST4, LTF, LYZ, PIP, PIGR, ALDOA | P01036, P02788, P61626, P12273, P01833 |
| GO:0048871 | Multicellular organismal homeostasis | 0.0049 | CST4, LTF, LYZ, PIP, SFN, PIGR | P01036, P02788, P61626, P12273, P31947, P01833 |
| GO:0065008 | Regulation of biological quality | 0.0043 | CTSG, CST4, LTF, LYZ, PIP, SFN, PIGR, S100A9, SLC25A5, DSP, YWHAZ, ARPC4, CFL1, PHB, ALDOA | P08311, P01036, P02788, P61626, P12273, P31947, P01833, P06702, P05141, P15924, P63104, P59998, P23528, P35232, P04075 |
| GO:2000117 | Negative regulation of cysteine-type endopeptidase activity | 0.00028 | CST4, LTF, SERPINB3, CSTB, CST1, CST5, SFN | P01036, P02788, P29508, Q76LA1, P01037, P28325, P31947 |
| GO:0052548 | Regulation of endopeptidase activity | 0.00028 | CST4, LTF, SERPINB3, CSTB, CST1, CST5, SFN, S100A9 | P01036, P02788, P29508, Q76LA1, P01037, P28325, P31947, P06702 |
| GO:0051336 | Regulation of hydrolase activity | 0.0024 | CST4, LTF, SERPINB3, CSTB, CST1, CST5, SFN, S100A9, PHB | P01036, P02788, P29508, P04080, P01037, P28325, P31947, P06702, P35232 |
| GO:0001580 | Detection of chemical stimulus involved in sensory perception of bitter taste | 0.00039 | CST4, PIP, CST1, PIGR | P01036, P12273, P01037, P01833 |
| GO:0019730 | Antimicrobial humoral response | 0.0015 | CTSG, LTF, LYZ, S100A9, DMBT1 | P08311, P02788, P61626, P06702, Q9UGM3 |
| GO:0050829 | Defense response to Gram-negative bacterium | 0.0056 | CTSG, LTF, LYZ, DMBT1 | P01036, P02788, P61626, Q9UGM3 |
| GO:0051702 | Biological process involved in interaction with symbiont | 0.0081 | CTSG, LTF, CFL1, PHB | P01036, P02788, P06702, P05141 |
| Reactome Pathway | ||||
| HSA-168249 | Innate Immune System | 3.13 × 10−6 | CTSG, LTF, LYZ, SERPINB3, CSTB, PIGR, S100A9, DSP, ARPC4, CFL1, MUC5B, ALDOA | P01036, P02788, P61626, P29508, P04080, P01833, P06702, P15924, P59998, P06702, Q9HC84, P15924, |
| HSA-6798695 | Neutrophil degranulation | 5.02 × 10−6 | CTSG, LTF, LYZ, SERPINB3, CSTB, PIGR, S100A9, DSP, ALDOA | P01036, P02788, P61626, P29508, P04080, P01833, P06702, P15924, P15924, |
| HSA-6803157 | Antimicrobial peptides | 0.0028 | CTSG, LTF, LYZ, S100A9 | P01036, P02788, P61626, P06702, |
| HSA-195258 | RHO GTPase Effectors | 0.0142 | SFN, S100A9, YWHAZ, ARPC4, CFL1 | P31947, P06702, P63104, P59998, P06702 |
| HSA-6799990 | Metal sequestration by antimicrobial proteins | 0.0164 | LTF, S100A9 | P02788, P06702 |
| Term ID | Term | FDR * | Associated Genes | Uniprot KB Code |
|---|---|---|---|---|
| Gene Ontology (GO) Biological Process | ||||
| GO:0061844 | Antimicrobial humoral immune response mediated by antimicrobial peptide | 0.00014 | S100A9, DMBT1, GAPDH, DCD, MUC7, HTN1 | P06702, Q9UGM3, P04406, P81605, Q8TAX7, P15515 |
| GO:0031640 | Killing of cells of another organism | 0.0022 | GAPDH, DCD, MUC7, HTN1 | P04406, P81605, Q8TAX7, P15515 |
| GO:0050832 | Defense response to fungus | 0.0020 | S100A9, GAPDH, DCD, HTN1 | P06702, P04406, P81605, P04075 |
| GO:0098542 | Defense response to other organism | 0.0150 | ANXA3, S100A9, DMBT1, GAPDH, DCD, MUC7, HTN1, VIM | P12429, P06702, Q9UGM3, P04406, P81605, Q8TAX7, P15515, P08670 |
