Milk-Derived Small Extracellular Vesicles Promote Osteogenic Differentiation and Inhibit Inflammation via microRNA-21
Abstract
:1. Introduction
2. Results
2.1. Expression of miR-21 in CAP
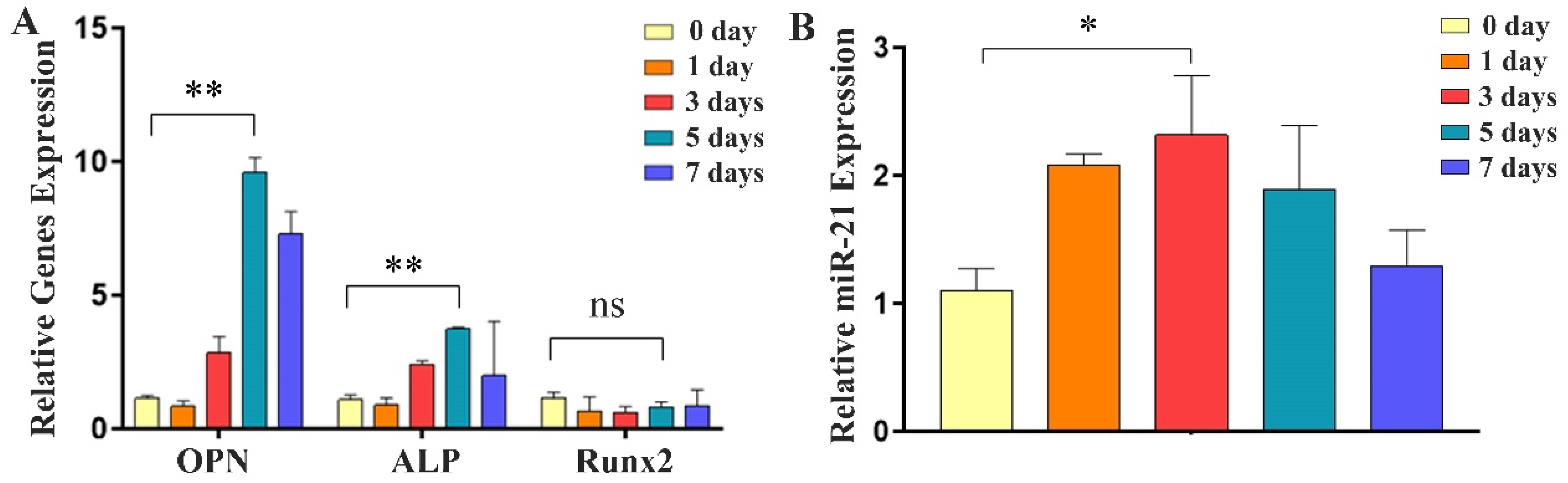
2.2. Expression of Osteogenic Factors and miR-21 during Differentiation of MC3T3-E1 Cells
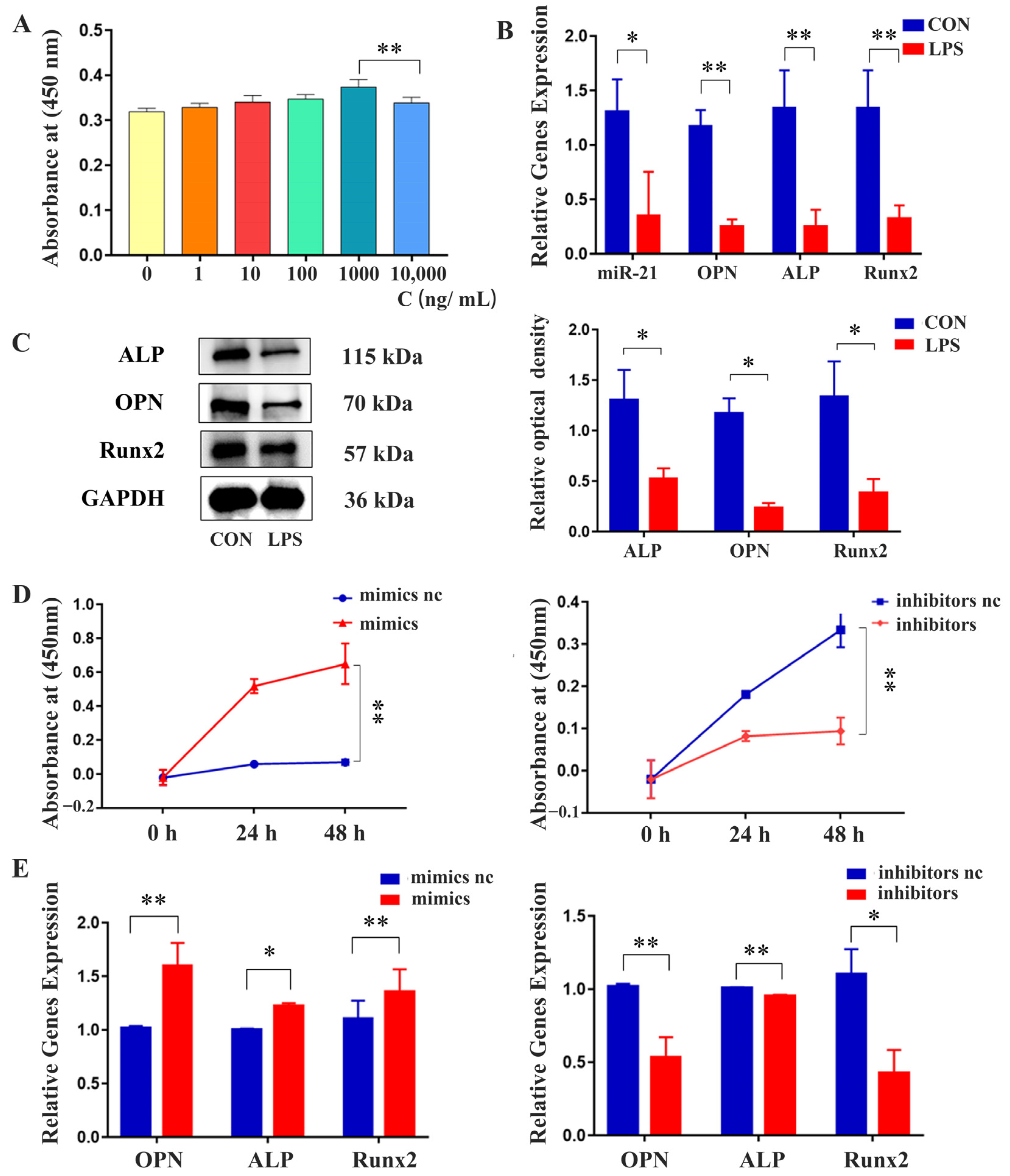
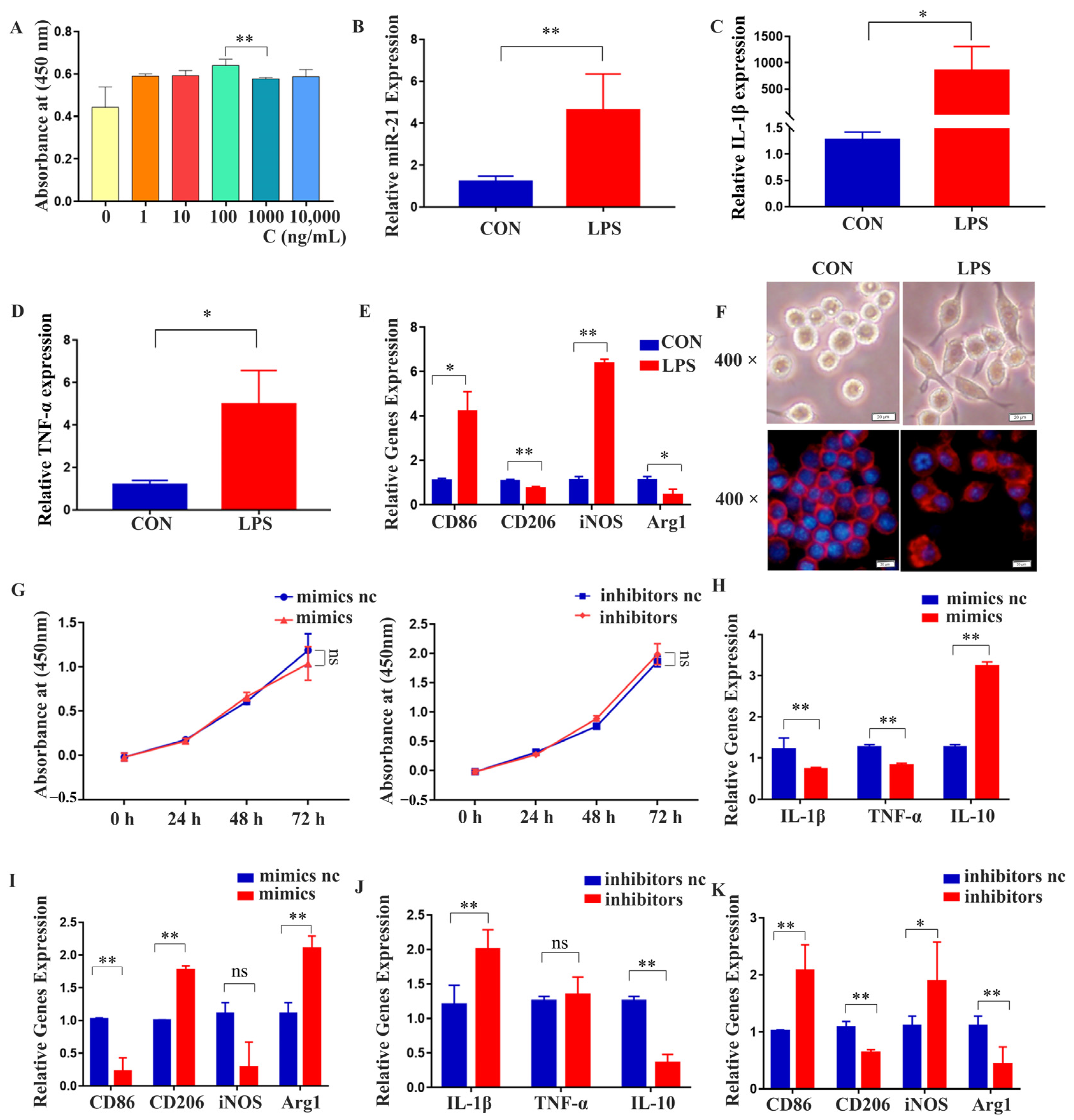
2.3. Effects of miR-21 on the Proliferation and Osteogenic Differentiation of MC3T3-E1 Cells under LPS Stimulation
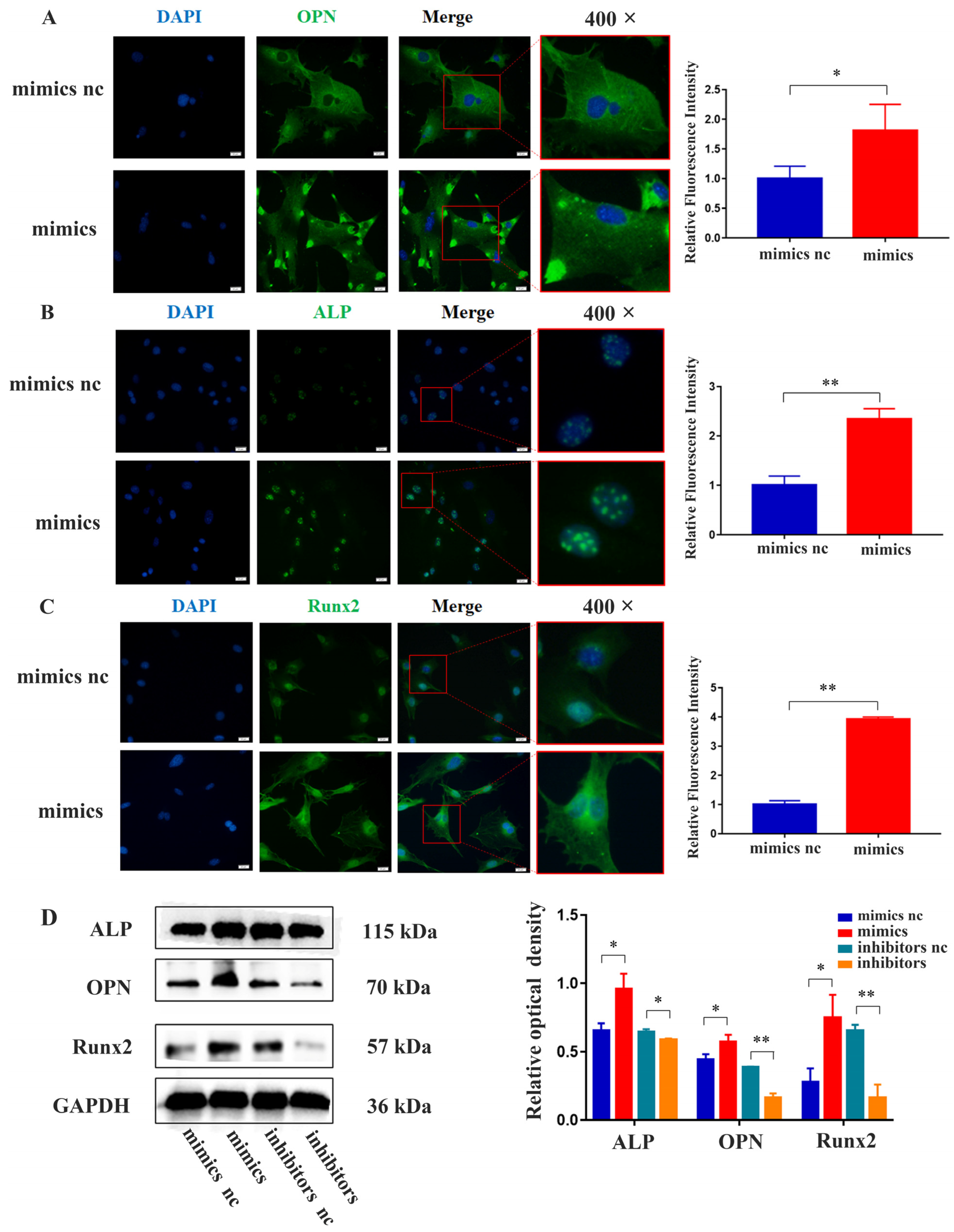
2.4. miR-21 Promoted the Expression of Osteogenic Proteins in MC3T3-E1 Cells under LPS Stimulation
2.5. miR-21 Regulated the Polarization and Expression of Inflammatory Factors in RAW 264.7 Cells under LPS Stimulation
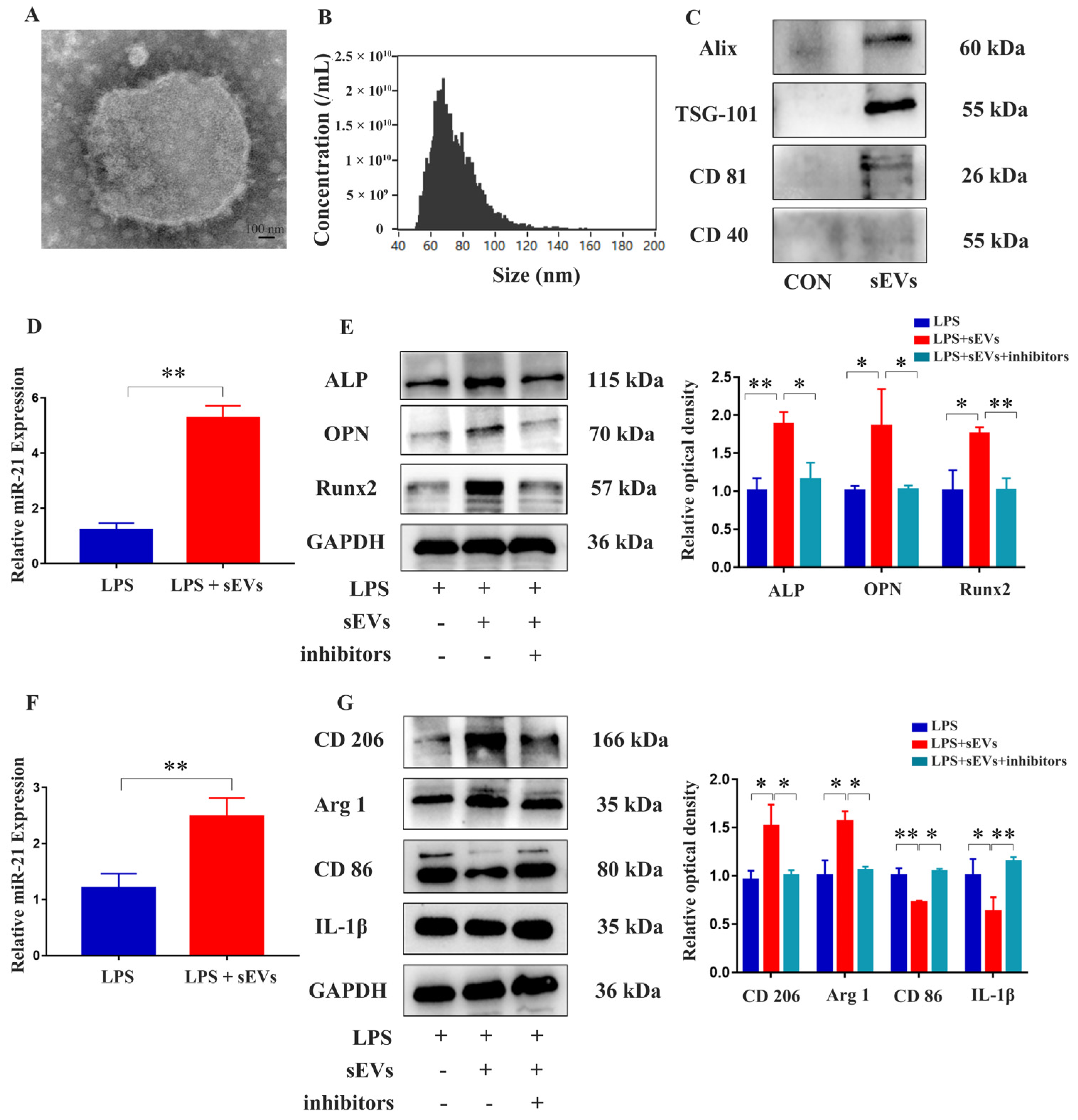
2.6. Isolation and Identification of Milk-Derived sEVs and Their Effects on MC3T3-E1 Cells and RAW 264.7 Cells
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Hematoxylin and Eosin (HE) Staining
4.3. Cell Culture and Induction
4.4. Cell Transfection Assay
4.5. Cell Proliferation Assay
4.6. Isolation and Characterization of sEVs Derived from Milk
4.7. Quantitative Reverse Transcription–Polymerase Chain Reaction (qRT-PCR)
4.8. Immunofluorescence (IF)
4.9. Western Blotting
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, X.; Wan, Q.; Cheng, L.; Xu, R. Mechanisms of bone remodeling and therapeutic strategies in chronic apical periodontitis. Front. Cell. Infect. Microbiol. 2022, 12, 908859. [Google Scholar] [CrossRef] [PubMed]
- Tibúrcio-Machado, C.S.; Michelon, C.; Zanatta, F.B.; Gomes, M.S.; Marin, J.A.; Bier, C.A. The global prevalence of apical periodontitis: A systematic review and meta-analysis. Int. Endod. J. 2021, 54, 712–735. [Google Scholar] [CrossRef]
- Gomes, B.; Herrera, D.R. Etiologic role of root canal infection in apical periodontitis and its relationship with clinical symptomatology. Braz. Oral. Res. 2018, 32 (Suppl. S1), e69. [Google Scholar] [CrossRef]
