The Interaction between the Host Genome, Epigenome, and the Gut–Skin Axis Microbiome in Atopic Dermatitis
Abstract
:1. Introduction
2. Genetic Predisposition to AD
3. Gut Microbiome, Immunity, and AD
3.1. Mode of Delivery and the Infant’s Gut Microbiota: Impact on the Risk of AD
3.2. Environmental Factors, the Gut Microbiome, and AD
3.3. Gut Virome, Mycobiome, and AD
4. Microbial Metabolites, Probiotics, and AD
4.1. Microbial Metabolites and Their Potential Role in AD
4.2. Prebiotics and Probiotics in AD
5. The Role of Genetic Predisposition in the Gut Microbiome Composition of AD Patients
6. The Role of the Gut Microbiome in Gene Expression and Epigenetic Regulation of AD
6.1. Non-Coding RNA Binding
6.2. DNA Methylation
7. Summary
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weidinger, S.; Beck, L.A.; Bieber, T.; Kabashima, K.; Irvine, A.D. Atopic dermatitis. Nat. Rev. Dis. Primers 2018, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Eichenfield, L.F.; Tom, W.L.; Berger, T.G.; Krol, A.; Paller, A.S.; Schwarzenberger, K.; Bergman, J.N.; Chamlin, S.L.; Cohen, D.E.; Cooper, K.D.; et al. Guidelines of care for the management of atopic dermatitis: Section 2. Management and treatment of atopic dermatitis with topical therapies. J. Am. Acad. Dermatol. 2014, 71, 116–132. [Google Scholar] [CrossRef] [PubMed]
- Margolis, D.J.; Gupta, J.; Apter, A.J.; Ganguly, T.; Hoffstad, O.; Papadopoulos, M.; Rebbeck, T.R.; Mitra, N. Filaggrin-2 variation is associated with more persistent atopic dermatitis in African American subjects. J. Allergy Clin. Immunol. 2014, 133, 784–789. [Google Scholar] [CrossRef]
- Wollenberg, A.; Barbarot, S.; Bieber, T.; Christen-Zaech, S.; Deleuran, M.; Fink-Wagner, A.; Gieler, U.; Girolomoni, G.; Lau, S.; Muraro, A.; et al. Consensus-based European guidelines for treatment of atopic eczema (atopic dermatitis) in adults and children: Part I. J. Eur. Acad. Dermatol. Venereol. JEADV 2018, 32, 657–682. [Google Scholar] [CrossRef]
- De Bruyn Carlier, T.; Badloe, F.M.S.; Ring, J.; Gutermuth, J.; Kortekaas Krohn, I. Autoreactive T cells and their role in atopic dermatitis. J. Autoimmun. 2021, 120, 102634. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Mempel, M.; Schober, W.; Behrendt, H.; Ring, J. Gender difference, sex hormones, and immediate type hypersensitivity reactions. Allergy 2008, 63, 1418–1427. [Google Scholar] [CrossRef]
- Hadi, H.A.; Tarmizi, A.I.; Khalid, K.A.; Gajdács, M.; Aslam, A.; Jamshed, S. The Epidemiology and Global Burden of Atopic Dermatitis: A Narrative Review. Life 2021, 11, 936. [Google Scholar] [CrossRef]
- Bylund, S.; Kobyletzki, L.B.; Svalstedt, M.; Svensson, A. Prevalence and Incidence of Atopic Dermatitis: A Systematic Review. Acta Derm. Venereol. 2020, 100, adv00160. [Google Scholar] [CrossRef]
- Nedoszytko, B.; Reszka, E.; Gutowska-Owsiak, D.; Trzeciak, M.; Lange, M.; Jarczak, J.; Niedoszytko, M.; Jablonska, E.; Romantowski, J.; Strapagiel, D.; et al. Genetic and Epigenetic Aspects of Atopic Dermatitis. Int. J. Mol. Sci. 2020, 21, 6484. [Google Scholar] [CrossRef]
- Martin, M.J.; Estravis, M.; Garcia-Sanchez, A.; Davila, I.; Isidoro-Garcia, M.; Sanz, C. Genetics and Epigenetics of Atopic Dermatitis: An Updated Systematic Review. Genes 2020, 11, 442. [Google Scholar] [CrossRef]
- de Sousa, T.R.; Fagundes, B.O.; Nascimento, A.; Fernandes, L.A.; Sgnotto, F.d.R.; Orfali, R.L.; Aoki, V.; Duarte, A.J.d.S.; Sanabani, S.S.; Victor, J.R. IgG from Adult Atopic Dermatitis (AD) Patients Induces Thymic IL-22 Production and CLA Expression on CD4+ T Cells: Possible Epigenetic Implications Mediated by miRNA. Int. J. Mol. Sci. 2022, 23, 6867. [Google Scholar] [CrossRef] [PubMed]
- Abreu, D.; Kim, B.S. Innate Immune Regulation of Dermatitis. Immunol. Allergy Clin. N. Am. 2021, 41, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, H.; Zhang, J.; Jin, K.; Ma, L.; Wang, Y.; Yang, S.; Wang, X.; Shen, Q.; Zhou, T.; et al. Comparison of Gut Viral Communities in Atopic Dermatitis and Healthy Children. Front. Med. 2022, 9, 835467. [Google Scholar] [CrossRef]
- Kantor, R.; Silverberg, J.I. Environmental risk factors and their role in the management of atopic dermatitis. Expert Rev. Clin. Immunol. 2017, 13, 15–26. [Google Scholar] [CrossRef]
