Functional Meta-Analysis of the Proteomic Responses of Arabidopsis Seedlings to the Spaceflight Environment Reveals Multi-Dimensional Sources of Variability across Spaceflight Experiments
Abstract
:1. Introduction
2. Results
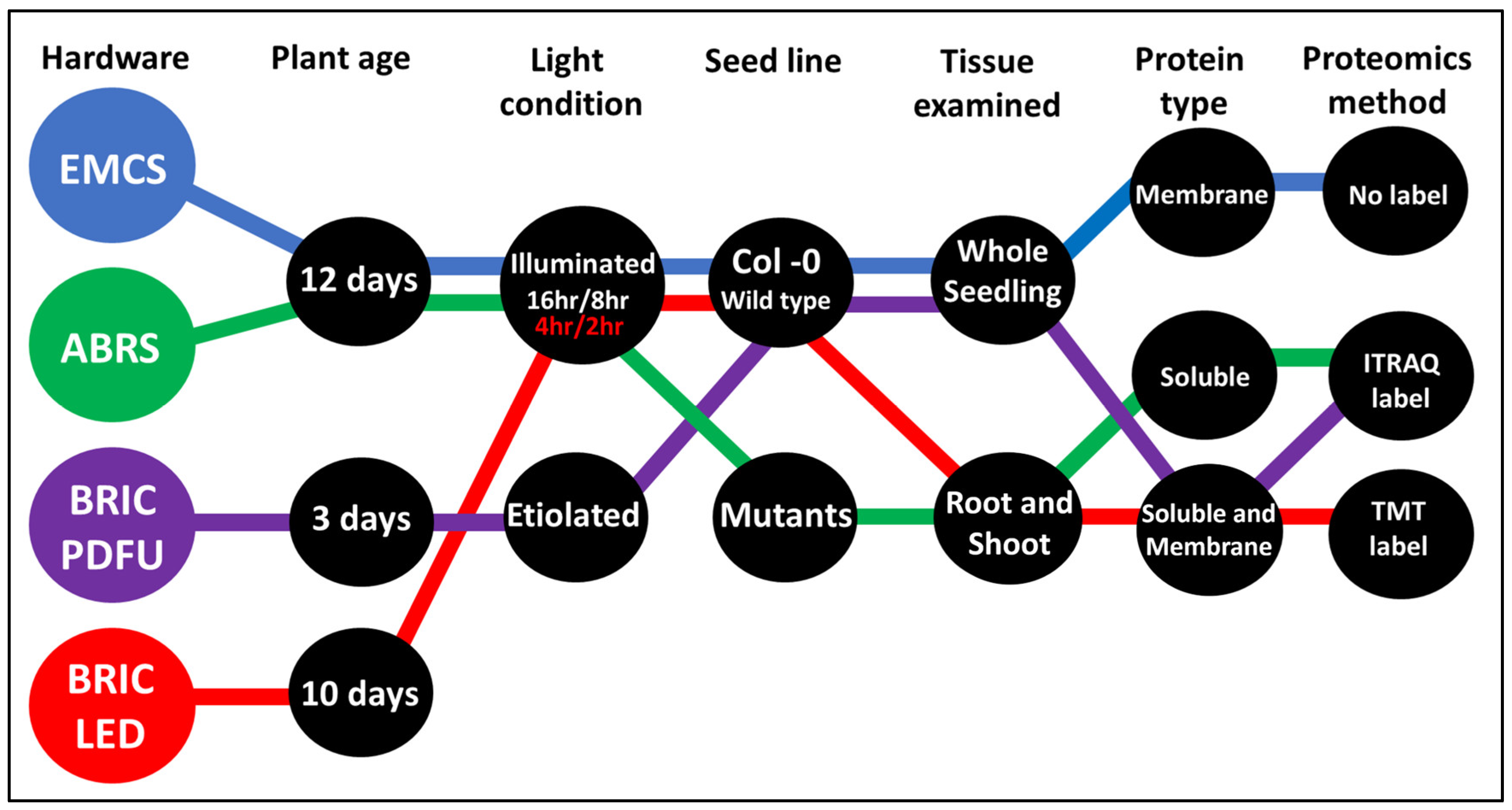
2.1. Curating the Proteome of Different Arabidopsis Spaceflight Experiments
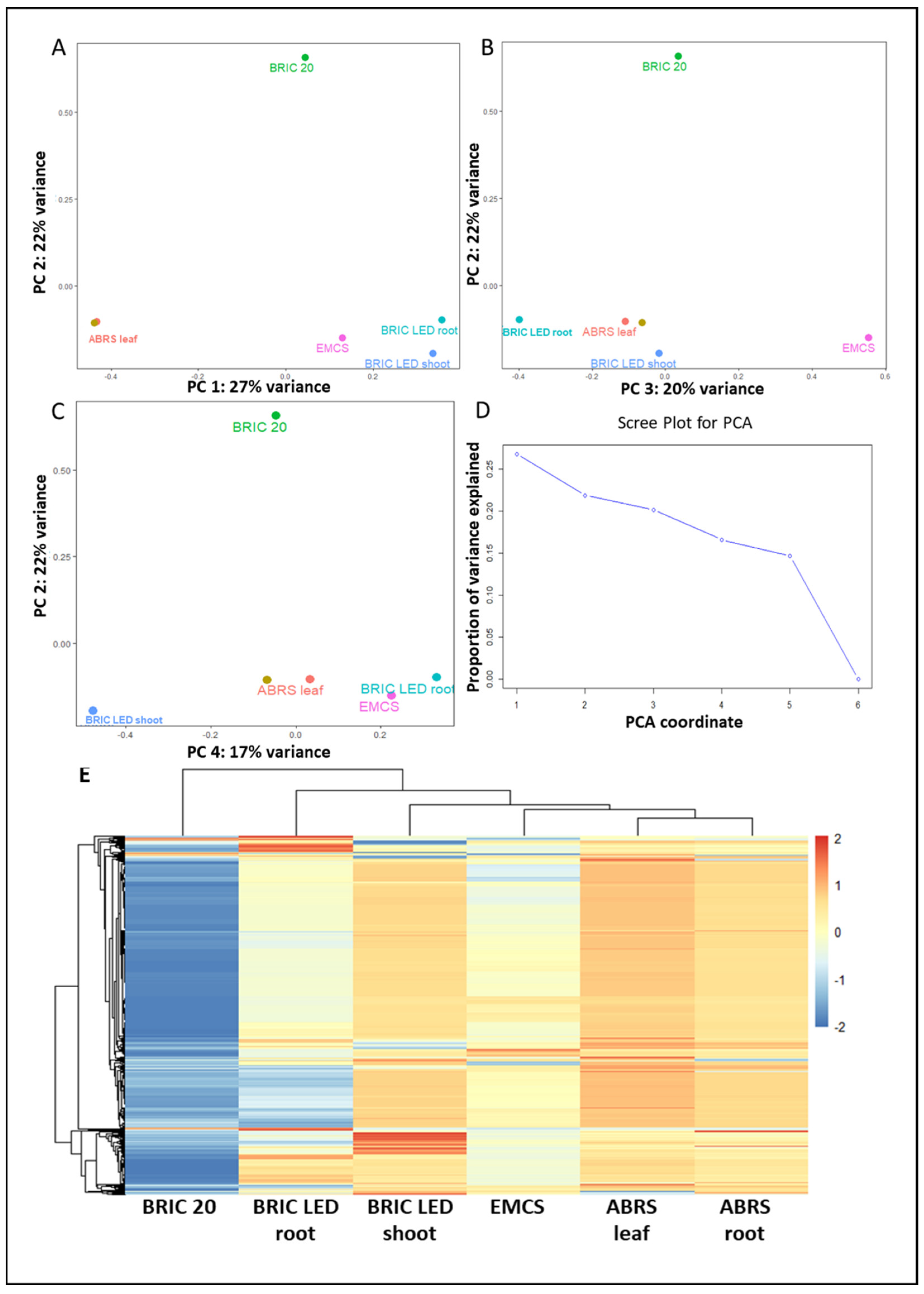
2.2. Principal Component Analysis
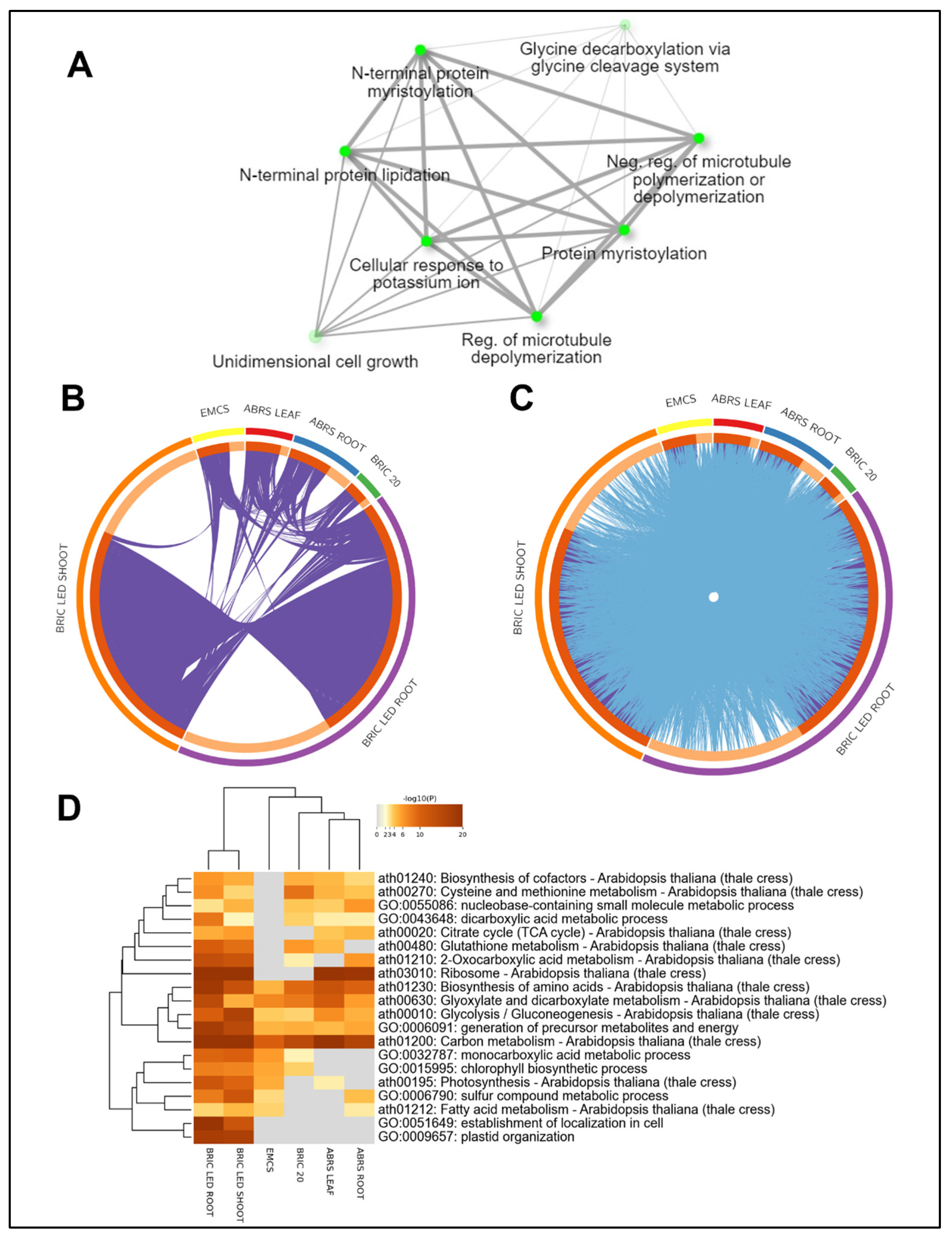
2.3. The Direct Intersection of the Four Proteomics Datasets
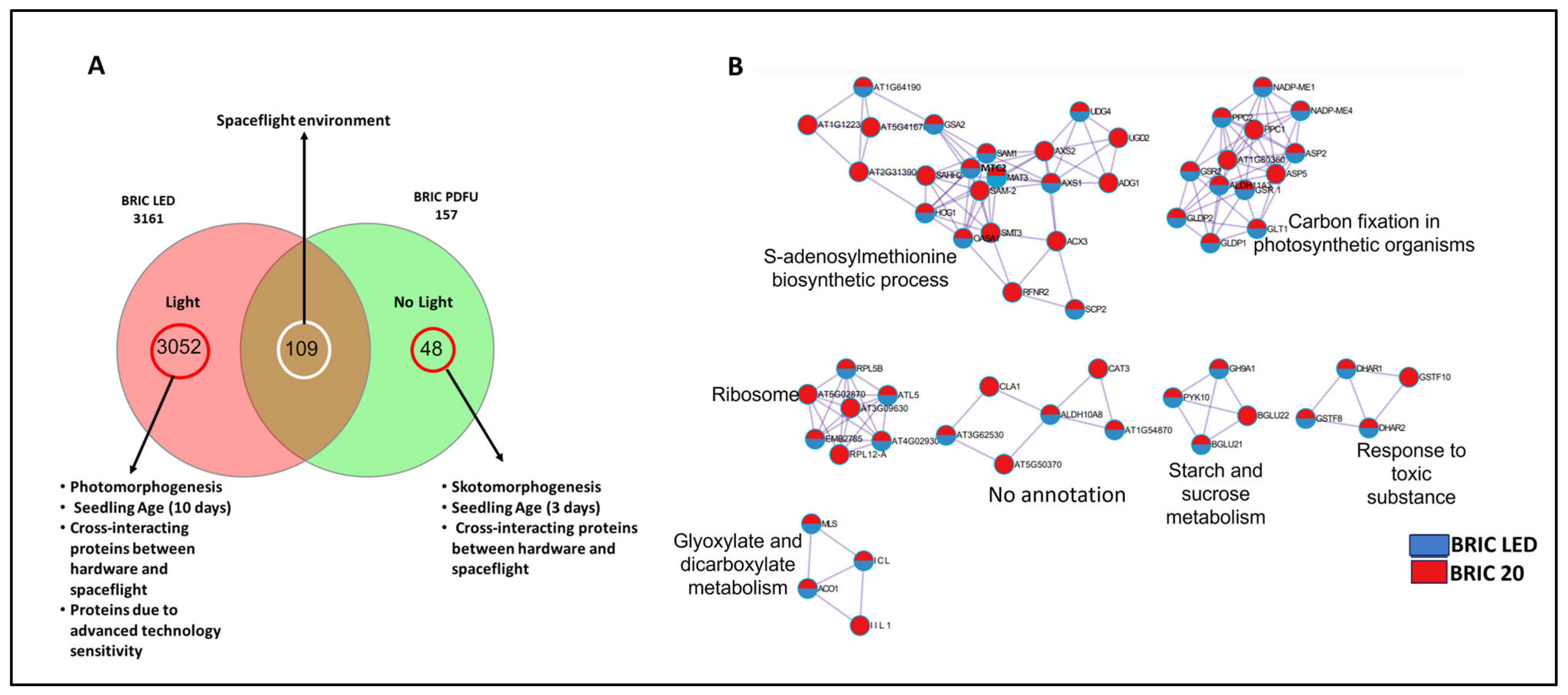
2.4. Comparing BRIC PDFU and BRIC LED Datasets: Impact of Light Conditions
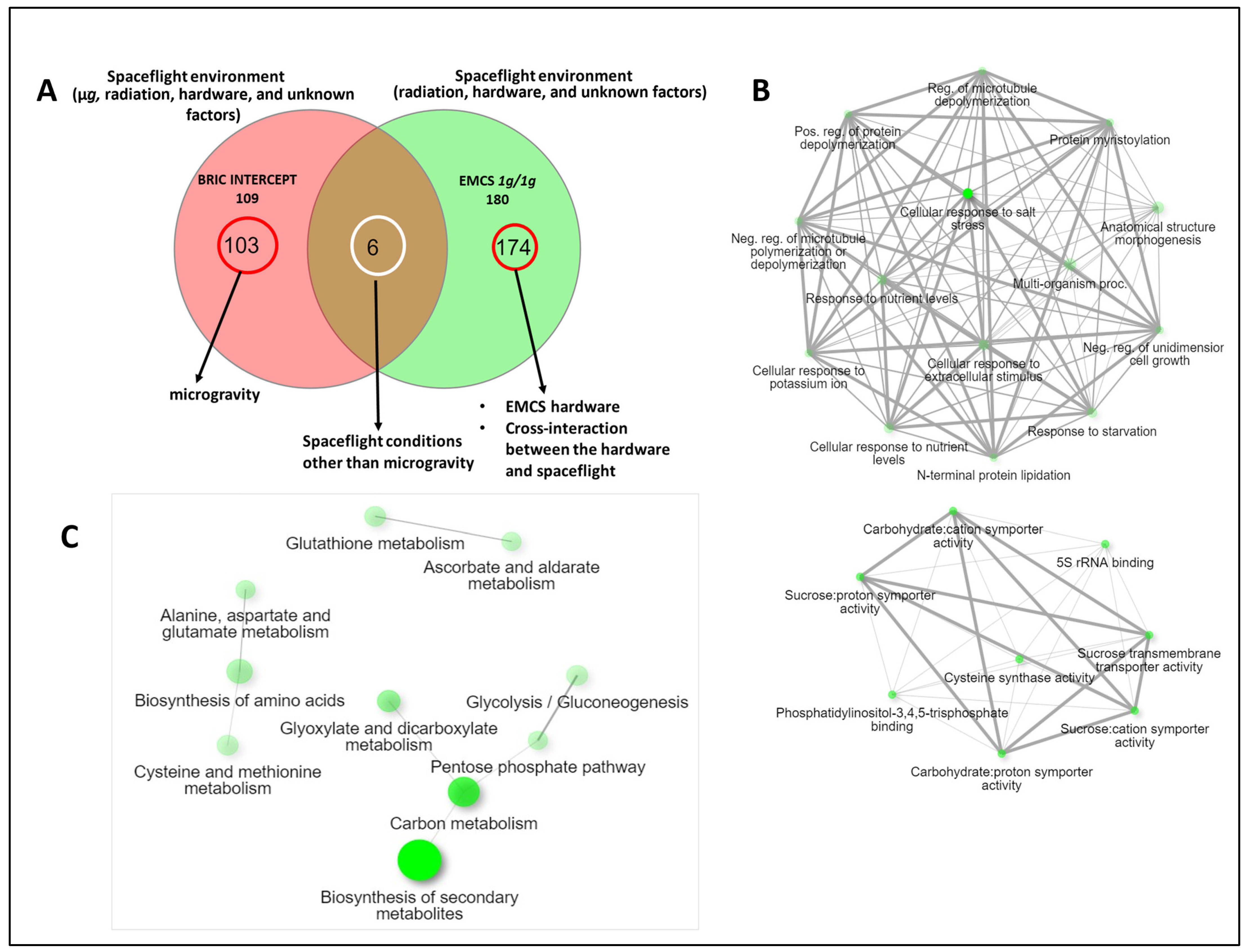
2.5. Eliminating Microgravity from the Comparison
3. Discussion
3.1. The Determinant of the Arabidopsis Proteomic Response to the Spaceflight Environment Is Multidimensional
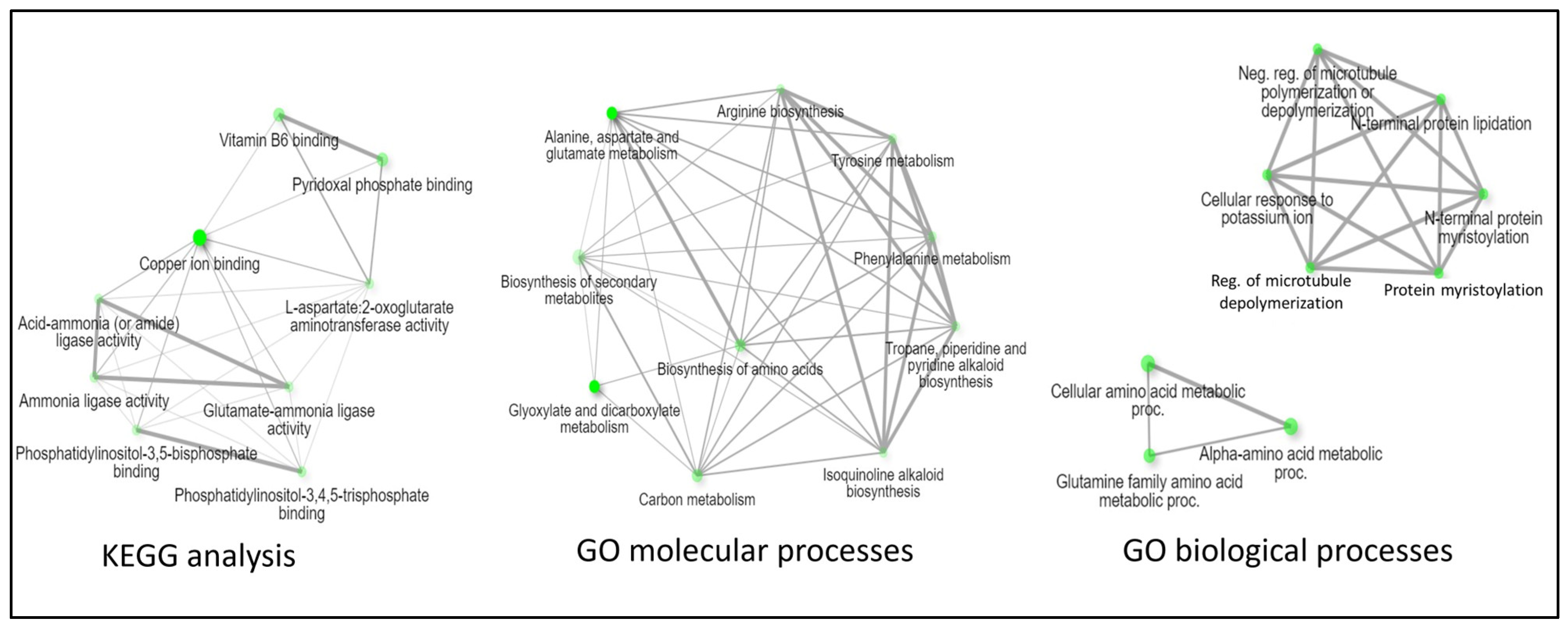
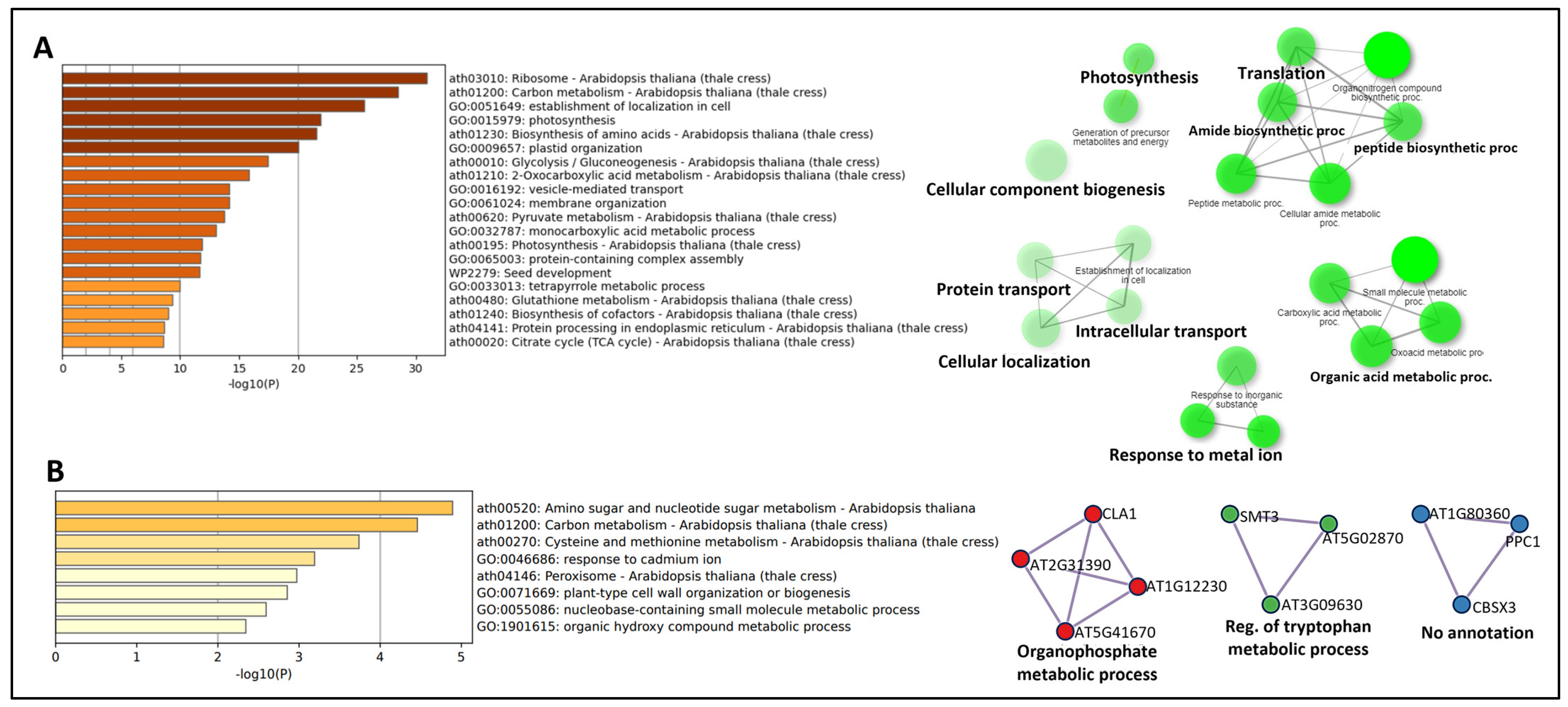
3.2. Protein Intersections and Functional Implications
3.3. Integration of Light Signals in Spaceflight Leads to Altered Metabolism
3.4. Disentangling Microgravity from Other Plant Spaceflight Responses
4. Materials and Methods
Accessing and Analyzing Individual Proteomics Datasets
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carillo, P.; Morrone, B.; Fusco, G.M.; De Pascale, S.; Rouphael, Y. Challenges for a Sustainable Food Production System on Board of the International Space Station: A Technical Review. Agronomy 2020, 10, 687. [Google Scholar] [CrossRef]
- Hughes, A.M.; Kiss, J.Z. -Omics studies of plant biology in spaceflight: A critical review of recent experiments. Front. Astron. Space Sci. 2022, 9, 964657. [Google Scholar] [CrossRef]
- Vandenbrink, J.P.; Kiss, J.Z. Plant responses to gravity. Semin. Cell Dev. Biol. 2019, 92, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Jan, R.; Kim, K.-M. Gravistimulation effects on Oryza sativa amino acid profile, growth pattern and expression of OsPIN genes. Sci. Rep. 2020, 10, 17303. [Google Scholar] [CrossRef]