- Gan, G.; Lu, B.; Zhang, R.; Luo, Y.; Chen, S.; Lei, H.; Li, Y.; Cai, Z.; Huang, X. Chronic apical periodontitis exacerbates atherosclerosis in apolipoprotein E-deficient mice and leads to changes in the diversity of gut microbiota. Int. Endod. J. 2022, 55, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Berggren, K.; Broström, A.; Firestone, A.; Wright, B.; Josefsson, E.; Lindmark, U. Oral health problems linked to obstructive sleep apnea are not always recognized within dental care-As described by dental professionals. Clin. Exp. Dent. Res. 2022, 8, 84–95. [Google Scholar] [CrossRef]
- Maniaci, A.; Ferlito, S.; Lechien, J.R.; Di Luca, M.; Iannella, G.; Cammaroto, G.; Cannavicci, A.; Pollicina, I.; Stilo, G.; Di Mauro, P.; et al. Anxiety, depression and sleepiness in OSA patients treated with barbed reposition pharyngoplasty: A prospective study. Eur. Arch. Oto-Rhino-Laryngol.—Head. Neck Surg. 2022, 279, 4189–4198. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.; Martinho, F.C.; Barbosa, D.S.; Antunes, L.S.; Póvoa, H.C.C.; Baltus, T.H.L.; Morelli, N.R.; Vargas, H.O.; Nunes, S.O.V.; Anderson, G.; et al. Increased Root Canal Endotoxin Levels are Associated with Chronic Apical Periodontitis, Increased Oxidative and Nitrosative Stress, Major Depression, Severity of Depression, and a Lowered Quality of Life. Mol. Neurobiol. 2018, 55, 2814–2827. [Google Scholar] [CrossRef]
- Xu, R.; Guo, D.; Zhou, X.; Sun, J.; Zhou, Y.; Fan, Y.; Zhou, X.; Wan, M.; Du, W.; Zheng, L. Disturbed bone remodelling activity varies in different stages of experimental, gradually progressive apical periodontitis in rats. Int. J. Oral. Sci. 2019, 11, 27. [Google Scholar] [CrossRef]
- Wang, S.; Xiong, G.; Ning, R.; Pan, Z.; Xu, M.; Zha, Z.; Liu, N. LncRNA MEG3 promotes osteogenesis of hBMSCs by regulating miR-21-5p/SOD3 axis. Acta Biochim. Pol. 2022, 69, 71–77. [Google Scholar] [CrossRef]