- Alves, E.; Gregorio, J.; Rijo, P.; Rosado, C.; Monteiro Rodrigues, L. Kefir and the Gut–skin Axis. Int. J. Environ. Res. Public Health 2022, 19, 13791. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, S.; Zhang, D.; Li, W.; Duan, Z.; Lu, S. Effects of Residential Environment and Lifestyle on Atopic Eczema Among Preschool Children in Shenzhen, China. Front. Public Health 2022, 10, 844832. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.-J.; Lee, S.-Y.; Park, Y.-M.; Kim, B.-S.; Lee, M.-J.; Kim, J.-H.; Jeong, S.; Lee, S.-H.; Park, M.J.; Rhee, E.-S.; et al. Interactions Between IL-17 Variants and Streptococcus in the Gut Contribute to the Development of Atopic Dermatitis in Infancy. Allergy Asthma Immunol. Res. 2021, 13, 404–419. [Google Scholar] [CrossRef] [PubMed]
- Diepgen, T. Epidemiology and job-related problems for the eczema patient. Acta Derm. Venereol. Suppl. 2005, 85, 41–44. [Google Scholar] [CrossRef]
- Esparza-Gordillo, J.; Weidinger, S.; Folster-Holst, R.; Bauerfeind, A.; Ruschendorf, F.; Patone, G.; Rohde, K.; Marenholz, I.; Schulz, F.; Kerscher, T.; et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat. Genet. 2009, 41, 596–601. [Google Scholar] [CrossRef]
- Sun, L.-D.; Xiao, F.-L.; Li, Y.; Zhou, W.-M.; Tang, H.-Y.; Tang, X.-F.; Zhang, H.; Schaarschmidt, H.; Zuo, X.-B.; Foelster-Holst, R.; et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat. Genet. 2011, 43, 690–694. [Google Scholar] [CrossRef]
- Paternoster, L.; Standl, M.; Chen, C.M.; Ramasamy, A.; Bonnelykke, K.; Duijts, L.; Ferreira, M.A.; Alves, A.C.; Thyssen, J.P.; Albrecht, E.; et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat. Genet. 2011, 44, 187–192. [Google Scholar] [CrossRef]
- EArly Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat. Genet. 2015, 47, 1449–1456. [Google Scholar] [CrossRef]
- Esparza-Gordillo, J.; Schaarschmidt, H.; Liang, L.; Cookson, W.; Bauerfeind, A.; Lee-Kirsch, M.A.; Nemat, K.; Henderson, J.; Paternoster, L.; Harper, J.I.; et al. A functional IL-6 receptor (IL6R) variant is a risk factor for persistent atopic dermatitis. J. Allergy Clin. Immunol. 2013, 132, 371–377. [Google Scholar] [CrossRef]
- van Beijsterveldt, C.E.; Boomsma, D.I. Genetics of parentally reported asthma, eczema and rhinitis in 5-yr-old twins. Eur. Respir. J. 2007, 29, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Canfora, E.E.; Blaak, E.E. Acetate: A diet-derived key metabolite in energy metabolism: Good or bad in context of obesity and glucose homeostasis? Curr. Opin. Clin. Nutr. Metab. Care 2017, 20, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Landman, C.; Quevrain, E. Gut microbiota: Description, role and pathophysiologic implications. La Rev. De Med. Interne 2016, 37, 418–423. [Google Scholar] [CrossRef]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef]
- Wells, J.M.; Rossi, O.; Meijerink, M.; van Baarlen, P. Epithelial crosstalk at the microbiota-mucosal interface. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4607–4614. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef]
- Saffouri, G.B.; Shields-Cutler, R.R.; Chen, J.; Yang, Y.; Lekatz, H.R.; Hale, V.L.; Cho, J.M.; Battaglioli, E.J.; Bhattarai, Y.; Thompson, K.J.; et al. Small intestinal microbial dysbiosis underlies symptoms associated with functional gastrointestinal disorders. Nat. Commun. 2019, 10, 2012. [Google Scholar] [CrossRef]
- Emoto, T.; Yamashita, T.; Sasaki, N.; Hirota, Y.; Hayashi, T.; So, A.; Kasahara, K.; Yodoi, K.; Matsumoto, T.; Mizoguchi, T.; et al. Analysis of Gut Microbiota in Coronary Artery Disease Patients: A Possible Link between Gut Microbiota and Coronary Artery Disease. J. Atheroscler. Thromb. 2016, 23, 908–921. [Google Scholar] [CrossRef]
- Islam, F.; Mitra, S.; Nafady, M.H.; Rahman, M.T.; Tirth, V.; Akter, A.; Emran, T.B.; Mohamed, A.A.; Algahtani, A.; El-Kholy, S.S. Neuropharmacological and Antidiabetic Potential of Lannea coromandelica (Houtt.) Merr. Leaves Extract: An Experimental Analysis. Evid. Based Complement. Altern. Med. Ecam 2022, 2022, 6144733. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Watanabe, S.; Narisawa, Y.; Arase, S.; Okamatsu, H.; Ikenaga, T.; Tajiri, Y.; Kumemura, M. Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. J. Allergy Clin. Immunol. 2003, 111, 587–591. [Google Scholar] [CrossRef]
- Song, H.; Yoo, Y.; Hwang, J.; Na, Y.C.; Kim, H.S. Faecalibacterium prausnitzii subspecies–level dysbiosis in the human gut microbiome underlying atopic dermatitis. J. Allergy Clin. Immunol. 2016, 137, 852–860. [Google Scholar] [CrossRef]
- Fieten, K.B.; Totte, J.E.E.; Levin, E.; Reyman, M.; Meijer, Y.; Knulst, A.; Schuren, F.; Pasmans, S. Fecal Microbiome and Food Allergy in Pediatric Atopic Dermatitis: A Cross-Sectional Pilot Study. Int. Arch. Allergy Immunol. 2018, 175, 77–84. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019, 574, 117–121. [Google Scholar] [CrossRef]