- Medina, F.J.; Manzano, A.; Villacampa, A.; Ciska, M.; Herranz, R. Understanding Reduced Gravity Effects on Early Plant Development Before Attempting Life-Support Farming in the Moon and Mars. Front. Astron. Space Sci. 2021, 8, 729154. [Google Scholar] [CrossRef]
- Haveman, N.; Paul, A.; Ferl, R. Plant Biology and a New Approach to Space Farming. In In-Space Manufacturing and Resources; Wiley: Hoboken, NJ, USA, 2022; pp. 67–87. [Google Scholar] [CrossRef]
- Beisel, N.S.; Noble, J.; Barbazuk, W.B.; Paul, A.-L.; Ferl, R.J. Spaceflight-induced alternative splicing during seedling development in Arabidopsis thaliana. NPJ Microgravity 2019, 5, 9. [Google Scholar] [CrossRef]
- Paul, A.-L.; Sng, N.J.; Zupanska, A.K.; Krishnamurthy, A.; Schultz, E.R.; Ferl, R.J. Genetic dissection of the Arabidopsis spaceflight transcriptome: Are some responses dispensable for the physiological adaptation of plants to spaceflight? PLoS ONE 2017, 12, e0180186. [Google Scholar] [CrossRef]
- Zapalska-Sozoniuk, M.; Chrobak, L.; Kowalczyk, K.; Kankofer, M. Is it useful to use several “omics” for obtaining valuable results? Mol. Biol. Rep. 2019, 46, 3597–3606. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Kruse, C.P.S.; Meyers, A.D.; Basu, P.; Hutchinson, S.; Luesse, D.R.; Wyatt, S.E. Spaceflight induces novel regulatory responses in Arabidopsis seedling as revealed by combined proteomic and transcriptomic analyses. BMC Plant Biol. 2020, 20, 237. [Google Scholar] [CrossRef]
- Paul, A.-L.; Zupanska, A.K.; Schultz, E.R.; Ferl, R.J. Organ-specific remodeling of the Arabidopsis transcriptome in response to spaceflight. BMC Plant Biol. 2013, 13, 112. [Google Scholar] [CrossRef]
- Ferl, R.J.; Koh, J.; Denison, F.; Paul, A.-L. Spaceflight Induces Specific Alterations in the Proteomes of Arabidopsis. Astrobiology 2015, 15, 32–56. [Google Scholar] [CrossRef]
- Mazars, C.; Brière, C.; Grat, S.; Pichereaux, C.; Rossignol, M.; Pereda-Loth, V.; Eche, B.; Boucheron-Dubuisson, E.; Le Disquet, I.; Medina, F.-J.; et al. Microsome-associated proteome modifications of Arabidopsis seedlings grown on board the International Space Station reveal the possible effect on plants of space stresses other than microgravity. Plant Signal. Behav. 2014, 9, e29637. [Google Scholar] [CrossRef]