- Hur, J.; Rhee, C.K.; Lee, S.Y.; Kim, Y.K.; Kang, J.Y. MicroRNA-21 inhibition attenuates airway inflammation and remodelling by modulating the transforming growth factor β-Smad7 pathway. Korean J. Intern. Med. 2021, 36, 706–720. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Li, X.; Zhang, Z.; Zhao, X.; Wang, C.; Wei, F. MicroRNA-21 promotes bone reconstruction in maxillary bone defects. J. Oral. Rehabil. 2020, 47 (Suppl. S1), 4–11. [Google Scholar] [CrossRef] [PubMed]
- Geng, Z.; Yu, Y.; Li, Z.; Ma, L.; Zhu, S.; Liang, Y.; Cui, Z.; Wang, J.; Yang, X.; Liu, C. miR-21 promotes osseointegration and mineralization through enhancing both osteogenic and osteoclastic expression. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 111, 110785. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Du, J.; Zhou, J.; Zhong, B.; Ba, L.; Zhang, J.; Liu, Y.; Liu, S. Elevated microRNA-21 Is a Brake of Inflammation Involved in the Development of Nasal Polyps. Front. Immunol. 2021, 12, 530488. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Su, L.; Duan, X.; Chen, X.; Hays, A.; Upadhyayula, S.; Shivde, J.; Wang, H.; Li, Y.; Huang, D.; et al. MicroRNA-21 down-regulates inflammation and inhibits periodontitis. Mol. Immunol. 2018, 101, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.A.; Garlet, G.P.; Fukada, S.Y.; Silva, J.S.; Cunha, F.Q. Chemokines in oral inflammatory diseases: Apical periodontitis and periodontal disease. J. Dent. Res. 2007, 86, 306–319. [Google Scholar] [CrossRef]
- Verweij, F.J.; Balaj, L.; Boulanger, C.M.; Carter, D.R.F.; Compeer, E.B.; D’Angelo, G.; El Andaloussi, S.; Goetz, J.G.; Gross, J.C.; Hyenne, V.; et al. The power of imaging to understand extracellular vesicle biology in vivo. Nat. Methods 2021, 18, 1013–1026. [Google Scholar] [CrossRef]
- Bernardi, S.; Balbi, C. Extracellular Vesicles: From Biomarkers to Therapeutic Tools. Biology 2020, 9, 258. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Fang, F.; Sun, M.; Zhang, Y.; Hu, M.; Zhang, J. Extracellular vesicles as bioactive nanotherapeutics: An emerging paradigm for regenerative medicine. Theranostics 2022, 12, 4879–4903. [Google Scholar] [CrossRef]