- Jakobsson, H.E.; Abrahamsson, T.R.; Jenmalm, M.C.; Harris, K.; Quince, C.; Jernberg, C.; Bjorksten, B.; Engstrand, L.; Andersson, A.F. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 2014, 63, 559–566. [Google Scholar] [CrossRef]
- Arboleya, S.; Suarez, M.; Fernandez, N.; Mantecon, L.; Solis, G.; Gueimonde, M.; de Los Reyes-Gavilan, C.G. C-section and the Neonatal Gut Microbiome Acquisition: Consequences for Future Health. Ann. Nutr. Metab. 2018, 73 (Suppl. 3), 17–23. [Google Scholar] [CrossRef]
- Negele, K.; Heinrich, J.; Borte, M.; von Berg, A.; Schaaf, B.; Lehmann, I.; Wichmann, H.E.; Bolte, G.; for the LISA Study Group. Mode of delivery and development of atopic disease during the first 2 years of life. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2004, 15, 48–54. [Google Scholar] [CrossRef]
- Debley, J.S.; Smith, J.M.; Redding, G.J.; Critchlow, C.W. Childhood asthma hospitalization risk after cesarean delivery in former term and premature infants. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2005, 94, 228–233. [Google Scholar] [CrossRef]
- Laubereau, B.; Filipiak-Pittroff, B.; von Berg, A.; Grubl, A.; Reinhardt, D.; Wichmann, H.E.; Koletzko, S.; for the GINI Study Group. Caesarean section and gastrointestinal symptoms, atopic dermatitis, and sensitisation during the first year of life. Arch. Dis. Child. 2004, 89, 993–997. [Google Scholar] [CrossRef]
- Mack, M.R.; Brestoff, J.R.; Berrien-Elliott, M.M.; Trier, A.M.; Yang, T.B.; McCullen, M.; Collins, P.L.; Niu, H.; Bodet, N.D.; Wagner, J.A.; et al. Blood natural killer cell deficiency reveals an immunotherapy strategy for atopic dermatitis. Sci. Transl. Med. 2020, 12, eaay1005. [Google Scholar] [CrossRef]
- Mubanga, M.; Lundholm, C.; D’Onofrio, B.M.; Stratmann, M.; Hedman, A.; Almqvist, C. Association of Early Life Exposure to Antibiotics With Risk of Atopic Dermatitis in Sweden. JAMA Netw. Open 2021, 4, e215245. [Google Scholar] [CrossRef]
- Apfelbacher, C.J.; Diepgen, T.L.; Schmitt, J. Determinants of eczema: Population-based cross-sectional study in Germany. Allergy 2011, 66, 206–213. [Google Scholar] [CrossRef]
- Schultz Larsen, F. Atopic dermatitis: A genetic-epidemiologic study in a population-based twin sample. J. Am. Acad. Dermatol. 1993, 28, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Loset, M.; Brown, S.J.; Saunes, M.; Hveem, K. Genetics of Atopic Dermatitis: From DNA Sequence to Clinical Relevance. Dermatology 2019, 235, 355–364. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Seo, J.-H.; Kwon, J.-W.; Yu, J.; Kim, B.-J.; Lee, S.-Y.; Kim, H.-B.; Kim, W.-K.; Kim, K.-W.; Shin, Y.-J.; et al. Exposure to gene-environment interactions before 1 year of age may favor the development of atopic dermatitis. Int. Arch. Allergy Immunol. 2012, 157, 363–371. [Google Scholar] [CrossRef]
- Jedrychowski, W.; Perera, F.; Maugeri, U.; Mrozek-Budzyn, D.; Miller, R.L.; Flak, E.; Mroz, E.; Jacek, R.; Spengler, J.D. Effects of prenatal and perinatal exposure to fine air pollutants and maternal fish consumption on the occurrence of infantile eczema. Int. Arch. Allergy Immunol. 2011, 155, 275–281. [Google Scholar] [CrossRef]
- Odhiambo, J.A.; Williams, H.C.; Clayton, T.O.; Robertson, C.F.; Asher, M.I.; ISAAC Phase Three Study Group. Global variations in prevalence of eczema symptoms in children from ISAAC Phase Three. J. Allergy Clin. Immunol. 2009, 124, 1251–1258 e1223. [Google Scholar] [CrossRef] [PubMed]
- Schultz Larsen, F.; Diepgen, T.; Svensson, A. The occurrence of atopic dermatitis in north Europe: An international questionnaire study. J. Am. Acad. Dermatol. 1996, 34, 760–764. [Google Scholar] [CrossRef]
- Belyhun, Y.; Amberbir, A.; Medhin, G.; Erko, B.; Hanlon, C.; Venn, A.; Britton, J.; Davey, G. Prevalence and risk factors of wheeze and eczema in 1-year-old children: The Butajira birth cohort, Ethiopia. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2010, 40, 619–626. [Google Scholar] [CrossRef]
- Shaw, T.E.; Currie, G.P.; Koudelka, C.W.; Simpson, E.L. Eczema prevalence in the United States: Data from the 2003 National Survey of Children’s Health. J. Investig. Dermatol. 2011, 131, 67–73. [Google Scholar] [CrossRef]
- Harada, Y.; Sakamoto, T.; Shinomura, T.; Takamoto, K.; Senda, T.; Tsuda, M. Total synthesis of a gene for octopus rhodopsin and its preliminary expression. J. Biochem. 1991, 110, 501–507. [Google Scholar] [CrossRef]
