Single-Cell RNA Sequencing of Donor-Reactive T Cells Reveals Role of Apoptosis in Donor-Specific Hyporesponsiveness of Kidney Transplant Recipients
Abstract
1. Introduction
2. Results
2.1. UMAP Clustering of Donor-Reactive T Cells
2.2. Differential Expression Analysis Determined T Cell Gene Expression Profile per Cluster
2.3. Gene Ontology Pathways Defined Cell Activity per Cluster
2.4. Differential Expression and GO Term Analysis between Post and Pre-Transplant Donor-stimulated Samples within Each Cluster
2.5. Diversity of Donor-Reactive TCR Repertoire Does Not Change over Time
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. PBMC Isolation
4.3. CD3+ T Cell Depletion of Allogeneic Stimuli
4.4. Sorting for CD137-Expressing Recipient T Cells
4.5. Combined Single Cell Transcriptomics and TRA/TRB Profiling
4.6. Amplification
4.7. Transcriptome and TCR Sequence Pre-Processing
4.8. Transcriptome Analysis
4.9. TCR Analysis
4.10. Shannon Diversity Calculations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mason, P.D.; Robinson, C.M.; Lechler, R.I. Detection of donor-specific hyporesponsiveness following late failure of human renal allografts. Kidney Int. 1996, 50, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Hornick, P.I.; Mason, P.D.; Yacoub, M.H.; Rose, M.L.; Batchelor, R.; Lechler, R.I. Assessment of the Contribution That Direct Allorecognition Makes to the Progression of Chronic Cardiac Transplant Rejection in Humans. Circulation 1998, 97, 1257–1263. [Google Scholar] [CrossRef] [PubMed][Green Version]
- de Haan, A.; Berg, A.P.v.D.; Hepkema, B.G.; van Dijk, E.; Haagsma, E.B.; The, T.H.; Slooff, M.J.; Lems, S.P.; de Leij, L.F.; Prop, J. Donor-specific hyporeactivity after liver transplantation: Prominent decreases in donor-specific cytotoxic T lymphocyte precursor frequencies independent of changes in helper T lymphocyte precursor frequencies or suppressor cell activity. Transplantation 1998, 66, 516–522. [Google Scholar] [CrossRef]
- Mestre, M.; Massip, E.; Bas, J.; Alsina, J.; Romeu, A.; Castelao, A.M.; Buendia, E.; Grinyó, J.M. Longitudinal study of the frequency of cytotoxic T cell precursors in kidney allograft recipients. Clin. Exp. Immunol. 1996, 104, 108–114. [Google Scholar] [CrossRef]
- DeBruyne, L.A.; Renlund, D.G.; Bishop, D.K. Evidence that human cardiac allograft acceptance is associated with a decrease in donor-reactive helper T lymphocytes. Transplantation 1995, 59, 778–783. [Google Scholar] [CrossRef] [PubMed]
- Ghobrial, I.I.; Morris, A.G.; Booth, L.J. Clinical significance of in vitro donor-specific hyporesponsiveness in renal allograft recipients as demonstrated by the MLR. Transpl. Int. 1994, 7, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Manickavasagar, R.; Thuraisingham, R. Post renal-transplant malignancy surveillance. Clin. Med. 2020, 20, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, J.; Beckmann, S.; Heller, K.; Hilgers, K.F.; Apel, H.; Spriewald, B.; Eckardt, K.-U.; Amann, K.U. Deceased Donor Kidney Transplantation in the Eurotransplant Senior Program (ESP): A Single-Center Experience from 2008 to 2013. Ann. Transplant. 2016, 21, 94–104. [Google Scholar] [CrossRef]
