Menstrual Tampons Are Reliable and Acceptable Tools to Self-Collect Vaginal Microbiome Samples
Abstract
:1. Introduction
2. Results
2.1. Tampon versus LVS
2.2. Fresh versus Frozen
2.3. Staphylococcus spp.
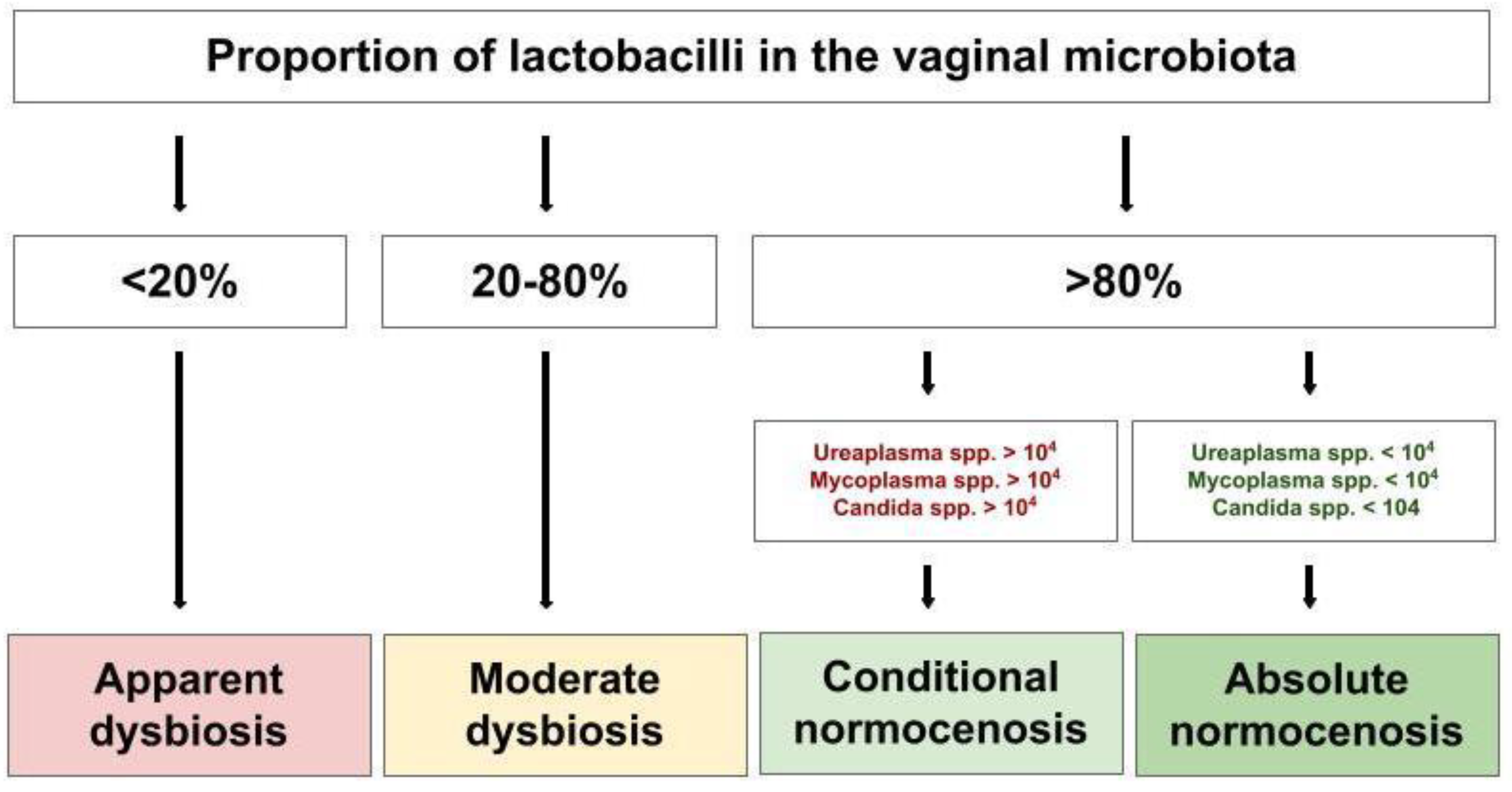
2.4. Diagnostic Conclusions
2.5. Tampon Acceptability
3. Discussion
4. Materials and Methods
4.1. Recruitment
4.2. Questionnaire
4.3. Sample Collection and Processing
4.4. DNA Extraction
4.5. Femoflor® 16 Real-Time PCR
4.6. Determination of Diagnostic Conclusions
4.7. Data Interpretation and Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- NHS Digital. Cervical Screening Programme, England-2020-21; NHS Digital: Leeds, UK, 2021. [Google Scholar]
- Trust, J.s.C.C. Embarrassment Preventing Young Women from Attending a Test That Could Save Their Life; Jo’s Cervical Cancer Trust: London, UK, 2017. [Google Scholar]
- Higgins, D.M.; Moore, M.; Alderton, L.; Weinberg, L.; Hickok, A.M.; Yale, A.; Wendel, K.A. Evaluation of a Statewide Online, At-Home Sexually Transmitted Infection and Human Immunodeficiency Virus Screening Program. Clin. Infect. Dis. 2023, 76, 2148–2153. [Google Scholar] [CrossRef] [PubMed]
- Verdoodt, F.; Jentschke, M.; Hillemanns, P.; Racey, C.S.; Snijders, P.J.; Arbyn, M. Reaching women who do not participate in the regular cervical cancer screening programme by offering self-sampling kits: A systematic review and meta-analysis of randomised trials. Eur. J. Cancer 2015, 51, 2375–2385. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kakkar, V.; Bhushan, I. Crosstalk between Vaginal Microbiome and Female Health: A review. Microb. Pathog. 2019, 136, 103696. [Google Scholar] [CrossRef] [PubMed]
- O’Hanlon, D.E.; Moench, T.R.; Cone, R.A. Vaginal pH and microbicidal lactic acid when lactobacilli dominate the microbiota. PLoS ONE 2013, 8, e80074. [Google Scholar] [CrossRef]
- Aldunate, M.; Srbinovski, D.; Hearps, A.C.; Latham, C.F.; Ramsland, P.A.; Gugasyan, R.; Cone, R.A.; Tachedjian, G. Antimicrobial and immune modulatory effects of lactic acid and short chain fatty acids produced by vaginal microbiota associated with eubiosis and bacterial vaginosis. Front. Physiol. 2015, 6, 164. [Google Scholar] [CrossRef]
- Huang, B.; Fettweis, J.M.; Brooks, J.P.; Jefferson, K.K.; Buck, G.A. The changing landscape of the vaginal microbiome. Clin. Lab. Med. 2014, 34, 747–761. [Google Scholar] [CrossRef]