- Kim, B.-J.; Lee, S.-Y.; Kim, H.-B.; Lee, E.; Hong, S.-J. Environmental changes, microbiota, and allergic diseases. Allergy Asthma Immunol. Res. 2014, 6, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Karlsson, C.; Olsson, C.; Adlerberth, I.; Wold, A.E.; Strachan, D.P.; Martricardi, P.M.; Aberg, N.; Perkin, M.R.; Tripodi, S.; et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J. Allergy Clin. Immunol. 2008, 121, 129–134. [Google Scholar] [CrossRef]
- Adlerberth, I.; Carlsson, B.; de Man, P.; Jalil, F.; Khan, S.R.; Larsson, P.; Mellander, L.; Svanborg, C.; Wold, A.E.; Hanson, L.A. Intestinal colonization with Enterobacteriaceae in Pakistani and Swedish hospital-delivered infants. Acta Paediatr. 1991, 80, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Abrahamsson, T.R.; Jakobsson, H.E.; Andersson, A.F.; Bjorksten, B.; Engstrand, L.; Jenmalm, M.C. Low diversity of the gut microbiota in infants with atopic eczema. J. Allergy Clin. Immunol. 2012, 129, 434–440.E2. [Google Scholar] [CrossRef] [PubMed]
- Nylund, L.; Nermes, M.; Isolauri, E.; Salminen, S.; de Vos, W.M.; Satokari, R. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy 2015, 70, 241–244. [Google Scholar] [CrossRef]
- Reichardt, N.; Duncan, S.H.; Young, P.; Belenguer, A.; McWilliam Leitch, C.; Scott, K.P.; Flint, H.J.; Louis, P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014, 8, 1323–1335. [Google Scholar] [CrossRef]
- Macia, L.; Thorburn, A.N.; Binge, L.C.; Marino, E.; Rogers, K.E.; Maslowski, K.M.; Vieira, A.T.; Kranich, J.; Mackay, C.R. Microbial influences on epithelial integrity and immune function as a basis for inflammatory diseases. Immunol. Rev. 2012, 245, 164–176. [Google Scholar] [CrossRef]
- De Benedetto, A.; Rafaels, N.M.; McGirt, L.Y.; Ivanov, A.I.; Georas, S.N.; Cheadle, C.; Berger, A.E.; Zhang, K.; Vidyasagar, S.; Yoshida, T.; et al. Tight junction defects in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2011, 127, 773–786.E7. [Google Scholar] [CrossRef] [PubMed]
- Rey, F.E.; Faith, J.J.; Bain, J.; Muehlbauer, M.J.; Stevens, R.D.; Newgard, C.B.; Gordon, J.I. Dissecting the in vivo metabolic potential of two human gut acetogens. J. Biol. Chem. 2010, 285, 22082–22090. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef]
- Suez, J.; Zmora, N.; Zilberman-Schapira, G.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Zur, M.; Regev-Lehavi, D.; Ben-Zeev Brik, R.; Federici, S.; et al. Post-Antibiotic Gut Mucosal Microbiome Reconstitution Is Impaired by Probiotics and Improved by Autologous FMT. Cell 2018, 174, 1406–1423 e1416. [Google Scholar] [CrossRef]
- Qu, Z.; Tian, P.; Yang, B.; Zhao, J.; Wang, G.; Chen, W. Fecal microbiota transplantation for diseases: Therapeutic potential, methodology, risk management in clinical practice. Life Sci. 2022, 304, 120719. [Google Scholar] [CrossRef]
- Mashiah, J.; Karady, T.; Fliss-Isakov, N.; Sprecher, E.; Slodownik, D.; Artzi, O.; Samuelov, L.; Ellenbogen, E.; Godneva, A.; Segal, E.; et al. Clinical efficacy of fecal microbial transplantation treatment in adults with moderate-to-severe atopic dermatitis. Immun. Inflamm. Dis. 2022, 10, e570. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, K.; Kim, W. Gut microbiota restoration through fecal microbiota transplantation: A new atopic dermatitis therapy. Exp. Mol. Med. 2021, 53, 907–916. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, Z.; Ma, Y.; Miao, L.; Zhao, K.; Wang, D.; Wang, M.; Ruan, H.; Xu, F.; Zhou, Q.; et al. Fecal microbiota transplantation affects the recovery of AD-skin lesions and enhances gut microbiota homeostasis. Int. Immunopharmacol. 2023, 118, 110005. [Google Scholar] [CrossRef]
- Moro, G.; Arslanoglu, S.; Stahl, B.; Jelinek, J.; Wahn, U.; Boehm, G. A mixture of prebiotic oligosaccharides reduces the incidence of atopic dermatitis during the first six months of age. Arch. Dis. Child. 2006, 91, 814–819. [Google Scholar] [CrossRef]
- Weston, S.; Halbert, A.; Richmond, P.; Prescott, S.L. Effects of probiotics on atopic dermatitis: A randomised controlled trial. Arch. Dis. Child. 2005, 90, 892–897. [Google Scholar] [CrossRef]
- Valenta, R.; Natter, S.; Seiberler, S.; Roschanak, M.; Mothes, N.; Mahler, V.; Eibensteiner, P. Autoallergy: A pathogenetic factor in atopic dermatitis? Curr. Probl. Dermatol. 1999, 28, 45–50. [Google Scholar] [CrossRef]
- Schmid-Grendelmeier, P.; Fluckiger, S.; Disch, R.; Trautmann, A.; Wuthrich, B.; Blaser, K.; Scheynius, A.; Crameri, R. IgE-mediated and T cell-mediated autoimmunity against manganese superoxide dismutase in atopic dermatitis. J. Allergy Clin. Immunol. 2005, 115, 1068–1075. [Google Scholar] [CrossRef]