- Du, S.; Guan, Y.; Xie, A.; Yan, Z.; Gao, S.; Li, W.; Rao, L.; Chen, X.; Chen, T. Extracellular vesicles: A rising star for therapeutics and drug delivery. J. Nanobiotechnol. 2023, 21, 231. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Jia, Y.; Yu, L.; Ma, T.; Xu, W.; Qian, H.; Sun, Y.; Shi, H. Small extracellular vesicles isolation and separation: Current techniques, pending questions and clinical applications. Theranostics 2022, 12, 6548–6575. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Q.; Tong, K.; Zhu, J.; Wang, H.; Chen, B.; Chen, L. BMSC-derived exosomes promote tendon-bone healing after anterior cruciate ligament reconstruction by regulating M1/M2 macrophage polarization in rats. Stem Cell Res. Ther. 2022, 13, 295. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, D.; Wang, H.; Chen, K.; Wang, S.; Xu, J.; Ji, P. Exosomes from adipose-derived stem cells regulate M1/M2 macrophage phenotypic polarization to promote bone healing via miR-451a/MIF. Stem Cell Res. Ther. 2022, 13, 149. [Google Scholar] [CrossRef]
- Yan, C.; Chen, J.; Wang, C.; Yuan, M.; Kang, Y.; Wu, Z.; Li, W.; Zhang, G.; Machens, H.G.; Rinkevich, Y.; et al. Milk exosomes-mediated miR-31-5p delivery accelerates diabetic wound healing through promoting angiogenesis. Drug Deliv. 2022, 29, 214–228. [Google Scholar] [CrossRef]
- Feng, X.; Chen, X.; Zheng, X.; Zhu, H.; Qi, Q.; Liu, S.; Zhang, H.; Che, J. Latest Trend of Milk Derived Exosomes: Cargos, Functions, and Applications. Front. Nutr. 2021, 8, 747294. [Google Scholar] [CrossRef]
- Vashisht, M.; Rani, P.; Onteru, S.K.; Singh, D. Curcumin Encapsulated in Milk Exosomes Resists Human Digestion and Possesses Enhanced Intestinal Permeability in Vitro. Appl. Biochem. Biotechnol. 2017, 183, 993–1007. [Google Scholar] [CrossRef]