- Borgdorff, H.; van der Veer, C.; van Houdt, R.; Alberts, C.J.; de Vries, H.J.; Bruisten, S.M.; Snijder, M.B.; Prins, M.; Geerlings, S.E.; Schim van der Loeff, M.F.; et al. The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands. PLoS ONE 2017, 12, e0181135. [Google Scholar] [CrossRef]
- Atashili, J.; Poole, C.; Ndumbe, P.M.; Adimora, A.A.; Smith, J.S. Bacterial vaginosis and HIV acquisition: A meta-analysis of published studies. Aids 2008, 22, 1493–1501. [Google Scholar] [CrossRef]
- Brotman, R.M. Vaginal microbiome and sexually transmitted infections: An epidemiologic perspective. J. Clin. Investig. 2011, 121, 4610–4617. [Google Scholar] [CrossRef]
- Haahr, T.; Jensen, J.S.; Thomsen, L.; Duus, L.; Rygaard, K.; Humaidan, P. Abnormal vaginal microbiota may be associated with poor reproductive outcomes: A prospective study in IVF patients. Hum. Reprod. 2016, 31, 795–803. [Google Scholar] [CrossRef]
- Brown, R.G.; Marchesi, J.R.; Lee, Y.S.; Smith, A.; Lehne, B.; Kindinger, L.M.; Terzidou, V.; Holmes, E.; Nicholson, J.K.; Bennett, P.R.; et al. Vaginal dysbiosis increases risk of preterm fetal membrane rupture, neonatal sepsis and is exacerbated by erythromycin. BMC Med. 2018, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Sezer, O.; Soyer Çalışkan, C.; Celik, S.; Kilic, S.S.; Kuruoglu, T.; Unluguzel Ustun, G.; Yurtcu, N. Assessment of vaginal and endometrial microbiota by real-time PCR in women with unexplained infertility. J. Obstet. Gynaecol. Res. 2022, 48, 129–139. [Google Scholar] [CrossRef]
- Mitra, A.; MacIntyre, D.A.; Mahajan, V.; Lee, Y.S.; Smith, A.; Marchesi, J.R.; Lyons, D.; Bennett, P.R.; Kyrgiou, M. Comparison of vaginal microbiota sampling techniques: Cytobrush versus swab. Sci. Rep. 2017, 7, 9802. [Google Scholar] [CrossRef] [PubMed]
- Adamson, P.C.; Huchko, M.J.; Moss, A.M.; Kinkel, H.F.; Medina-Marino, A. Acceptability and Accuracy of Cervical Cancer Screening Using a Self-Collected Tampon for HPV Messenger-RNA Testing among HIV-Infected Women in South Africa. PLoS ONE 2015, 10, e0137299. [Google Scholar] [CrossRef] [PubMed]