- Velthuis, J.H.L.; Mol, W.M.; Weimar, W.; Baan, C.C. CD4+ CD25bright+ Regulatory T Cells Can Mediate Donor Nonreactivity in Long-Term Immunosuppressed Kidney Allograft Patients. Am. J. Transplant. 2006, 6, 2955–2964. [Google Scholar] [CrossRef]
- Game, D.S.; Hernandez-Fuentes, M.P.; Chaudhry, A.N.; Lechler, R.I. CD4+CD25+ Regulatory T Cells Do Not Significantly Contribute to Direct Pathway Hyporesponsiveness in Stable Renal Transplant Patients. J. Am. Soc. Nephrol. 2003, 14, 1652–1661. [Google Scholar] [CrossRef]
- Hornick, P.I.; Brookes, P.A.; Mason, P.D.; Taylor, K.M.; Yacoub, M.H.; Rose, M.L.; Batchelor, R.; Lechler, R.I. Optimizing a limiting dilution culture system for quantifying the frequency of interleukin-2-producing alloreactive t helper lymphocytes1. Transplantation 1997, 64, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.F.; Hernandez-Fuentes, M.; Baker, R.; Chaudhry, A.; Lechler, R.I. Reversibility with Interleukin-2 Suggests that T Cell Anergy Contributes to Donor-Specific Hyporesponsiveness in Renal Transplant Patients. J. Am. Soc. Nephrol. 2002, 13, 2983–2989. [Google Scholar] [CrossRef] [PubMed]
- van der List, A.C.J.; Litjens, N.H.R.; Klepper, M.; Betjes, M.G.H. Expression of Senescence Marker TIGIT Identifies Polyfunctional Donor-Reactive CD4+ T Cells Preferentially Lost After Kidney Transplantation. Front. Immunol. 2021, 12, 656846. [Google Scholar] [CrossRef]
- van der List, A.C.J.; Litjens, N.H.R.; Klepper, M.; Prevoo, F.; Betjes, M.G.H. Progressive Loss of Donor-Reactive CD4+ Effector Memory T Cells due to Apoptosis Underlies Donor-Specific Hyporesponsiveness in Stable Renal Transplant Recipients. J. Immunol. 2022, 209, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Morris, H.; DeWolf, S.; Robins, H.; Sprangers, B.; LoCascio, S.A.; Shonts, B.A.; Kawai, T.; Wong, W.; Yang, S.; Zuber, J.; et al. Tracking donor-reactive T cells: Evidence for clonal deletion in tolerant kidney transplant patients. Sci. Transl. Med. 2015, 7, 272ra10. [Google Scholar] [CrossRef]
- Dziubianau, M.; Hecht, J.; Kuchenbecker, L.; Sattler, A.; Stervbo, U.; Rödelsperger, C.; Nickel, P.; Neumann, A.U.; Robinson, P.N.; Mundlos, S.; et al. TCR Repertoire Analysis by Next Generation Sequencing Allows Complex Differential Diagnosis of T Cell–Related Pathology. Am. J. Transplant. 2013, 13, 2842–2854. [Google Scholar] [CrossRef]
- Emerson, R.O.; Mathew, J.M.; Konieczna, I.M.; Robins, H.S.; Leventhal, J.R. Defining the Alloreactive T Cell Repertoire Using High-Throughput Sequencing of Mixed Lymphocyte Reaction Culture. PLoS ONE 2014, 9, e111943. [Google Scholar] [CrossRef][Green Version]
- Savage, T.M.; Shonts, B.A.; Lau, S.; Obradovic, A.; Robins, H.; Shaked, A.; Shen, Y.; Sykes, M. Deletion of donor-reactive T cell clones after human liver transplant. Am. J. Transplant. 2019, 20, 538–545. [Google Scholar] [CrossRef]