- Li, D.; Yao, X.; Yue, J.; Fang, Y.; Cao, G.; Midgley, A.C.; Nishinari, K.; Yang, Y. Advances in Bioactivity of MicroRNAs of Plant-Derived Exosome-Like Nanoparticles and Milk-Derived Extracellular Vesicles. J. Agric. Food Chem. 2022, 70, 6285–6299. [Google Scholar] [CrossRef]
- do Nascimento, I.V.; Rodrigues, M.I.Q.; Isaias, P.H.C.; Barros-Silva, P.G.; Sousa, F.B.; Nunes Alves, A.P.N.; Mota, M.R.L. Chronic systemic corticosteroid therapy influences the development of pulp necrosis and experimental apical periodontitis, exacerbating the inflammatory process and bone resorption in rats. Int. Endod. J. 2022, 55, 646–659. [Google Scholar] [CrossRef]
- Huang, L.; Wu, H.; Wu, Y.; Song, F.; Zhang, L.; Li, Z.; Sun, H.; Huang, C. Pcsk9 Knockout Aggravated Experimental Apical Periodontitis via LDLR. J. Dent. Res. 2022, 101, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Amaral, R.R.; Braga, T.; Siqueira, J.F., Jr.; Rôças, I.N.; da Costa Rachid, C.T.C.; Oliveira, A.G.G.; de Souza Côrtes, M.I.; Love, R.M. Root Canal Microbiome Associated with Asymptomatic Apical Periodontitis as Determined by High-Throughput Sequencing. J. Endod. 2022, 48, 487–495. [Google Scholar] [CrossRef]
- Sureshbabu, N.M.; Ranganath, A.; Jacob, B. Concentrated Growth Factor—Surgical Management of Large Periapical Lesion Using a Novel Platelet Concentrate in Combination with Bone Graft. Ann. Maxillofac. Surg. 2020, 10, 246–250. [Google Scholar]
- Nara, K.; Kawashima, N.; Noda, S.; Fujii, M.; Hashimoto, K.; Tazawa, K.; Okiji, T. Anti-inflammatory roles of microRNA 21 in lipopolysaccharide-stimulated human dental pulp cells. J. Cell. Physiol. 2019, 234, 21331–21341. [Google Scholar] [CrossRef]
- Sikora, M.; Śmieszek, A.; Pielok, A.; Marycz, K. MiR-21-5p regulates the dynamic of mitochondria network and rejuvenates the senile phenotype of bone marrow stromal cells (BMSCs) isolated from osteoporotic SAM/P6 mice. Stem Cell Res. Ther. 2023, 14, 54. [Google Scholar] [CrossRef]