- Kato, A.; Fukai, K.; Oiso, N.; Hosomi, N.; Murakami, T.; Ishii, M. Association of SPINK5 gene polymorphisms with atopic dermatitis in the Japanese population. Br. J. Dermatol. 2003, 148, 665–669. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, J.; Newton, G.K.; Perrior, T.R.; Robinson, C. Allergen Delivery Inhibitors: A Rationale for Targeting Sentinel Innate Immune Signaling of Group 1 House Dust Mite Allergens through Structure-Based Protease Inhibitor Design. Mol. Pharmacol. 2018, 94, 1007–1030. [Google Scholar] [CrossRef]
- Chapman, M.D.; Pomes, A.; Breiteneder, H.; Ferreira, F. Nomenclature and structural biology of allergens. J. Allergy Clin. Immunol. 2007, 119, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Tsai, M.; Piliponsky, A.M. The development of allergic inflammation. Nature 2008, 454, 445–454. [Google Scholar] [CrossRef]
- Mok, K.; Suratanon, N.; Roytrakul, S.; Charoenlappanit, S.; Patumcharoenpol, P.; Chatchatee, P.; Vongsangnak, W.; Nakphaichit, M. ITS2 Sequencing and Targeted Meta-Proteomics of Infant Gut Mycobiome Reveal the Functional Role of Rhodotorula sp. during Atopic Dermatitis Manifestation. J. Fungi 2021, 7, 748. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Inan, M.S.; Rasoulpour, R.J.; Yin, L.; Hubbard, A.K.; Rosenberg, D.W.; Giardina, C. The luminal short-chain fatty acid butyrate modulates NF-κB activity in a human colonic epithelial cell line. Gastroenterology 2000, 118, 724–734. [Google Scholar] [CrossRef]
- Segain, J.P.; Raingeard de la Bletiere, D.; Bourreille, A.; Leray, V.; Gervois, N.; Rosales, C.; Ferrier, L.; Bonnet, C.; Blottiere, H.M.; Galmiche, J.P. Butyrate inhibits inflammatory responses through NFκB inhibition: Implications for Crohn’s disease. Gut 2000, 47, 397–403. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Schulthess, J.; Pandey, S.; Capitani, M.; Rue-Albrecht, K.C.; Arnold, I.; Franchini, F.; Chomka, A.; Ilott, N.E.; Johnston, D.G.W.; Pires, E.; et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 2019, 50, 432–445 e437. [Google Scholar] [CrossRef]
- Kim, C.H. Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell. Mol. Immunol. 2021, 18, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Stec, A.; Sikora, M.; Maciejewska, M.; Paralusz-Stec, K.; Michalska, M.; Sikorska, E.; Rudnicka, L. Bacterial Metabolites: A Link between Gut Microbiota and Dermatological Diseases. Int. J. Mol. Sci. 2023, 24, 3494. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef]
- Szelest, M.; Walczak, K.; Plech, T. A New Insight into the Potential Role of Tryptophan-Derived AhR Ligands in Skin Physiological and Pathological Processes. Int. J. Mol. Sci. 2021, 22, 1104. [Google Scholar] [CrossRef]
- Tsuji, G.; Hashimoto-Hachiya, A.; Kiyomatsu-Oda, M.; Takemura, M.; Ohno, F.; Ito, T.; Morino-Koga, S.; Mitoma, C.; Nakahara, T.; Uchi, H.; et al. Aryl hydrocarbon receptor activation restores filaggrin expression via OVOL1 in atopic dermatitis. Cell Death Dis. 2017, 8, e2931. [Google Scholar] [CrossRef] [PubMed]
- Paller, A.S.; Stein Gold, L.; Soung, J.; Tallman, A.M.; Rubenstein, D.S.; Gooderham, M. Efficacy and patient-reported outcomes from a phase 2b, randomized clinical trial of tapinarof cream for the treatment of adolescents and adults with atopic dermatitis. J. Am. Acad. Dermatol. 2021, 84, 632–638. [Google Scholar] [CrossRef]
- Stein Gold, L.; Rubenstein, D.S.; Peist, K.; Jain, P.; Tallman, A.M. Tapinarof cream 1% once daily and benvitimod 1% twice daily are 2 distinct topical medications. J. Am. Acad. Dermatol. 2021, 85, e201–e202. [Google Scholar] [CrossRef]
- Chittim, C.L.; Martinez Del Campo, A.; Balskus, E.P. Gut bacterial phospholipase Ds support disease-associated metabolism by generating choline. Nat. Microbiol. 2019, 4, 155–163. [Google Scholar] [CrossRef]
- Sikora, M.; Kiss, N.; Stec, A.; Giebultowicz, J.; Samborowska, E.; Jazwiec, R.; Dadlez, M.; Olszewska, M.; Rudnicka, L. Trimethylamine N-Oxide, a Gut Microbiota-Derived Metabolite, Is Associated with Cardiovascular Risk in Psoriasis: A Cross-Sectional Pilot Study. Dermatol. Ther. 2021, 11, 1277–1289. [Google Scholar] [CrossRef]
- Lin, R.-J.; Qiu, L.-H.; Guan, R.-Z.; Hu, S.-J.; Liu, Y.-Y.; Wang, G.-J. Protective effect of probiotics in the treatment of infantile eczema. Exp. Ther. Med. 2015, 9, 1593–1596. [Google Scholar] [CrossRef]
- Ciprandi, G.; Vizzaccaro, A.; Cirillo, I.; Tosca, M.A. Bacillus clausii exerts immuno-modulatory activity in allergic subjects: A pilot study. Eur. Ann. Allergy Clin. Immunol. 2005, 37, 129–134. [Google Scholar]