- Tiiti, T.A.; Mashishi, T.L.; Nkwinika, V.V.; Benoy, I.; Selabe, S.G.; Bogers, J.; Lebelo, R.L. High-risk human papillomavirus detection in self-collected vaginal samples compared with healthcare worker collected cervical samples among women attending gynecology clinics at a tertiary hospital in Pretoria, South Africa. Virol. J. 2021, 18, 192. [Google Scholar] [CrossRef] [PubMed]
- Bakkum-Gamez, J.N.; Wentzensen, N.; Maurer, M.J.; Hawthorne, K.M.; Voss, J.S.; Kroneman, T.N.; Famuyide, A.O.; Clayton, A.C.; Halling, K.C.; Kerr, S.E.; et al. Detection of endometrial cancer via molecular analysis of DNA collected with vaginal tampons. Gynecol. Oncol. 2015, 137, 14–22. [Google Scholar] [CrossRef]
- Knox, J.; Tabrizi, S.N.; Miller, P.; Petoumenos, K.; Law, M.; Chen, S.; Garland, S.M. Evaluation of self-collected samples in contrast to practitioner-collected samples for detection of Chlamydia trachomatis, Neisseria gonorrhoeae, and Trichomonas vaginalis by polymerase chain reaction among women living in remote areas. Sex. Transm. Dis. 2002, 29, 647–654. [Google Scholar] [CrossRef]
- McLarty, J.W.; Williams, D.L.; Loyd, S.; Hagensee, M.E. Cervical Human Papillomavirus Testing with Two Home Self-Collection Methods Compared With a Standard Clinically Collected Sampling Method. Sex. Transm. Dis. 2019, 46, 670–675. [Google Scholar] [CrossRef]
- Kimmitt, P.T.; Tabrizi, S.N.; Crosatti, M.; Garland, S.M.; Schober, P.C.; Rajakumar, K.; Chapman, C.A. Pilot study of the utility and acceptability of tampon sampling for the diagnosis of Neisseria gonorrhoeae and Chlamydia trachomatis infections by duplex realtime polymerase chain reaction in United Kingdom sex workers. Int. J. STD AIDS 2010, 21, 279–282. [Google Scholar] [CrossRef]
- Romo, L.F.; Berenson, A.B. Tampon use in adolescence: Differences among European American, African American and Latina women in practices, concerns, and barriers. J. Pediatr. Adolesc. Gynecol. 2012, 25, 328–333. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Chiaruzzi, M.; Barbry, A.; Muggeo, A.; Tristan, A.; Jacquemond, I.; Badiou, C.; Cluzeau, L.; Bourdeau, S.; Durand, T.; Engelmann, A.; et al. Vaginal Tampon Colonization by Staphylococcus aureus in Healthy Women. Appl. Env. Microbiol. 2020, 86, e01249-20. [Google Scholar] [CrossRef] [PubMed]
- Femoflor 16 REAL-TIME PCR Detection Kit, USER MANUAL. Available online: https://dna-technology.com/sites/default/files/femoflor16_en.pdf (accessed on 12 September 2023).