- Barker, R.; Kruse, C.P.S.; Johnson, C.; Saravia-Butler, A.; Fogle, H.; Chang, H.-S.; Trane, R.M.; Kinscherf, N.; Villacampa, A.; Manzano, A.; et al. Meta-analysis of the space flight and microgravity response of the Arabidopsis plant transcriptome. NPJ Microgravity 2023, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Kodaira, S.; Naito, M.; Uchihori, Y.; Hashimoto, H.; Yano, H.; Yamagishi, A. Space Radiation Dosimetry at the Exposure Facility of the International Space Station for the Tanpopo Mission. Astrobiology 2021, 21, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Tejos, R.; Rodriguez-Furlan, C.; Adamowski, M.; Sauer, M.; Norambuena, L.; Friml, J. PATELLINS are regulators of auxin-mediated PIN1 relocation and plant development in Arabidopsis thaliana. J. Cell Sci. 2017, 131, jcs204198. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, M.; Marti, L.; Ferrari, S.; Tanaka-Takada, N.; Maeshima, M.; Ott, T.; De Lorenzo, G.; Mattei, B. The plasma membrane–associated Ca2+-binding protein, PCaP1, is required for oligogalacturonide and flagellin-induced priming and immunity. Plant Cell Environ. 2021, 44, 3078–3093. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Xia, F.; Xiao, S.; Li, J. TRAF proteins as key regulators of plant development and stress responses. J. Integr. Plant Biol. 2021, 64, 431–448. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Greguš, M.; Kostas, J.C.; Ray, S.; Abbatiello, S.E.; Ivanov, A.R. Improved Sensitivity of Ultralow Flow LC–MS-Based Proteomic Profiling of Limited Samples Using Monolithic Capillary Columns and FAIMS Technology. Anal. Chem. 2020, 92, 14702–14712. [Google Scholar] [CrossRef] [PubMed]
- Sweet, S.; Chain, D.; Yu, W.; Martin, P.; Rebelatto, M.; Chambers, A.; Cecchi, F.; Kim, Y.J. The addition of FAIMS increases targeted proteomics sensitivity from FFPE tumor biopsies. Sci. Rep. 2022, 12, 13876. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, A.; Branca, R.M.; Lehtiö, J.; Forshed, J. Quantitative accuracy in mass spectrometry based proteomics of complex samples: The impact of labeling and precursor interference. J. Proteom. 