- De Simone, M.; Rossetti, G.; Pagani, M. Single Cell T Cell Receptor Sequencing: Techniques and Future Challenges. Front. Immunol. 2018, 9, 1638. [Google Scholar] [CrossRef]
- Litjens, N.H.R.; Langerak, A.W.; van der List, A.C.J.; Klepper, M.; de Bie, M.; Azmani, Z.; Dekker, A.T.D.; Brouwer, R.W.W.; Betjes, M.G.H.; Van Ijcken, W.F.J. Validation of a Combined Transcriptome and T Cell Receptor Alpha/Beta (TRA/TRB) Repertoire Assay at the Single Cell Level for Paucicellular Samples. Front. Immunol. 2020, 11, 1999. [Google Scholar] [CrossRef]
- Litjens, N.H.R.; A de Wit, E.; Baan, C.C.; Betjes, M.G.H. Activation-induced CD137 is a fast assay for identification and multi-parameter flow cytometric analysis of alloreactive T cells. Clin. Exp. Immunol. 2013, 174, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Goda, C.; Kanaji, T.; Kanaji, S.; Tanaka, G.; Arima, K.; Ohno, S.; Izuhara, K. Involvement of IL-32 in activation-induced cell death in T cells. Int. Immunol. 2006, 18, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Windgassen, D.; Papoutsakis, E.T. A global transcriptional view of apoptosis in human T-cell activation. BMC Med. Genom. 2008, 1, 53. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yue, H. Thymidine phosphorylase: A potential new target for treating cardiovascular disease. Trends Cardiovasc. Med. 2018, 28, 157–171. [Google Scholar] [CrossRef]
- Takeda, M.; Tezuka, T.; Kim, M.; Choi, J.; Oichi, Y.; Kobayashi, H.; Harada, K.H.; Mizushima, T.; Taketani, S.; Koizumi, A.; et al. Moyamoya disease patient mutations in the RING domain of RNF213 reduce its ubiquitin ligase activity and enhance NFκB activation and apoptosis in an AAA+ domain-dependent manner. Biochem. Biophys. Res. Commun. 2020, 525, 668–674. [Google Scholar] [CrossRef]
- DeWolf, S.; Grinshpun, B.; Savage, T.; Lau, S.P.; Obradovic, A.; Shonts, B.; Yang, S.; Morris, H.; Zuber, J.; Winchester, R.; et al. Quantifying size and diversity of the human T cell alloresponse. J. Clin. Investig. 2018, 3, e121256. [Google Scholar] [CrossRef]
- Aschauer, C.; Jelencsics, K.; Hu, K.; Heinzel, A.; Gregorich, M.G.; Vetter, J.; Schaller, S.; Winkler, S.M.; Weinberger, J.; Pimenov, L.; et al. Prospective Tracking of Donor-Reactive T-Cell Clones in the Circulation and Rejecting Human Kidney Allografts. Front. Immunol. 2021, 12, 750005. [Google Scholar] [CrossRef]
- Tian, G.; Li, M.; Lv, G. Analysis of T-Cell Receptor Repertoire in Transplantation: Fingerprint of T Cell-mediated Alloresponse. Front. Immunol. 2022, 12, 778559. [Google Scholar] [CrossRef]
- Qian, S.; Lu, L.; Fu, F.; Li, Y.; Li, W.; E Starzl, T.; Fung, J.J.; Thomson, A.W. Apoptosis within spontaneously accepted mouse liver allografts: Evidence for deletion of cytotoxic T cells and implications for tolerance induction. J. Immunol. 1997, 158, 4654–4661. [Google Scholar] [CrossRef]