- Kurita, T.; Li, X.; Bhawal, U.K. Crosstalk between microRNA-21-5p and the transcription factor Dec1 maintains osteoblast function. Biochem. Biophys. Res. Commun. 2022, 632, 32–39. [Google Scholar] [CrossRef]
- Zi, J.; Wang, F.; Liu, Z.; Wang, Y.; Qiu, L.; Zhu, G.; Li, H. Impact of Toll-like Receptor 4 Expression on Inflammatory Responses Related to Premature Membrane Rupture Induced by Lipopolysaccharide. Discov. Med. 2023, 35, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yue, Y.; Chan, W.; Wei, W.; Liu, X.; Wang, M.; Li, J.; Hao, L. RGS10 negatively regulates apical periodontitis via TFEB-mediated autophagy in BABL/c mice model and in vitro. Int. Endod. J. 2023, 56, 854–868. [Google Scholar] [CrossRef]
- Zhang, F.; Gluck, J.M.; Brown, A.C.; Zaharoff, D.A.; King, M.W. Heparin Affinity-Based IL-4 Delivery to Modulate Macrophage Phenotype and Endothelial Cell Activity In Vitro. ACS Appl. Mater. Interfaces 2023. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Holden, J.A.; Attard, T.J.; Laughton, K.M.; Mansell, A.; O’Brien-Simpson, N.M.; Reynolds, E.C. Porphyromonas gingivalis lipopolysaccharide weakly activates M1 and M2 polarized mouse macrophages but induces inflammatory cytokines. Infect. Immun. 2014, 82, 4190–4203. [Google Scholar] [CrossRef] [PubMed]
- Veloso, P.; Fernández, A.; Terraza-Aguirre, C.; Álvarez, C.; Vernal, R.; Escobar, A.; Hernández, M. Macrophages skew towards M1 profile through reduced CD163 expression in symptomatic apical periodontitis. Clin. Oral. Investig. 2020, 24, 4571–4581. [Google Scholar] [CrossRef]
- Marycz, K.; Smieszek, A.; Marcinkowska, K.; Sikora, M.; Turlej, E.; Sobierajska, P.; Patej, A.; Bienko, A.; Wiglusz, R.J. Nanohydroxyapatite (nHAp) Doped with Iron Oxide Nanoparticles (IO), miR-21 and miR-124 Under Magnetic Field Conditions Modulates Osteoblast Viability, Reduces Inflammation and Inhibits the Growth of Osteoclast—A Novel Concept for Osteoporosis Treatment: Part 1. Int. J. Nanomed. 2021, 16, 3429–3456. [Google Scholar]