2014, 96, 133–144. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; Da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A Web-Based Tool for the Analysis of Sets through Venn Diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Basu, P.; Kruse, C.P.; Luesse, D.R.; Wyatt, S.E. Growth in spaceflight hardware results in alterations to the transcriptome and proteome. Life Sci. Space Res. 2017, 15, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.-L.; Zupanska, A.K.; Ostrow, D.T.; Zhang, Y.; Sun, Y.; Li, J.-L.; Shanker, S.; Farmerie, W.G.; Amalfitano, C.E.; Ferl, R.J.; et al. Spaceflight Transcriptomes: Unique Responses to a Novel Environment. Astrobiology 2012, 12, 40–56. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Sokolowska, E.M.; Di Vittori, V.; de Souza, L.P.; Kuhalskaya, A.; Brotman, Y.; Alseekh, S.; Fernie, A.R.; Skirycz, A. Multi-omics analysis of early leaf development in Arabidopsis thaliana. Patterns 2021, 2, 100235. [Google Scholar] [CrossRef]
- Paradiso, R.; Proietti, S. Light-Quality Manipulation to Control Plant Growth and Photomorphogenesis in Greenhouse Horticulture: The State of the Art and the Opportunities of Modern LED Systems. J. Plant Growth Regul. 2022, 41, 742–780. [Google Scholar] [CrossRef]
- Bąchor, R.; Waliczek, M.; Stefanowicz, P.; Szewczuk, Z. Trends in the Design of New Isobaric Labeling Reagents for Quantitative Proteomics. Molecules 2019, 24, 701. [Google Scholar] [CrossRef]
- Cheng, Z.; Teo, G.; Krueger, S.; Rock, T.M.; Koh, H.W.; Choi, H.; Vogel, C. Differential dynamics of the mammalian mRNA and protein expression response to misfolding stress. Mol. Syst. Biol. 2016, 12, 855. [Google Scholar] [CrossRef]
- Czolpinska, M.; Rurek, M. Plant Glycine-Rich Proteins in Stress Response: An Emerging, Still Prospective Story. Front. Plant Sci. 2018, 9, 302. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Cheng, K.; Li, J.; Deng, Z.; Zhang, C.; Zhu, H. Roles of Plant Glycine-Rich RNA-Binding Proteins in Development and Stress Responses. Int. J. Mol. Sci. 2021, 22, 5849. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, A.F.; Bocca, S.N.; Ramos, R.L.B.; Barrôco, R.M.; Magioli, C.; Jorge, V.C.; Coutinho, T.C.; Rangel-Lima, C.M.; De Rycke, R.; Inzé, D.; et al. AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta 2007, 225, 1339–1351. [Google Scholar] [CrossRef]
- López-Calcagno, P.E.; Fisk, S.; Brown, K.L.; Bull, S.E.; South, P.F.; Raines, C.A. Overexpressing the H-protein of the glycine cleavage system increases biomass yield in glasshouse and field-grown transgenic tobacco plants. Plant Biotechnol. J. 2019, 17, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Hu, H.; Zhang, S.-B. Photorespiration plays an important role in the regulation of photosynthetic electron flow under fluctuating light in tobacco plants grown under full sunlight. Front. Plant Sci. 2015, 6, 621. [Google Scholar] [CrossRef] [PubMed]
- Engel, N.; van den Daele, K.; Kolukisaoglu, U.; Morgenthal, K.; Weckwerth, W.; Pärnik, T.; Keerberg, O.; Bauwe, H. Deletion of Glycine Decarboxylase in Arabidopsis Is Lethal under Nonphotorespiratory Conditions. Plant Physiol. 2007, 144, 1328–1335. [Google Scholar] [CrossRef] [PubMed]
- Khedkar, G.; Prakash, B.; Khedkar, C.; Chopade, B. Nucleic Acids. In Encyclopedia of Food and Health; Elsevier: Amsterdam, The Netherlands, 2016; pp. 84–92. [Google Scholar] [CrossRef]
- Cui, J.; Xia, W.; Wei, S.; Zhang, M.; Wang, W.; Zeng, D.; Liu, M.; Sun, Y.; Lu, W. Photosynthetic Performance of Rice Seedlings Originated from Seeds Exposed to Spaceflight Conditions. Photochem. Photobiol. 2019, 95, 1205–1212. [Google Scholar] [CrossRef]
- Caumon, H.; Vernoux, T. A matter of time: Auxin signaling dynamics and the regulation of auxin responses during plant development. J. Exp. Bot. 2023, 74, 3887–3902. [Google Scholar] [CrossRef]
- Ferl, R.J.; Paul, A.-L. The effect of spaceflight on the gravity-sensing auxin gradient of roots: GFP reporter gene microscopy on orbit. NPJ Microgravity 2016, 2, 15023. [Google Scholar] [CrossRef]