- de Roock, S.; van Elk, M.; van Dijk, M.E.; Timmerman, H.M.; Rijkers, G.T.; Prakken, B.J.; Hoekstra, M.O.; de Kleer, I.M. Lactic acid bacteria differ in their ability to induce functional regulatory T cells in humans. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2010, 40, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; Li, B.; et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018, 562, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Kepert, I.; Fonseca, J.; Muller, C.; Milger, K.; Hochwind, K.; Kostric, M.; Fedoseeva, M.; Ohnmacht, C.; Dehmel, S.; Nathan, P.; et al. D-tryptophan from probiotic bacteria influences the gut microbiome and allergic airway disease. J. Allergy Clin. Immunol. 2017, 139, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- Wickens, K.; Black, P.N.; Stanley, T.V.; Mitchell, E.; Fitzharris, P.; Tannock, G.W.; Purdie, G.; Crane, J.; Probiotic Study Group. A differential effect of 2 probiotics in the prevention of eczema and atopy: A double-blind, randomized, placebo-controlled trial. J. Allergy Clin. Immunol. 2008, 122, 788–794. [Google Scholar] [CrossRef]
- Kalliomaki, M.; Salminen, S.; Arvilommi, H.; Kero, P.; Koskinen, P.; Isolauri, E. Probiotics in primary prevention of atopic disease: A randomised placebo-controlled trial. Lancet 2001, 357, 1076–1079. [Google Scholar] [CrossRef]
- Wickens, K.; Black, P.; Stanley, T.V.; Mitchell, E.; Barthow, C.; Fitzharris, P.; Purdie, G.; Crane, J. A protective effect of Lactobacillus rhamnosus HN001 against eczema in the first 2 years of life persists to age 4 years. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2012, 42, 1071–1079. [Google Scholar] [CrossRef]
- Kalliomaki, M.; Salminen, S.; Poussa, T.; Arvilommi, H.; Isolauri, E. Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet 2003, 361, 1869–1871. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kwon, J.H.; Ahn, S.H.; Lee, S.I.; Han, Y.S.; Choi, Y.O.; Lee, S.Y.; Ahn, K.M.; Ji, G.E. Effect of probiotic mix (Bifidobacterium bifidum, Bifidobacterium lactis, Lactobacillus acidophilus) in the primary prevention of eczema: A double-blind, randomized, placebo-controlled trial. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2010, 21, e386–e393. [Google Scholar] [CrossRef]
- Ro, A.D.B.; Simpson, M.R.; Ro, T.B.; Storro, O.; Johnsen, R.; Videm, V.; Oien, T. Reduced Th22 cell proportion and prevention of atopic dermatitis in infants following maternal probiotic supplementation. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2017, 47, 1014–1021. [Google Scholar] [CrossRef]
- Simpson, M.R.; Dotterud, C.K.; Storro, O.; Johnsen, R.; Oien, T. Perinatal probiotic supplementation in the prevention of allergy related disease: 6 year follow up of a randomised controlled trial. BMC Dermatol. 2015, 15, 13. [Google Scholar] [CrossRef] [PubMed]
- Fiocchi, A.; Pawankar, R.; Cuello-Garcia, C.; Ahn, K.; Al-Hammadi, S.; Agarwal, A.; Beyer, K.; Burks, W.; Canonica, G.W.; Ebisawa, M.; et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ. J. 2015, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- He, J.Q.; Chan-Yeung, M.; Becker, A.B.; Dimich-Ward, H.; Ferguson, A.C.; Manfreda, J.; Watson, W.T.; Sandford, A.J. Genetic variants of the IL13 and IL4 genes and atopic diseases in at-risk children. Genes Immun. 2003, 4, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Quaranta, M.; Knapp, B.; Garzorz, N.; Mattii, M.; Pullabhatla, V.; Pennino, D.; Andres, C.; Traidl-Hoffmann, C.; Cavani, A.; Theis, F.J.; et al. Intraindividual genome expression analysis reveals a specific molecular signature of psoriasis and eczema. Sci. Transl. Med. 2014, 6, 244ra90. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.A.; Chadwick, V.S.; Murray, A. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J. Med. Microbiol. 2005, 54, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Zoetendal, E.G.; Akkermans, A.D.L.; Vliet, W.M.A.-V.; de Visser, J.A.G.M.; de Vos, W.M. The Host Genotype Affects the Bacterial Community in the Human Gastronintestinal Tract. Microb. Ecol. Health Dis. 2001, 13, 129–134. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Davenport, E.R.; Beaumont, M.; Jackson, M.A.; Knight, R.; Ober, C.; Spector, T.D.; Bell, J.T.; Clark, A.G.; Ley, R.E. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016, 19, 731–743. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Qin, Y.; Wade, P.A. Crosstalk between the microbiome and epigenome: Messages from bugs. J. Biochem. 2018, 163, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Rakyan, V.K.; Hildmann, T.; Novik, K.L.; Lewin, J.; Tost, J.; Cox, A.V.; Andrews, T.D.; Howe, K.L.; Otto, T.; Olek, A.; et al. DNA methylation profiling of the human major histocompatibility complex: A pilot study for the human epigenome project. PLoS Biol. 2004, 2, e405. [Google Scholar] [CrossRef] [PubMed]