- Madhyastha, R.; Madhyastha, H.; Nurrahmah, Q.I.; Purbasari, B.; Maruyama, M.; Nakajima, Y. MicroRNA 21 Elicits a Pro-inflammatory Response in Macrophages, with Exosomes Functioning as Delivery Vehicles. Inflammation 2021, 44, 1274–1287. [Google Scholar] [CrossRef] [PubMed]
- Lechien, J.R.; Hans, S.; De Marrez, L.G.; Dequanter, D.; Rodriguez, A.; Muls, V.; Ben Abdelouahed, F.; Evrard, L.; Maniaci, A.; Saussez, S.; et al. Prevalence and Features of Laryngopharyngeal Reflux in Patients with Primary Burning Mouth Syndrome. Laryngoscope 2021, 131, E2627–E2633. [Google Scholar] [CrossRef]
- Zabolotnyi, D.; Kizim, Y.; Zabolotna, D.; Sulaieva, O.; Kizim, V. Laryngopharyngeal Reflux Alters Macrophage Polarization in Human Papilloma Virus-Negative Squamous Cell Carcinoma of the Larynx in Males. Clin. Exp. Otorhinolaryngol. 2021, 14, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Shi, C.; Yu, X.; Yang, Q.; Wu, S.; Liu, R.; Liu, T.; Wang, L.; Niu, W. Milk-derived small extracellular vesicles: Nanomaterials to promote bone formation. J. Nanobiotechnol. 2022, 20, 370. [Google Scholar] [CrossRef]
- Tong, L.; Hao, H.; Zhang, Z.; Lv, Y.; Liang, X.; Liu, Q.; Liu, T.; Gong, P.; Zhang, L.; Cao, F.; et al. Milk-derived extracellular vesicles alleviate ulcerative colitis by regulating the gut immunity and reshaping the gut microbiota. Theranostics 2021, 11, 8570–8586. [Google Scholar] [CrossRef]
- Lee, K.S.; Lee, J.; Kim, H.K.; Yeom, S.H.; Woo, C.H.; Jung, Y.J.; Yun, Y.E.; Park, S.Y.; Han, J.; Kim, E.; et al. Extracellular vesicles from adipose tissue-derived stem cells alleviate osteoporosis through osteoprotegerin and miR-21-5p. J. Extracell. Vesicles 2021, 10, e12152. [Google Scholar] [CrossRef]
- Verma, N.; Sangwan, P.; Tewari, S.; Duhan, J. Effect of Different Concentrations of Sodium Hypochlorite on Outcome of Primary Root Canal Treatment: A Randomized Controlled Trial. J. Endod. 2019, 45, 357–363. [Google Scholar] [CrossRef] [PubMed]

| Primers | Base Sequence |
|---|---|
| U6 F | 5′-GTGAAGGTCGGAGTCAACG-3′ |