| GO:0042742 | Defense response to bacterium | 0.0215 | ANXA3, S100A9, DMBT1, DCD, HTN1 | P12429, P06702, Q9UGM3, Q9UGM3, P81605, P04075 |
| GO:0044419 | Biological process involved in interspecies interaction between organisms | 0.0316 | ANXA3, SERPINB3, S100A9, DMBT1, GAPDH, DCD, MUC7, HTN1, VIM | P12429, P29508, P06702, Q9UGM3, P04406, P81605, Q8TAX7, P15515, P08670 |
| GO:0052548 | Regulation of endopeptidase activity | 0.00019 | CST4, SERPINB3, CST1, CST5, S100A9, SERPINB1, GAPDH, SMR3B | P01036, P29508, P01037, P28325, P06702, P30740, P04406, P02814 |
| GO:0006096 | Glycolytic process | 0.00091 | PGK1, GAPDH, LDHA, ALDOA | P00558, P04406, P00338, P04075 |
| GO:0035606 | Peptidyl-cysteine S-trans-nitrosylation | 0.0063 | S100A9, GAPDH | P06702, P04406 |
| GO:0001580 | Detection of chemical stimulus involved in sensory perception of bitter taste | 0.0113 | CST4, CST1, PIGR | P01036, P01037, P01833 |
| GO:0006955 | Immune response | 0.0150 | ANXA3, PIGR, S100A9, DMBT1, GAPDH, DCD, MUC7, HTN1, VIM | P12429, P01833, P06702, Q9UGM3, P04406, P81605, Q8TAX7, P15515, P08670 |
| GO:0035425 | Autocrine signaling | 0.0253 | SERPINB3, S100A9 | P29508, P06702 |
| Reactome Pathway | ||||
| HSA-168249 | Innate Immune System | 7.93 × 10−5 | CAPZA1, SERPINB3, PIGR, S100A9, DSP, SERPINB1, DCD, MUC7, HTN1, MUC5B, ALDOA | P52907, P29508, P01833, P01833, P15924, P30740, P81605, Q8TAX7, P15515, Q9HC84, P04075, |
| HSA-70263 | Gluconeogenesis | 0.0107 | PGK1, GAPDH, ALDOA | P00558, P04406, P04075 |
| HSA-6798695 | Neutrophil degranulation | 0.0179 | SERPINB3, PIGR, S100A9, DSP, SERPINB1, ALDOA | P01833, P01833, P06702, P15924, P30740, P04075 |
| Term ID | Term | FDR * | Associated Genes | Uniprot KB code |
|---|---|---|---|---|
| Gene Ontology (GO) Biological Process | ||||
| GO:0030216 | Keratinocyte differentiation | 0.00015 | CNFN, FLG, HRNR, ANXA1, DSP, CASP14 | Q9BYD5, P20930, Q86YZ3, P04083, P15924, P31944 |
| GO:0030855 | Epithelial cell differentiation | 0.00072 | CNFN, FLG, HRNR, DMBT1, PGK1, ANXA1, DSP, CASP14 | Q9BYD5, P20930, Q86YZ3, Q9UGM3, P00558, P04083, P15924, P31944 |
| GO:0018149 | Peptide cross-linking | 0.0121 | FLG, ANXA1, DSP | P20930, P04083, P15924 |
| GO:0001580 | Detection of chemical stimulus involved in sensory perception of bitter taste | 0.00063 | PIP, AZGP1, CST1, PIGR | P12273, P25311, P01037, P01833 |
| GO:0051346 | Negative regulation of hydrolase activity | 0.0032 | SERPINB3, CST1, CST5, ANXA1, SERPINB1, SMR3B | P29508, P01037, P28325, P04083, P30740, P02814 |
| GO:0052548 | Regulation of endopeptidase activity | 0.0072 | SERPINB3, CST1, CST5, S100A9, SERPINB1, SMR3B | P30740, P01037, P28325, P06702, P02814 |
| GO:0035425 | Autocrine signaling | 0.0371 | SERPINB3, S100A9 | P29508, P06702 |
| GO:0048871 | Multicellular organismal homeostasis | 0.0413 | PIP, AZGP1, PIGR, FLG, HRNR | P12273, P25311, P01833, P11362, Q86YZ3 |
| Reactome Pathway | ||||