- Steger, U.; Denecke, C.; Sawitzki, B.; Karim, M.; Jones, N.D.; Wood, K.J. Exhaustive Differentiation of Alloreactive CD8+ T Cells: Critical for Determination of Graft Acceptance or Rejection. Transplantation 2008, 85, 1339–1347. [Google Scholar] [CrossRef]
- Tay, S.S.; Lu, B.; Sierro, F.; Benseler, V.; McGuffog, C.M.; Bishop, G.A.; Cowan, P.J.; McCaughan, G.W.; Dwyer, K.M.; Bowen, D.G.; et al. Differential migration of passenger leukocytes and rapid deletion of naive alloreactive CD8 T cells after mouse liver transplantation. Liver Transplant. 2013, 19, 1224–1235. [Google Scholar] [CrossRef] [PubMed]
- Wekerle, T.; Kurtz, J.; Sayegh, M.H.; Ito, H.; Wells, A.D.; Bensinger, S.; Shaffer, J.; Turka, L.A.; Sykes, M. Peripheral Deletion After Bone Marrow Transplantation with Costimulatory Blockade Has Features of Both Activation-Induced Cell Death and Passive Cell Death. J. Immunol. 2001, 166, 2311–2316. [Google Scholar] [CrossRef] [PubMed]
- Cippà, P.E.; Gabriel, S.S.; Chen, J.; Bardwell, P.D.; Bushell, A.; Guimezanes, A.; Kraus, A.K.; Wekerle, T.; Wüthrich, R.P.; Fehr, T. Targeting apoptosis to induce stable mixed hematopoietic chimerism and long-term allograft survival without myelosuppressive conditioning in mice. Blood 2013, 122, 1669–1677. [Google Scholar] [CrossRef]
- Litjens, N.H.R.; van der List, A.C.J.; Klepper, M.; Prevoo, F.; Boer, K.; A Hesselink, D.; Betjes, M.G.H. Polyfunctional donor-reactive T cells are associated with acute T-cell-mediated rejection of the kidney transplant. Clin. Exp. Immunol. 2023. [Google Scholar] [CrossRef]
- Wang, X.; Shen, X.; Chen, S.; Liu, H.; Hong, N.; Zhong, H.; Chen, X.; Jin, W. Reinvestigation of Classic T Cell Subsets and Identification of Novel Cell Subpopulations by Single-Cell RNA Sequencing. J. Immunol. 2022, 208, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Kouno, T.; Ikawa, T.; Hayatsu, N.; Miyajima, Y.; Yabukami, H.; Terooatea, T.; Sasaki, T.; Suzuki, T.; Valentine, M.; et al. Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc. Natl. Acad. Sci. USA 2019, 116, 24242–24251. [Google Scholar] [CrossRef] [PubMed]
- Corselli, M.; Saksena, S.; Nakamoto, M.; Lomas, W.E.; Taylor, I.; Chattopadhyay, P.K. Single cell multiomic analysis of T cell exhaustion in vitro. Cytom. Part A 2021, 101, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Litjens, N.H.R.; van de Wetering, J.; van Besouw, N.M.; Betjes, M.G.H. The human alloreactive CD4+ T-cell repertoire is biased to a Th17 response and the frequency is inversely related to the number of HLA class II mismatches. Blood 2009, 114, 3947–3955. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 357–360. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Cunningham, F.; Achuthan, P.; Akanni, W.; Allen, J.; Amode, M.R.; Armean, I.M.; Bennett, R.; Bhai, J.; Billis, K.; Boddu, S.; et al. Ensembl 2019. Nucleic Acids Res. 2019, 47, D745–D751. [Google Scholar] [CrossRef]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinform. 2014, 47, 11.12.1–11.12.34. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2013, 30, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Lefranc, M. Nomenclature of the Human T Cell Receptor Genes. Curr. Protoc. Immunol. 2000, 40, A.1O.1–A.1O.23. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Ma, N.; Madden, T.L.; Ostell, J.M. IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013, 41, W34–W40. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef] [PubMed]