| U6 R | 5′-TGAGGTCAATGAAGGGGTC-3′ |
| miR-21-5p-F | 5′-TCGCCCGTAGCTTATCAGACT-3′ |
| miR-21-5p-R | 5′-CCAGTGCAGGGTCCGAGGT-3′ |
| m-OPN-F | 5′-GCTCAGCACCTGAATGTACC-3′ |
| m-OPN-R | 5′-CCTCGGCTCGATGGCTAGC-3′ |
| m-ALP-F | 5′-TGAATCGGAACAACCTGACTGA-3′ |
| m-ALP-R | 5′-GAGCCTGCTTGGCCTTACC-3′ |
| m-IL-1β-F | 5′-CCAGGATGAGGACATGAGCAC-3′ |
| m-IL-1β-R | 5′-TGTTGTTCATCTCGGAGCCTGTA-3′ |
| m-TNF-α-F | 5′-GGTGCCTATGTCTCAGCCTCT-3′ |
| m-TNF-α-R | 5′-ACGTGGGCTACAGGCTTGTC-3′ |
| m-CD86-F | 5′-TTTCCTCCAAACCTCTCAATTTCA-3′ |
| m-CD86-R | 5′-TGGGCCTGCTAGGCTGATT-3′ |
| m-CD206-F | 5′-AGCTTCATCTTCGGGCCTTT-3′ |
| m-CD206-R | 5′-CCCTTGGGTTGAGGATCCAT-3′ |
| m-Arg1-F | 5′-TGGGTGACTCCCTGCATATCT-3′ |
| m-Arg1-R | 5′-CACCTTGGTCTTGGAGCTTATTAAA-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Liu, S.; Wu, S.; Xia, M.; Liu, W.; Wang, L.; Dong, M.; Niu, W. Milk-Derived Small Extracellular Vesicles Promote Osteogenic Differentiation and Inhibit Inflammation via microRNA-21. Int. J. Mol. Sci. 2023, 24, 13873. https://doi.org/10.3390/ijms241813873
Liu R, Liu S, Wu S, Xia M, Liu W, Wang L, Dong M, Niu W. Milk-Derived Small Extracellular Vesicles Promote Osteogenic Differentiation and Inhibit Inflammation via microRNA-21. International Journal of Molecular Sciences. 2023; 24(18):13873. https://doi.org/10.3390/ijms241813873
Chicago/Turabian StyleLiu, Runyuan, Shuo Liu, Saixuan Wu, Meng Xia, Wanqing Liu, Lina Wang, Ming Dong, and Weidong Niu. 2023. "Milk-Derived Small Extracellular Vesicles Promote Osteogenic Differentiation and Inhibit Inflammation via microRNA-21" International Journal of Molecular Sciences 24, no. 18: 13873. https://doi.org/10.3390/ijms241813873
APA StyleLiu, R., Liu, S., Wu, S., Xia, M., Liu, W., Wang, L., Dong, M., & Niu, W. (2023). Milk-Derived Small Extracellular Vesicles Promote Osteogenic Differentiation and Inhibit Inflammation via microRNA-21. International Journal of Molecular Sciences, 24(18), 13873. https://doi.org/10.3390/ijms241813873