- Blackshaw, S.; Harpavat, S.; Trimarchi, J.; Cai, L.; Huang, H.; Kuo, W.P.; Weber, G.; Lee, K.; Fraioli, R.E.; Cho, S.H.; et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004, 2, E247. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Li, H.M.; Xiao, Y.J.; Min, Z.S.; Tan, C. Identification and interaction analysis of key genes and microRNAs in atopic dermatitis by bioinformatics analysis. Clin. Exp. Dermatol. 2019, 44, 257–264. [Google Scholar] [CrossRef]
- Chen, X.-F.; Zhang, L.-J.; Zhang, J.; Dou, X.; Shao, Y.; Jia, X.-J.; Zhang, W.; Yu, B. MiR-151a is involved in the pathogenesis of atopic dermatitis by regulating interleukin-12 receptor β2. Exp. Dermatol. 2018, 27, 427–432. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Rao, D.S.; Baltimore, D. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012, 30, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Liew, W.C.; Sundaram, G.M.; Quah, S.; Lum, G.G.; Tan, J.S.L.; Ramalingam, R.; Common, J.E.A.; Tang, M.B.Y.; Lane, E.B.; Thng, S.T.G.; et al. Belinostat resolves skin barrier defects in atopic dermatitis by targeting the dysregulated miR-335:SOX6 axis. J. Allergy Clin. Immunol. 2020, 146, 606–620 e612. [Google Scholar] [CrossRef]
- Li, M.; Chen, W.-D.; Wang, Y.-D. The roles of the gut microbiota–miRNA interaction in the host pathophysiology. Mol. Med. 2020, 26, 101. [Google Scholar] [CrossRef] [PubMed]
- Sonkoly, E.; Janson, P.; Majuri, M.L.; Savinko, T.; Fyhrquist, N.; Eidsmo, L.; Xu, N.; Meisgen, F.; Wei, T.; Bradley, M.; et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J. Allergy Clin. Immunol. 2010, 126, 581–589.E20. [Google Scholar] [CrossRef]
- Fife, B.T.; Bluestone, J.A. Control of peripheral T-cell tolerance and autoimmunity via the CTLA-4 and PD-1 pathways. Immunol. Rev. 2008, 224, 166–182. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Meng, W.; Ye, S.; Zhang, X.; Mo, X.; Liu, J.; Chen, D.; Lin, Y. MicroRNA-146a as a potential regulator involved in the pathogenesis of atopic dermatitis. Mol. Med. Rep. 2019, 20, 4645–4653. [Google Scholar] [CrossRef]
- Rebane, A.; Runnel, T.; Aab, A.; Maslovskaja, J.; Ruckert, B.; Zimmermann, M.; Plaas, M.; Karner, J.; Treis, A.; Pihlap, M.; et al. MicroRNA-146a alleviates chronic skin inflammation in atopic dermatitis through suppression of innate immune responses in keratinocytes. J. Allergy Clin. Immunol. 2014, 134, 836–847 e811. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Yang, Z.; Zeng, B.; Wang, C.; Wang, H.; Huang, P.; Pan, Y. MicroRNA-124 alleviates chronic skin inflammation in atopic eczema via suppressing innate immune responses in keratinocytes. Cell. Immunol. 2017, 319, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Viennois, E.; Chassaing, B.; Tahsin, A.; Pujada, A.; Wang, L.; Gewirtz, A.T.; Merlin, D. Host-derived fecal microRNAs can indicate gut microbiota healthiness and ability to induce inflammation. Theranostics 2019, 9, 4542–4557. [Google Scholar] [CrossRef] [PubMed]
- Hesson, L.B. Gut microbiota and obesity-related gastrointestinal cancer: A focus on epigenetics. Transl. Gastrointest. Cancer 2013, 2, 204–210. [Google Scholar]
- Kao, M.-S.; Huang, S.; Chang, W.-L.; Hsieh, M.-F.; Huang, C.-J.; Gallo, R.L.; Huang, C.-M. Microbiome precision editing: Using PEG as a selective fermentation initiator against methicillin-resistant Staphylococcus aureus. Biotechnol. J. 2017, 12, 4. [Google Scholar] [CrossRef]
- Hamidi, T.; Singh, A.K.; Chen, T. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics 2015, 7, 247–265. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Ladd-Acosta, C.; Wen, B.; Wu, Z.; Montano, C.; Onyango, P.; Cui, H.; Gabo, K.; Rongione, M.; Webster, M.; et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009, 41, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Sekigawa, I.; Ogasawara, H.; Mitsuishi, K.; Hira, K.; Ikeda, S.; Ogawa, H. Expression of DNMT-1 in patients with atopic dermatitis. Arch. Dermatol. Res. 2006, 298, 253–256. [Google Scholar] [CrossRef]