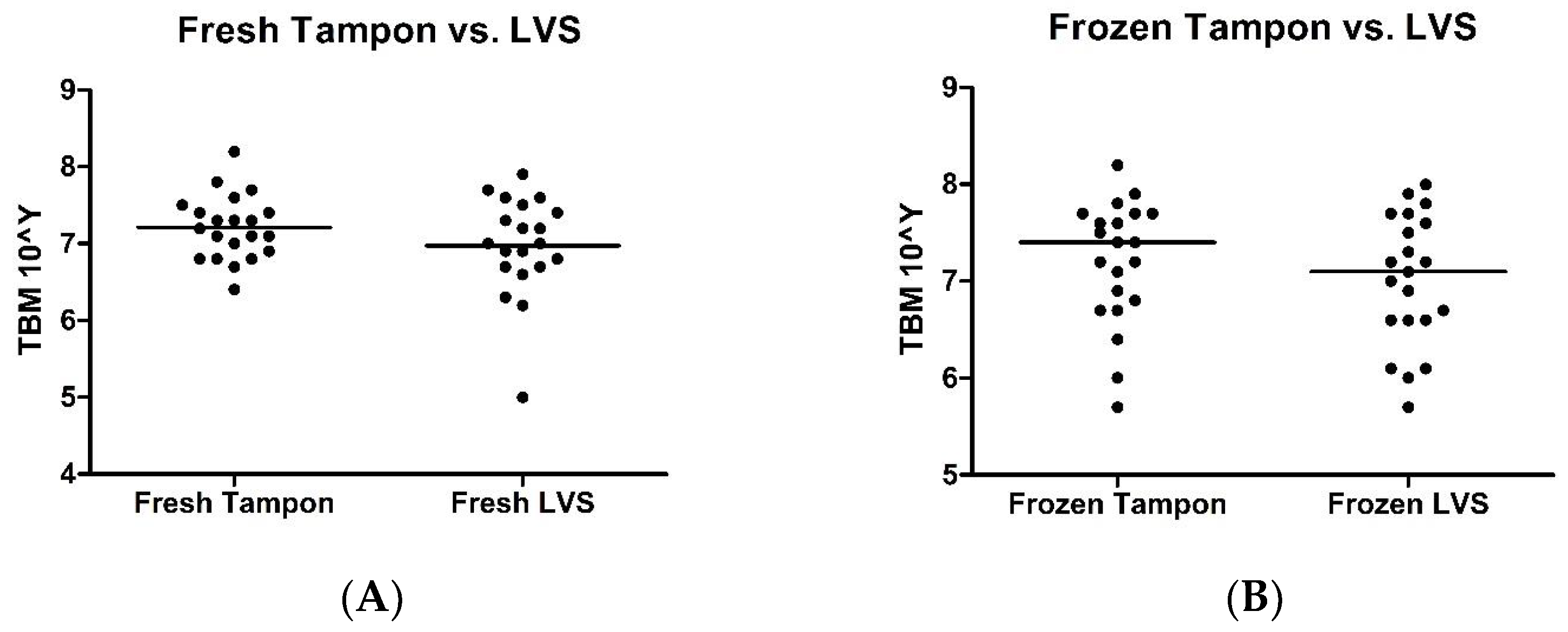
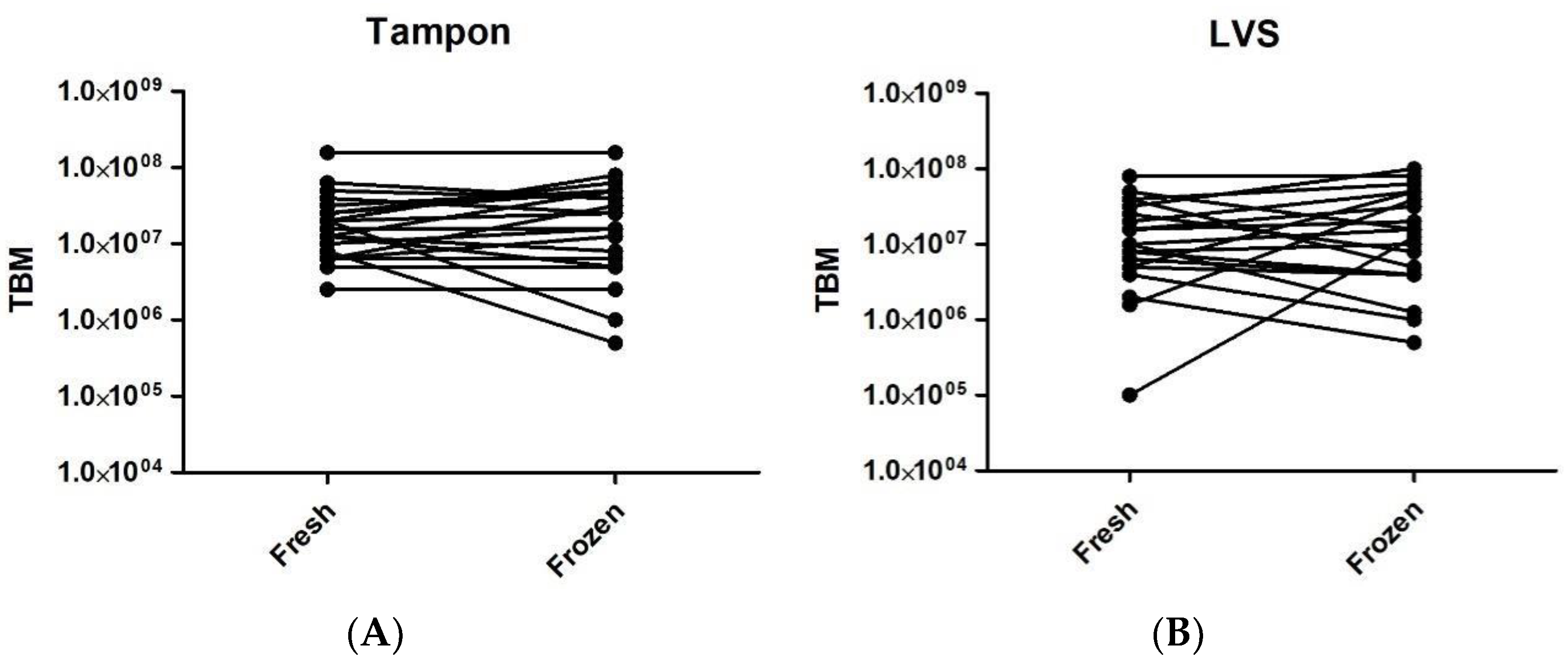
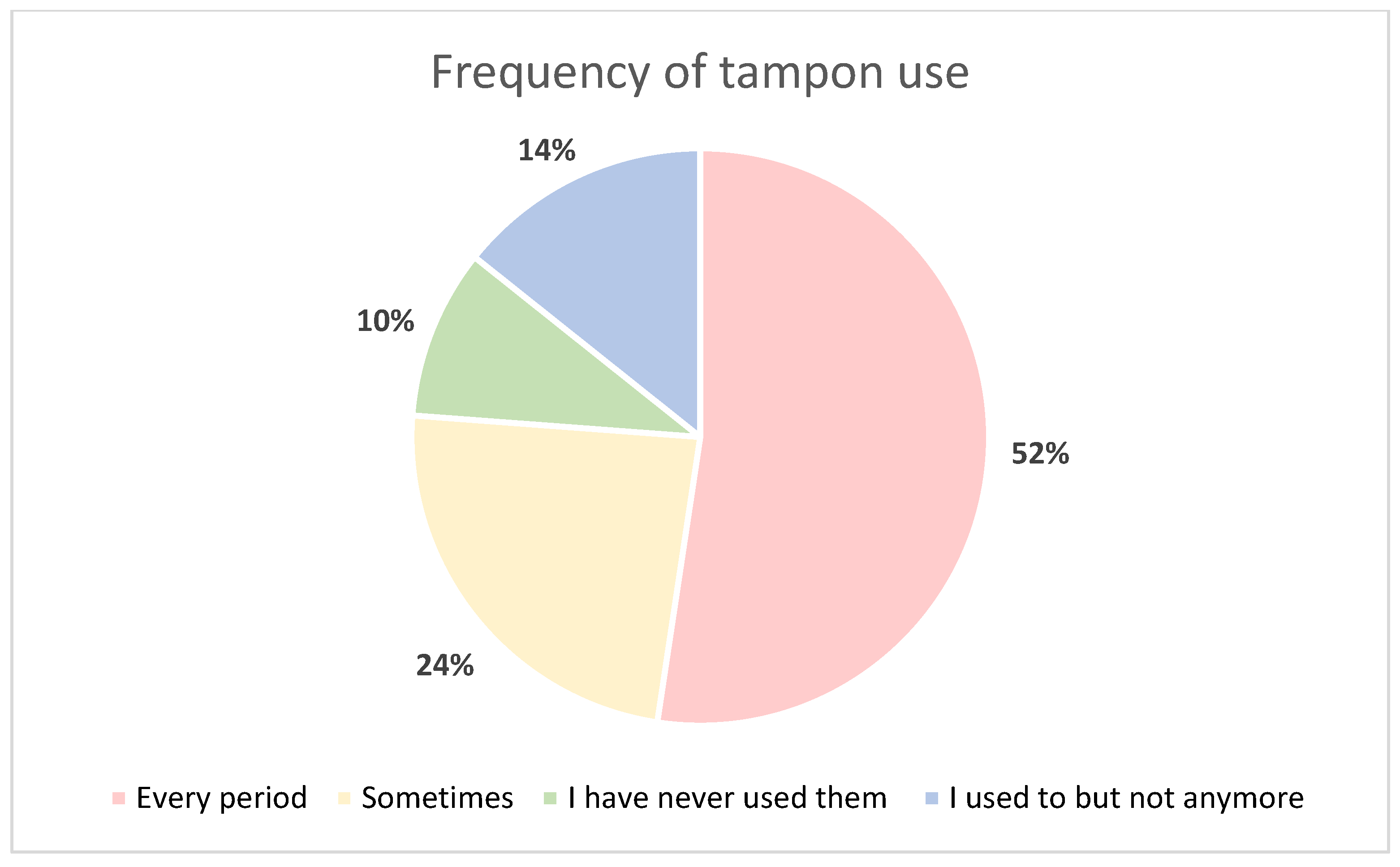

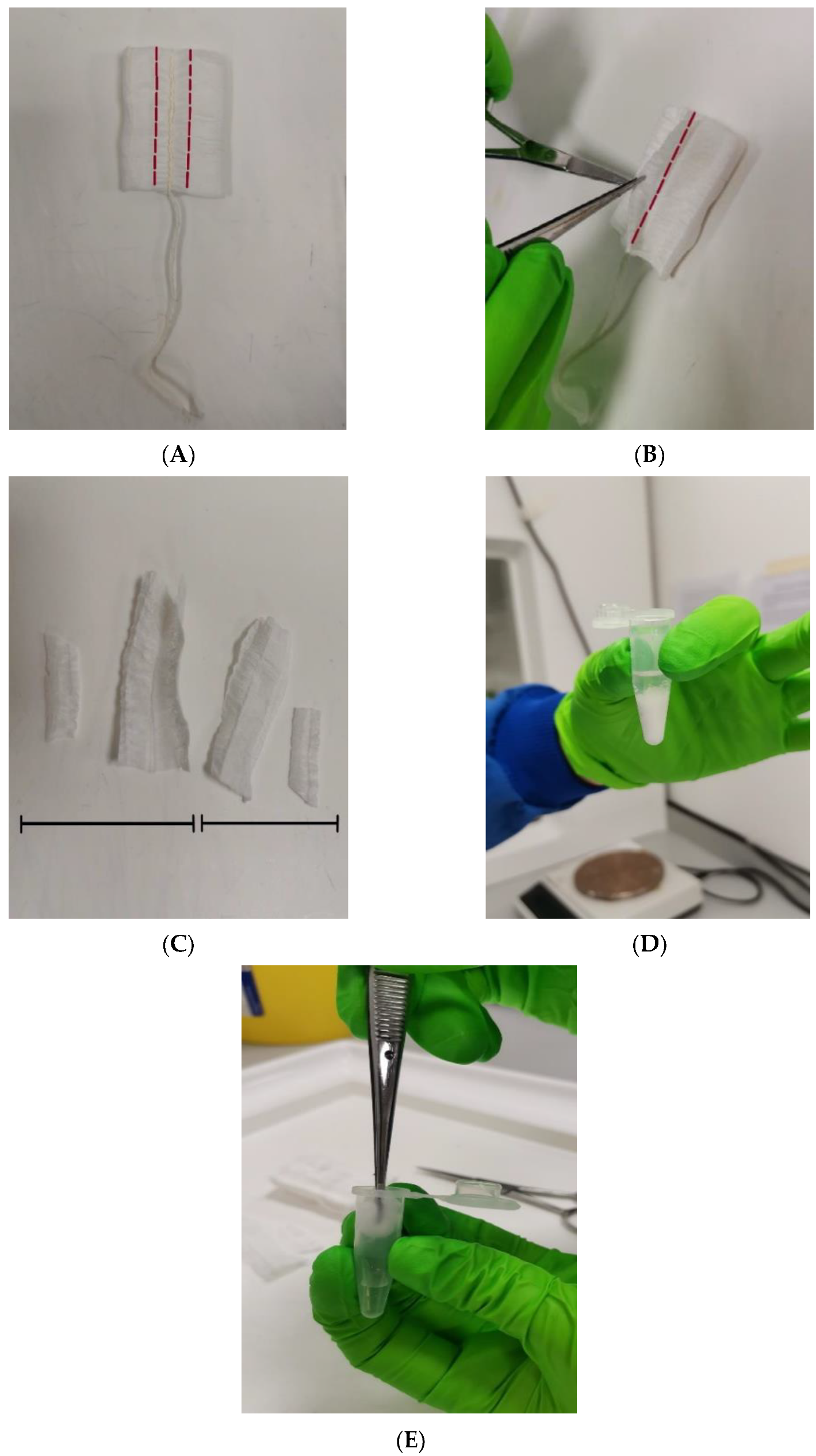
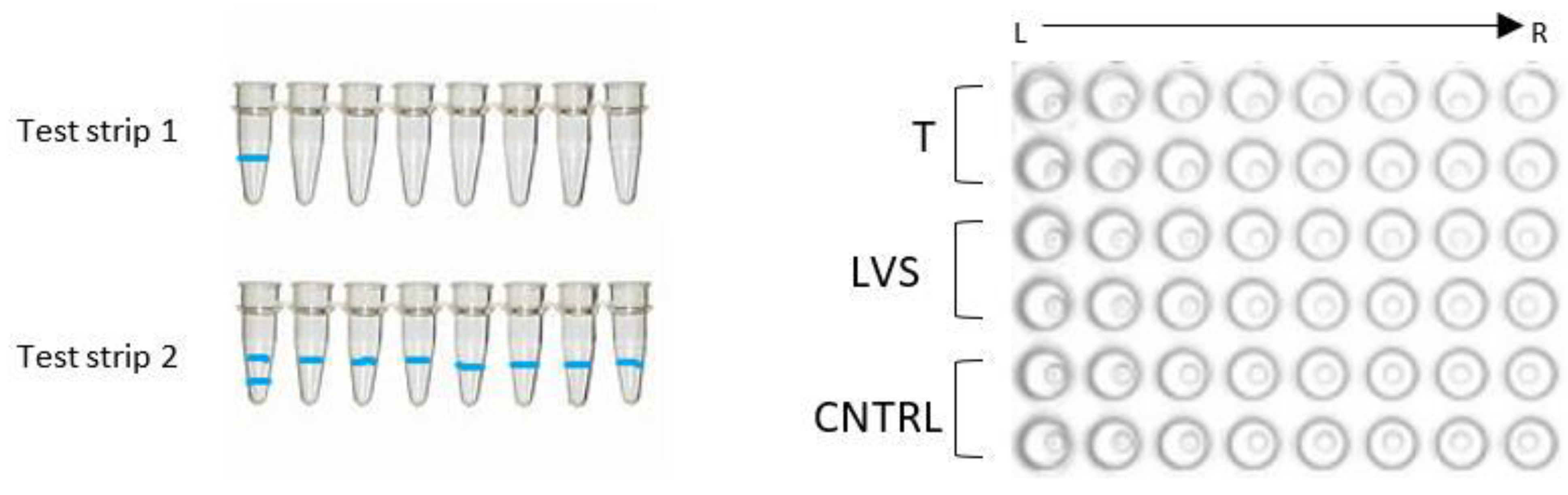
| Sample ID | Fresh Tampon | Frozen Tampon | Fresh LVS | Frozen LVS |
|---|---|---|---|---|
| VMF01 | 106.8 | 106.8 | 106.7 | 106.6 |
| VMF02 | 107.4 | 107.8 | 107.7 | 107.2 |
| VMF03 | 107.3 | 107.4 | 106.9 | 107.0 |
| VMF04 | 106.8 | 107.5 | 105.0 | 107.1 |
| VMF05 | 108.2 | 108.2 | 107.5 | 108.0 |
| VMF06 | 107.6 | 107.4 | 106.7 | 107.7 |
| VMF07 | 107.1 | 107.7 | 107.0 | 106.1 |