- Zappia, L.; Oshlack, A. Clustering trees: A visualization for evaluating clusterings at multiple resolutions. GigaScience 2018, 7, giy083. [Google Scholar] [CrossRef]
- Ntranos, V.; Yi, L.; Melsted, P.; Pachter, L. A discriminative learning approach to differential expression analysis for single-cell RNA-seq. Nat. Methods 2019, 16, 163–166. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]

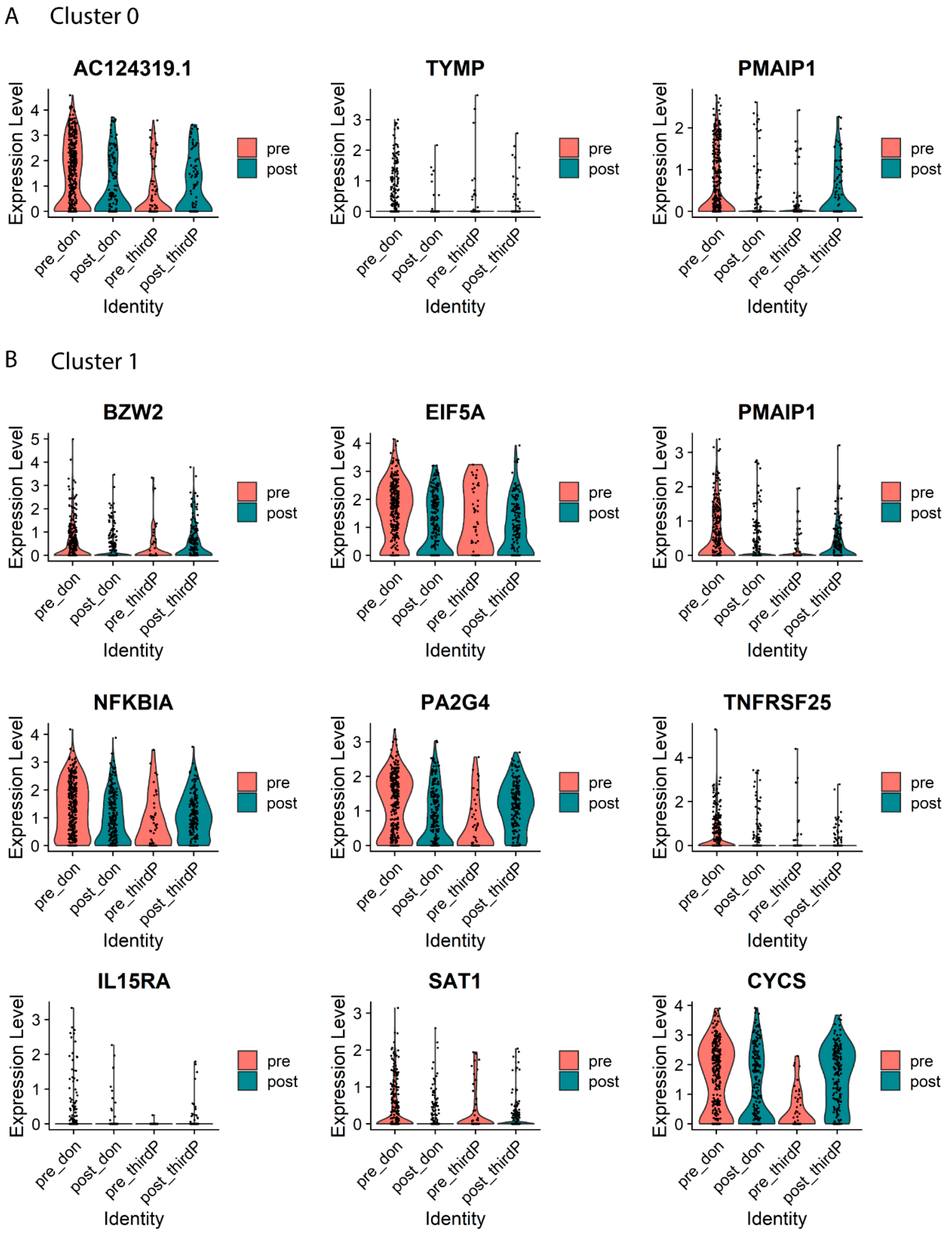

| Gene_ID | Symbol | Average log2FC | Adjusted p-Value | Description |
|---|---|---|---|---|
| ENSG00000173821 | RNF213 | −0.79 | 0.05 | ring finger protein 213 |
| ENSG00000025708 | TYMP | −0.66 | 0.002 | thymidine phosphorylase |
| ENSG00000141682 | PMAIP1 | −0.42 | 0.01 | phorbol-12-myristate-13-acetate-induced protein 1 |
| Gene_ID | Symbol | Average log2FC | Adjusted p-Value | Description |
|---|---|---|---|---|
| ENSG00000136261 | BZW2 | −0.75 | 5.9 × 10−3 | basic leucine zipper and W2 domains 2 |
| ENSG00000132507 | EIF5A | −0.73 | 4.8 × 10−5 | eukaryotic translation initiation factor 5A |
| ENSG00000141682 | PMAIP1 | −0.61 | 7.1 × 10−5 | Phorbol-12-myristate-13-acetate-induced protein 1 |
| ENSG00000100906 | NFKBIA | −0.57 | 1.1 × 10−2 | NFKB inhibitor alpha |
| ENSG00000170515 | PA2G4 | −0.54 | 4.6 × 10−4 | proliferation-associated 2G4 |
| ENSG00000215788 | TNFRSF25 | −0.47 | 1.1 × 10−2 | TNF receptor superfamily member 25 |
| ENSG00000049249 | TNFRSF9 | −0.47 | 1.5 × 10−2 | TNF receptor superfamily member 9 |
| ENSG00000134470 | IL15RA | −0.46 | 6.6 × 10−3 | interleukin 15 receptor subunit alpha |
| ENSG00000130066 | SAT1 | −0.44 | 3.8 × 10−2 | spermidine/spermine N1-acetyltransferase 1 |
| ENSG00000172115 | CYCS | −0.42 | 1.8 × 10−2 | cytochrome c, somatic |
| ENSG00000136045 | PWP1 | −0.41 | 1.5 × 10−2 | PWP1 homolog, endonuclein |
| ENSG00000134987 | WDR36 | −0.40 | 1.5 × 10−2 | WD repeat domain 36 |
| ENSG00000116717 | GADD45A | −0.39 | 4.1 × 10−3 | growth arrest and DNA damage inducible alpha |
| ENSG00000013306 | SLC25A39 | −0.38 | 3.1 × 10−2 | solute carrier family 25 member 39 |
| ENSG00000231925 | TAPBP | −0.37 | 5.4 × 10−3 | TAP binding protein |
| ENSG00000196396 | PTPN1 | −0.35 | 1.7 × 10−3 | protein tyrosine phosphatase non-receptor type 1 |
| ENSG00000143942 | CHAC2 | −0.33 | 4.1 × 10−5 | ChaC cation transport regulator homolog 2 |
| ENSG00000115946 | PNO1 | −0.28 | 3.1 × 10−2 | partner of NOB1 homolog |
| ENSG00000105447 | GRWD1 | −0.27 | 9.9 × 10−5 | glutamate rich WD repeat containing 1 |
| Category | Direction | Over- Represented p-Value | * DE Genes in Term | ** Total Genes in Term | Term |
|---|---|---|---|---|---|