| HSA-6798695 | Neutrophil degranulation | 6.70 × 10−5 | SERPINB3, TUBB4B, PIGR, S100A9, HRNR, DSP, SERPINB1, ALDOA | P29508, P68371, P01833, P06702, Q86YZ3, P15924, P30740, V9HWH1, P04075 |
| HSA-168256 | Immune System | 0.0020 | SERPINB3, P4HB, TUBB4B, PIGR, S100A9, HRNR, ANXA1, DSP, SERPINB1, MUC5B, ALDOA | P29508, P07237, P68371, P01833, P06702, Q86YZ3, P04083, P15924, P30740, Q9HC84, P04075 |
| Patient | Age/Gender | Diagnosis | Secondary Cutaneous Symptoms (SM+C) | Tryptase (μg/L) | Mutation |
|---|---|---|---|---|---|
| #9 | 45/M | SM | Y | 33.6 | M541L |
| #11 | 58/F | SM | Y | 20.2 | D816V |
| #13 | 35/F | SM | Y | 44.7 | |
| #15 | 38/F | SM | Y | 7.66 | D816V |
| #16 | 52/M | SM | Y | 37.5 | D816V |
| #18 | 55/M | SM | 27.1 | D816V | |
| #19 | 48/M | SM | Y | 16.2 | |
| #20 | 32/M | SM | Y | 53.5 | D816V |
| #22 | 60/F | SM | Y | 23.6 | |
| #24 | 49/M | SM | Y | 29.1 | D816V |
| #26 | 35/F | SM | Y | 16.5 | |
| #28 | 38/F | SM | Y | 26.2 | D816V |
| #29 | 57/M | SM | Y | 77.4 | D816V |
| #35 | 34/M | SM | Y | 28.7 | |
| #36 | 52/F | SM | Y | 16.1 | D816V |
| #41 | 38/M | SM | Y | 26.3 | |
| #49 | 51/M | SM | Y | 5.02 | |
| #51 | 29/M | SM | Y | 64.5 | |
| #59 | 67/F | SM | Y | 30.6 | D816V |
| #60 | 49/F | SM | Y | 15.8 | D816V |
| #63 | 56/M | SM | Y | 60.8 | D816V |
| #66 | 36/F | SM | Y | 6.63 | |
| #68 | 42/M | SM | 21.9 | D816V | |
| #68 | 54/F | SM | Y | 21.9 | D816V |
| #70 | 72/M | SM | Y | 59.7 | D816V |
| #71 | 35/M | SM | 16.5 | ||
| #72 | 44/F | SM | Y | 126 | D816V |
| #73 | 35/M | SM | Y | 40.8 | D816V |
| #74 | 56/M | SM | 5.67 | ||
| #75 | 64/F | SM | Y | 45.1 | |
| #76 | 79/F | SM | Y | 73.9 | D816V |
| #77 | 35/F | SM | Y | 44.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serrao, S.; Contini, C.; Guadalupi, G.; Olianas, A.; Lai, G.; Messana, I.; Castagnola, M.; Costanzo, G.; Firinu, D.; Del Giacco, S.; et al. Salivary Cystatin D Interactome in Patients with Systemic Mastocytosis: An Exploratory Study. Int. J. Mol. Sci. 2023, 24, 14613. https://doi.org/10.3390/ijms241914613
Serrao S, Contini C, Guadalupi G, Olianas A, Lai G, Messana I, Castagnola M, Costanzo G, Firinu D, Del Giacco S, et al. Salivary Cystatin D Interactome in Patients with Systemic Mastocytosis: An Exploratory Study. International Journal of Molecular Sciences. 2023; 24(19):14613. https://doi.org/10.3390/ijms241914613
Chicago/Turabian StyleSerrao, Simone, Cristina Contini, Giulia Guadalupi, Alessandra Olianas, Greca Lai, Irene Messana, Massimo Castagnola, Giulia Costanzo, Davide Firinu, Stefano Del Giacco, and et al. 2023. "Salivary Cystatin D Interactome in Patients with Systemic Mastocytosis: An Exploratory Study" International Journal of Molecular Sciences 24, no. 19: 14613. https://doi.org/10.3390/ijms241914613
APA StyleSerrao, S., Contini, C., Guadalupi, G., Olianas, A., Lai, G., Messana, I., Castagnola, M., Costanzo, G., Firinu, D., Del Giacco, S., Manconi, B., & Cabras, T. (2023). Salivary Cystatin D Interactome in Patients with Systemic Mastocytosis: An Exploratory Study. International Journal of Molecular Sciences, 24(19), 14613. https://doi.org/10.3390/ijms241914613