| VMF08 | 107.1 | 106.7 | 107.4 | 106.9 |
| VMF09 | 106.8 | 107.1 | 106.6 | 106.0 |
| VMF10 | 106.9 | 105.7 | 106.3 | 105.7 |
| VMF11 | 107.3 | 106.0 | 106.2 | 107.6 |
| VMF12 | 107.5 | 107.7 | 107.2 | 107.5 |
| VMF13 | 107.8 | 107.6 | 107.6 | 107.8 |
| VMF14 | 107.0 | 107.2 | 107.3 | 107.7 |
| VMF15 | 107.1 | 106.9 | 107.0 | 107.2 |
| VMF16 | 107.7 | 107.6 | 106.8 | 106.6 |
| VMF17 | 107.4 | 107.7 | 107.9 | 107.9 |
| VMF18 | 106.7 | 106.7 | 106.9 | 106.6 |
| VMF19 | 107.2 | 107.2 | 107.2 | 107.3 |
| VMF20 | 106.4 | 106.4 | - | 106.1 |
| VMF21 | 107.3 | 107.9 | 107.6 | 106.7 |
| Sample ID | Fresh Tampon | Frozen Tampon | Fresh LVS | Frozen LVS |
|---|---|---|---|---|
| VMF01 | AAnD | AAnD | AAnD | AAnD |
| VMF02 | AN | AN | AN | AN |
| VMF03 | AN | AN | AN | AN |
| VMF04 | AN | AN | - | AN |
| VMF05 | MAnD | AN | MAnD | MAnD |
| VMF06 | AN | AN | AN | AN |
| VMF07 | AN | AN | AN | AN |
| VMF08 | MAnD | MAnD | AAnD | AAnD |
| VMF09 | AN | AN | AN | AN |
| VMF10 | MAnD | - | CN +c | - |
| VMF11 | AN | - | AN | AN |
| VMF12 | MAnD | MAnD | AAnD | AAnD |
| VMF13 | MAnD | MAnD | AAnD | AAnD |
| VMF14 | AN | AN | AN | - |
| VMF15 | CN | AN | MAnD | MAnD |
| VMF16 | CN | CN | CN | CN |
| VMF17 | AN | AN | AN | AN |