| GO:0033140 | down | 1.02 × 10−5 | 2 | 3 | negative regulation of peptidyl-serine phosphorylation of STAT protein |
| GO:0033139 | down | 5.12 × 10−5 | 2 | 6 | regulation of peptidyl-serine phosphorylation of STAT protein |
| GO:2001244 | down | 1.35 × 10−4 | 3 | 52 | positive regulation of intrinsic apoptotic signaling pathway |
| GO:2001242 | down | 1.55 × 10−4 | 4 | 146 | regulation of intrinsic apoptotic signaling pathway |
| GO:0042501 | down | 1.62 × 10−4 | 2 | 10 | serine phosphorylation of STAT protein |
| GO:0019221 | down | 4.62 × 10−4 | 5 | 364 | cytokine-mediated signaling pathway |
| GO:0033209 | down | 5.64 × 10−4 | 3 | 89 | tumor necrosis factor-mediated signaling pathway |
| GO:0006364 | down | 7.43 × 10−4 | 4 | 219 | rRNA processing |
| GO:0006915 | down | 7.84 × 10−4 | 9 | 1447 | apoptotic process |
| GO:0033137 | down | 8.37 × 10−4 | 2 | 23 | negative regulation of peptidyl-serine phosphorylation |
| Sample | TRA | TRB | TRA AND TRB |
|---|---|---|---|
| C0-P0_pre_don | 0.92 | 0.87 | 0.99 |
| C0-P0_post_don | 0.90 | 0.92 | 0.98 |
| C1-P1_pre_don | 0.99 | 0.99 | 1.00 |
| C1-P1_post_don | 0.98 | 0.98 | 0.99 |
| C2-P2_pre_don | 0.78 | 0.79 | 0.94 |
| C2-P2_post_don | 0.94 | 0.69 | 0.93 |
| C0-P0_pre_thirdP | 0.87 | 0.92 | 0.97 |
| C0-P0_post_thirdP | 0.67 | 0.83 | 0.81 |
| C2-P2_pre_thirdP | 0.90 | 0.67 | 0.94 |
| C2-P2_post_thirdP | 0.78 | 0.61 | 0.79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van der List, A.C.J.; Litjens, N.H.R.; Brouwer, R.W.W.; Klepper, M.; den Dekker, A.T.; van Ijcken, W.F.J.; Betjes, M.G.H. Single-Cell RNA Sequencing of Donor-Reactive T Cells Reveals Role of Apoptosis in Donor-Specific Hyporesponsiveness of Kidney Transplant Recipients. Int. J. Mol. Sci. 2023, 24, 14463. https://doi.org/10.3390/ijms241914463
van der List ACJ, Litjens NHR, Brouwer RWW, Klepper M, den Dekker AT, van Ijcken WFJ, Betjes MGH. Single-Cell RNA Sequencing of Donor-Reactive T Cells Reveals Role of Apoptosis in Donor-Specific Hyporesponsiveness of Kidney Transplant Recipients. International Journal of Molecular Sciences. 2023; 24(19):14463. https://doi.org/10.3390/ijms241914463
Chicago/Turabian Stylevan der List, Amy C. J., Nicolle H. R. Litjens, Rutger W. W. Brouwer, Mariska Klepper, Alexander T. den Dekker, Wilfred F. J. van Ijcken, and Michiel G. H. Betjes. 2023. "Single-Cell RNA Sequencing of Donor-Reactive T Cells Reveals Role of Apoptosis in Donor-Specific Hyporesponsiveness of Kidney Transplant Recipients" International Journal of Molecular Sciences 24, no. 19: 14463. https://doi.org/10.3390/ijms241914463
APA Stylevan der List, A. C. J., Litjens, N. H. R., Brouwer, R. W. W., Klepper, M., den Dekker, A. T., van Ijcken, W. F. J., & Betjes, M. G. H. (2023). Single-Cell RNA Sequencing of Donor-Reactive T Cells Reveals Role of Apoptosis in Donor-Specific Hyporesponsiveness of Kidney Transplant Recipients. International Journal of Molecular Sciences, 24(19), 14463. https://doi.org/10.3390/ijms241914463