- Herranz, R.; Vandenbrink, J.P.; Villacampa, A.; Manzano, A.; Poehlman, W.L.; Feltus, F.A.; Kiss, J.Z.; Medina, F.J. RNAseq Analysis of the Response of Arabidopsis thaliana to Fractional Gravity Under Blue-Light Stimulation during Spaceflight. Front. Plant Sci. 2019, 10, 1529. [Google Scholar] [CrossRef]
- Jin, J.; Chen, H.; Cai, W. Transcriptome Analysis of Oryza sativa Calli under Microgravity. Microgravity Sci. Technol. 2015, 27, 437–453. [Google Scholar] [CrossRef]
- Xu, P.; Chen, H.; Jin, J.; Cai, W. Single-base resolution methylome analysis shows epigenetic changes in Arabidopsis seedlings exposed to microgravity spaceflight conditions on board the SJ-10 recoverable satellite. NPJ Microgravity 2018, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.O.; Haas, F.B.; Khan, S.; Bowden, L.; Ignatz, M.; Enfissi, E.M.A.; Gawthrop, F.; Griffiths, A.; Fraser, P.D.; Rensing, S.A.; et al. Rocket Science: The Effect of Spaceflight on Germination Physiology, Ageing, and Transcriptome of Eruca sativa Seeds. Life 2020, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Chew, S.Y.; Chee, W.J.Y.; Than, L.T.L. The glyoxylate cycle and alternative carbon metabolism as metabolic adaptation strategies of Candida glabrata: Perspectives from Candida albicans and Saccharomyces cerevisiae. J. Biomed. Sci. 2019, 26, 52. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Sutter, B.M.; Wang, Y.; Kuang, Z.; Tu, B.P. A Metabolic Function for Phospholipid and Histone Methylation. Mol. Cell 2017, 66, 180–193.e8. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.-L.; Haveman, N.; Califar, B.; Ferl, R.J. Epigenomic Regulators Elongator Complex Subunit 2 and Methyltransferase 1 Differentially Condition the Spaceflight Response in Arabidopsis. Front. Plant Sci. 2021, 12, 691790. [Google Scholar] [CrossRef]
- Zhou, M.; Sng, N.J.; LeFrois, C.E.; Paul, A.-L.; Ferl, R.J. Epigenomics in an extraterrestrial environment: Organ-specific alteration of DNA methylation and gene expression elicited by spaceflight in Arabidopsis thaliana. BMC Genom. 2019, 20, 205. [Google Scholar] [CrossRef]