| VMF18 | AN | N | AN | AN |
| VMF19 | MMD | MAnD | AN | CN +c |
| VMF20 | AAnD | AAnD | - | AAnD |
| VMF21 | AN | AN | AN | AN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turner, F.; Drury, J.; Hapangama, D.K.; Tempest, N. Menstrual Tampons Are Reliable and Acceptable Tools to Self-Collect Vaginal Microbiome Samples. Int. J. Mol. Sci. 2023, 24, 14121. https://doi.org/10.3390/ijms241814121
Turner F, Drury J, Hapangama DK, Tempest N. Menstrual Tampons Are Reliable and Acceptable Tools to Self-Collect Vaginal Microbiome Samples. International Journal of Molecular Sciences. 2023; 24(18):14121. https://doi.org/10.3390/ijms241814121
Chicago/Turabian StyleTurner, Florence, Josephine Drury, Dharani K. Hapangama, and Nicola Tempest. 2023. "Menstrual Tampons Are Reliable and Acceptable Tools to Self-Collect Vaginal Microbiome Samples" International Journal of Molecular Sciences 24, no. 18: 14121. https://doi.org/10.3390/ijms241814121
APA StyleTurner, F., Drury, J., Hapangama, D. K., & Tempest, N. (2023). Menstrual Tampons Are Reliable and Acceptable Tools to Self-Collect Vaginal Microbiome Samples. International Journal of Molecular Sciences, 24(18), 14121. https://doi.org/10.3390/ijms241814121