- Zhu, Z.; Lee, P.H.; Chaffin, M.D.; Chung, W.; Loh, P.R.; Lu, Q.; Christiani, D.C.; Liang, L. Author Correction: A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat. Genet. 2018, 50, 1753. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, W. Genome-Wide Integration of Genetic and Genomic Studies of Atopic Dermatitis: Insights into Genetic Architecture and Pathogenesis. J. Investig. Dermatol. 2022, 142, 2958–2967.E8. [Google Scholar] [CrossRef] [PubMed]
- Noh, Y.H.; Lee, J.; Seo, S.J.; Myung, S.C. Promoter DNA methylation contributes to human β-defensin-1 deficiency in atopic dermatitis. Anim. Cells Syst. 2018, 22, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-D.; Huang, Y.-H.; Guo, M.M.-H.; Chang, L.-S.; Chu, C.-H.; Bu, L.-F.; Chu, C.-L.; Lee, C.-H.; Liu, S.-F.; Kuo, H.-C. DNA Methylation Array Identifies Golli-MBP as a Biomarker for Disease Severity in Childhood Atopic Dermatitis. J. Investig. Dermatol. 2022, 142, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Hayakawa, K.; Fujishiro, M.; Ikeda, K.; Tsushima, H.; Hirai, T.; Kawasaki, M.; Tominaga, M.; Suga, Y.; Takamori, K.; et al. Social defeat stress exacerbates atopic dermatitis through downregulation of DNA methyltransferase 1 and upregulation of C-C motif chemokine receptor 7 in skin dendritic cells. Biochem. Biophys. Res. Commun. 2020, 529, 1073–1079. [Google Scholar] [CrossRef]
- Yi, J.Z.; McGee, J.S. Epigenetic-modifying therapies: An emerging avenue for the treatment of inflammatory skin diseases. Exp. Dermatol. 2021, 30, 1167–1176. [Google Scholar] [CrossRef]
- Levkovich, T.; Poutahidis, T.; Smillie, C.; Varian, B.J.; Ibrahim, Y.M.; Lakritz, J.R.; Alm, E.J.; Erdman, S.E. Probiotic bacteria induce a ‘glow of health’. PLoS ONE 2013, 8, e53867. [Google Scholar] [CrossRef]
- Gensollen, T.; Blumberg, R.S. Correlation between early-life regulation of the immune system by microbiota and allergy development. J. Allergy Clin. Immunol. 2017, 139, 1084–1091. [Google Scholar] [CrossRef]
- Nylund, L.; Satokari, R.; Nikkila, J.; Rajilic-Stojanovic, M.; Kalliomaki, M.; Isolauri, E.; Salminen, S.; de Vos, W.M. Microarray analysis reveals marked intestinal microbiota aberrancy in infants having eczema compared to healthy children in at-risk for atopic disease. BMC Microbiol. 2013, 13, 12. [Google Scholar] [CrossRef]
- Ismail, I.H.; Boyle, R.J.; Licciardi, P.V.; Oppedisano, F.; Lahtinen, S.; Robins-Browne, R.M.; Tang, M.L. Early gut colonization by Bifidobacterium breve and B. catenulatum differentially modulates eczema risk in children at high risk of developing allergic disease. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2016, 27, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Stobberingh, E.E.; Thijs, C.; Adams, H.; Vink, C.; van Ree, R.; van den Brandt, P.A. Molecular fingerprinting of the intestinal microbiota of infants in whom atopic eczema was or was not developing. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2006, 36, 1602–1608. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Lee, S.-Y.; Kang, M.-J.; Kim, K.; Won, S.; Kim, B.-J.; Choi, K.Y.; Kim, B.-S.; Cho, H.-J.; Kim, Y.; et al. Clostridia in the gut and onset of atopic dermatitis via eosinophilic inflammation. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2016, 117, 91–92.E1. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-Y.; Lee, E.; Park, Y.M.; Hong, S.-J. Microbiome in the Gut–skin Axis in Atopic Dermatitis. Allergy Asthma Immunol. Res. 2018, 10, 354–362. [Google Scholar] [CrossRef] [PubMed]
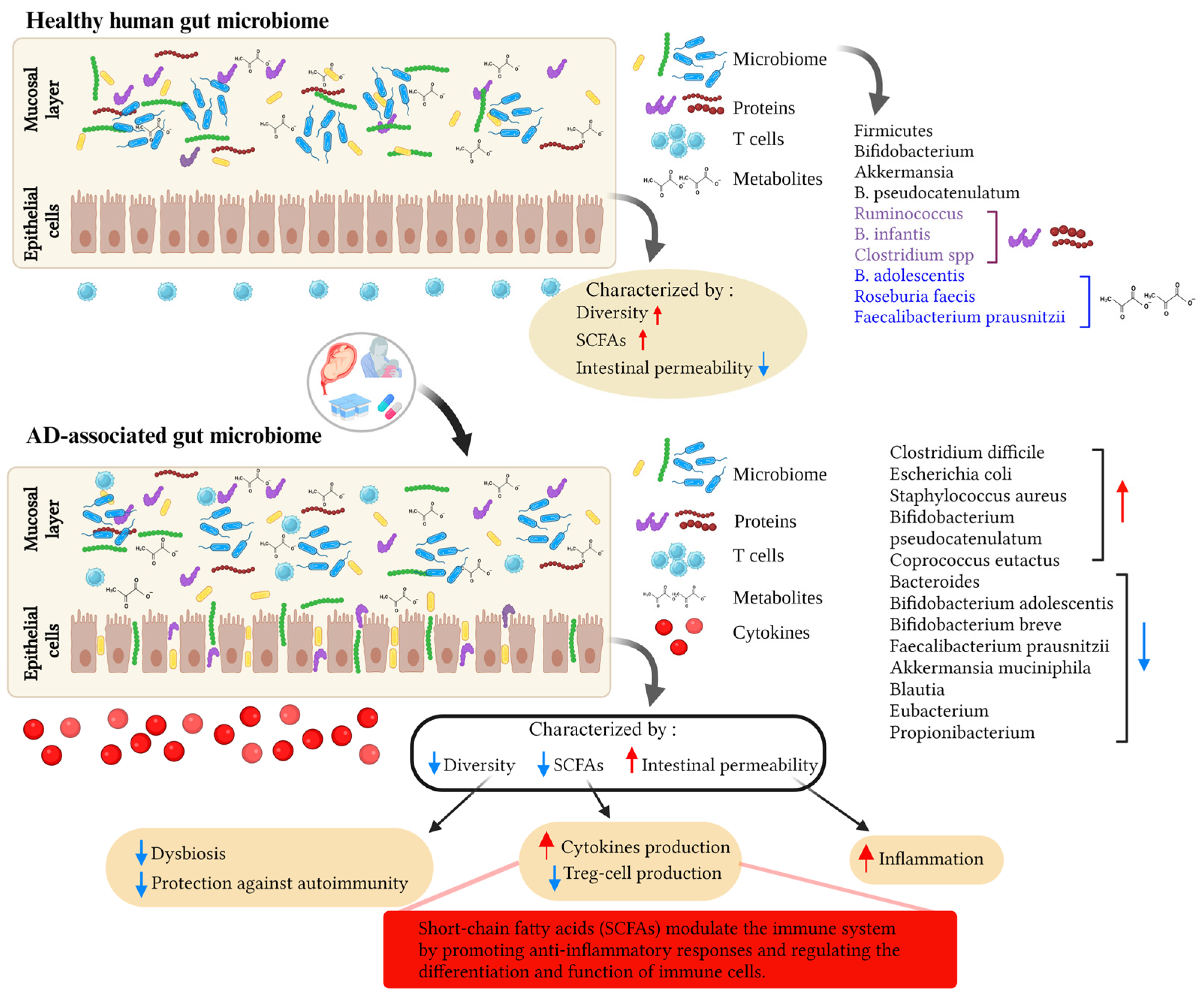
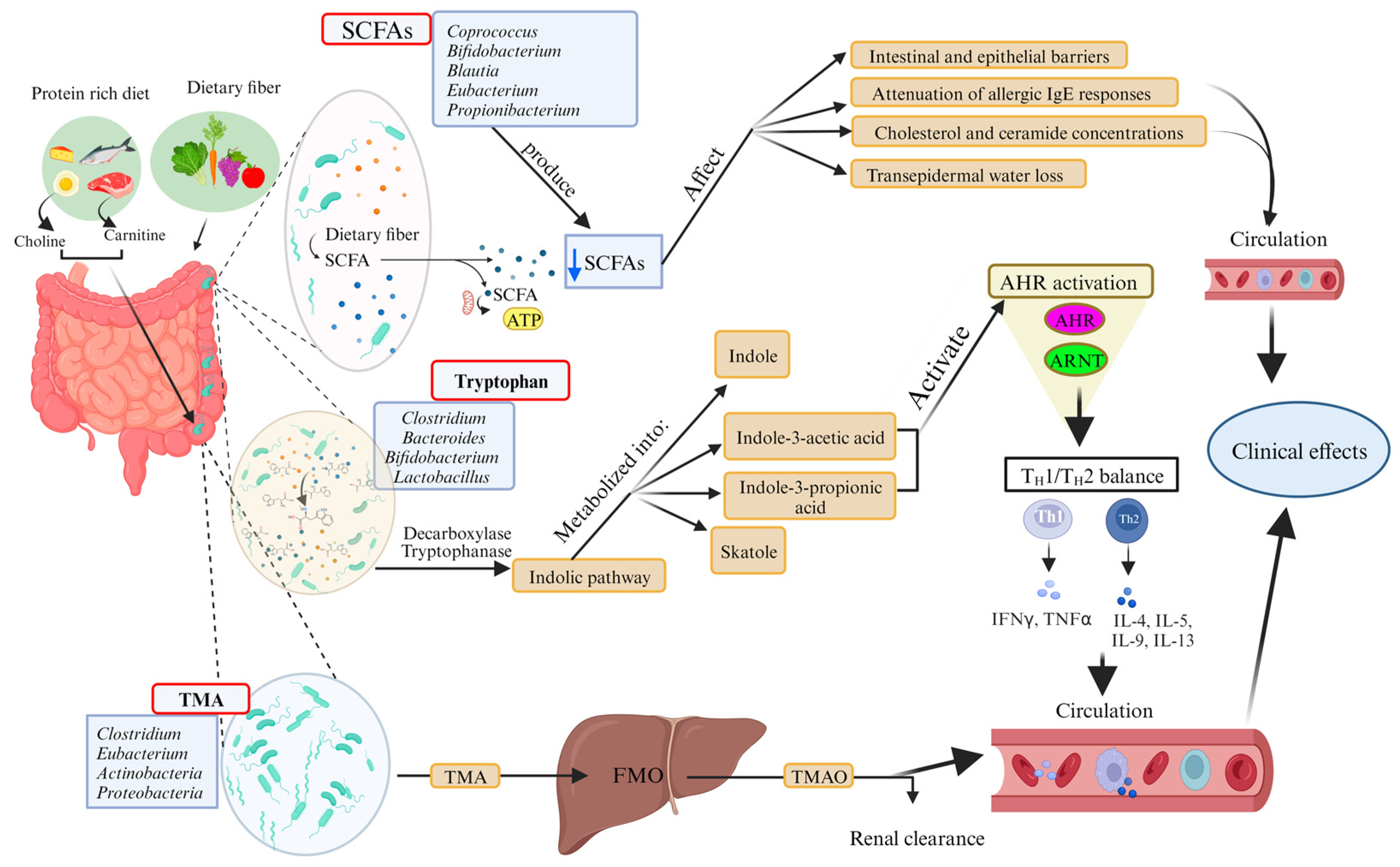

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pessôa, R.; Clissa, P.B.; Sanabani, S.S. The Interaction between the Host Genome, Epigenome, and the Gut–Skin Axis Microbiome in Atopic Dermatitis. Int. J. Mol. Sci. 2023, 24, 14322. https://doi.org/10.3390/ijms241814322
Pessôa R, Clissa PB, Sanabani SS. The Interaction between the Host Genome, Epigenome, and the Gut–Skin Axis Microbiome in Atopic Dermatitis. International Journal of Molecular Sciences. 2023; 24(18):14322. https://doi.org/10.3390/ijms241814322
Chicago/Turabian StylePessôa, Rodrigo, Patricia Bianca Clissa, and Sabri Saeed Sanabani. 2023. "The Interaction between the Host Genome, Epigenome, and the Gut–Skin Axis Microbiome in Atopic Dermatitis" International Journal of Molecular Sciences 24, no. 18: 14322. https://doi.org/10.3390/ijms241814322
APA StylePessôa, R., Clissa, P. B., & Sanabani, S. S. (2023). The Interaction between the Host Genome, Epigenome, and the Gut–Skin Axis Microbiome in Atopic Dermatitis. International Journal of Molecular Sciences, 24(18), 14322. https://doi.org/10.3390/ijms241814322