- Zhang, W.; Qin, W.; Li, H.; Wu, A.-M. Biosynthesis and Transport of Nucleotide Sugars for Plant Hemicellulose. Front. Plant Sci. 2021, 12, 723128. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Perez-Llorca, M.; Munné-Bosch, S.; Gibin, M.S.; Sato, F.; Pelozo, A.; Pattaro, M.C.; Giacomelli, M.E.; Rüggeberg, M.; et al. Cell wall structure and composition is affected by light quality in tomato seedlings. J. Photochem. Photobiol. B Biol. 2020, 203, 111745. [Google Scholar] [CrossRef]
- Frolov, A.; Didio, A.; Ihling, C.; Chantzeva, V.; Grishina, T.; Hoehenwarter, W.; Sinz, A.; Smolikova, G.; Bilova, T.; Medvedev, S. The effect of simulated microgravity on the Brassica napus seedling proteome. Funct. Plant Biol. 2018, 45, 440–452. [Google Scholar] [CrossRef]
- Kamal, K.Y.; van Loon, J.J.; Medina, F.J.; Herranz, R. Differential transcriptional profile through cell cycle progression in Arabidopsis cultures under simulated microgravity. Genomics 2019, 111, 1956–1965. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Yu, S.; Ferretti, D.; Schessner, J.P.; Rudolph, J.D.; Borner, G.H.H.; Cox, J. Expanding the Perseus Software for Omics Data Analysis With Custom Plugins. Curr. Protoc. Bioinform. 2020, 71, e105. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Alvarez, S.; Choudhury, S.R.; Pandey, S. Comparative Quantitative Proteomics Analysis of the ABA Response of Roots of Drought-Sensitive and Drought-Tolerant Wheat Varieties Identifies Proteomic Signatures of Drought Adaptability. J. Proteome Res. 2014, 13, 1688–1701. [Google Scholar] [CrossRef]
- Wang, H.; Alvarez, S.; Hicks, L.M. Comprehensive Comparison of iTRAQ and Label-free LC-Based Quantitative Proteomics Approaches Using Two Chlamydomonas reinhardtii Strains of Interest for Biofuels Engineering. J. Proteome Res. 2011, 11, 487–501. [Google Scholar] [CrossRef] [PubMed]
| Gene ID | Symbol | Name | ABRS | EMCS | BRIC PDFU | BRIC LED |
|---|---|---|---|---|---|---|
| Proteins shared by all the datasets | Log 2 Fold Change (L2FC) | |||||
| AT1G27090 | Glycine-rich protein | −0.66 | 0.29 | 0.23 | −0.37 | |
| AT1G72150 | PATL1 | Patellin-1 | 0.71 | −1.318 | −0.17 | 0.57 |
| AT4G20260 | PCAP1 | Plasma-membrane-associated cation-binding protein 1 | 0.73 | −0.79 | −0.44 | 0.49 |
| AT4G33010 | GLDP1 | Glycine decarboxylase P-protein 1 | −0.96 | −1.27 | −0.35 | −0.47 |
| AT5G26260 | Leucine-rich repeat receptor-like kinase1 | 0.59 | 0.56 | −0.414 | 0.8 | |
| Proteins shared by EMCS, BRIC PDFU, and BRIC LED but not ABRS | ||||||
| AT1G74470 | CHLP | Geranylgeranyl diphosphate reductase | 0.37 | 0.54 | −0.54 | |
| AT3G16470 | JAL35 | Jacalin-related lectin 35 | −1.53 | −0.46 | −0.35 | |
| AT3G22640 | PAP85 | Vicilin-like seed storage protein | −0.901 | 0.02 | 3.31 | |
| AT5G19550 | ASP2 | Aspartate aminotransferase | 1.13 | −0.14 | 0.31 | |
| AT5G37600 | GLN1-1 | Glutamine synthetase cytosolic isozyme 1-1 | 0.82 | −0.06 | 0.23 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olanrewaju, G.O.; Kruse, C.P.S.; Wyatt, S.E. Functional Meta-Analysis of the Proteomic Responses of Arabidopsis Seedlings to the Spaceflight Environment Reveals Multi-Dimensional Sources of Variability across Spaceflight Experiments. Int. J. Mol. Sci. 2023, 24, 14425. https://doi.org/10.3390/ijms241914425
Olanrewaju GO, Kruse CPS, Wyatt SE. Functional Meta-Analysis of the Proteomic Responses of Arabidopsis Seedlings to the Spaceflight Environment Reveals Multi-Dimensional Sources of Variability across Spaceflight Experiments. International Journal of Molecular Sciences. 2023; 24(19):14425. https://doi.org/10.3390/ijms241914425
Chicago/Turabian StyleOlanrewaju, Gbolaga O., Colin P. S. Kruse, and Sarah E. Wyatt. 2023. "Functional Meta-Analysis of the Proteomic Responses of Arabidopsis Seedlings to the Spaceflight Environment Reveals Multi-Dimensional Sources of Variability across Spaceflight Experiments" International Journal of Molecular Sciences 24, no. 19: 14425. https://doi.org/10.3390/ijms241914425
APA StyleOlanrewaju, G. O., Kruse, C. P. S., & Wyatt, S. E. (2023). Functional Meta-Analysis of the Proteomic Responses of Arabidopsis Seedlings to the Spaceflight Environment Reveals Multi-Dimensional Sources of Variability across Spaceflight Experiments. International Journal of Molecular Sciences, 24(19), 14425. https://doi.org/10.3390/ijms241914425






