LINC01605 Is a Novel Target of Mutant p53 in Breast and Ovarian Cancer Cell Lines
Abstract
1. Introduction
2. Results
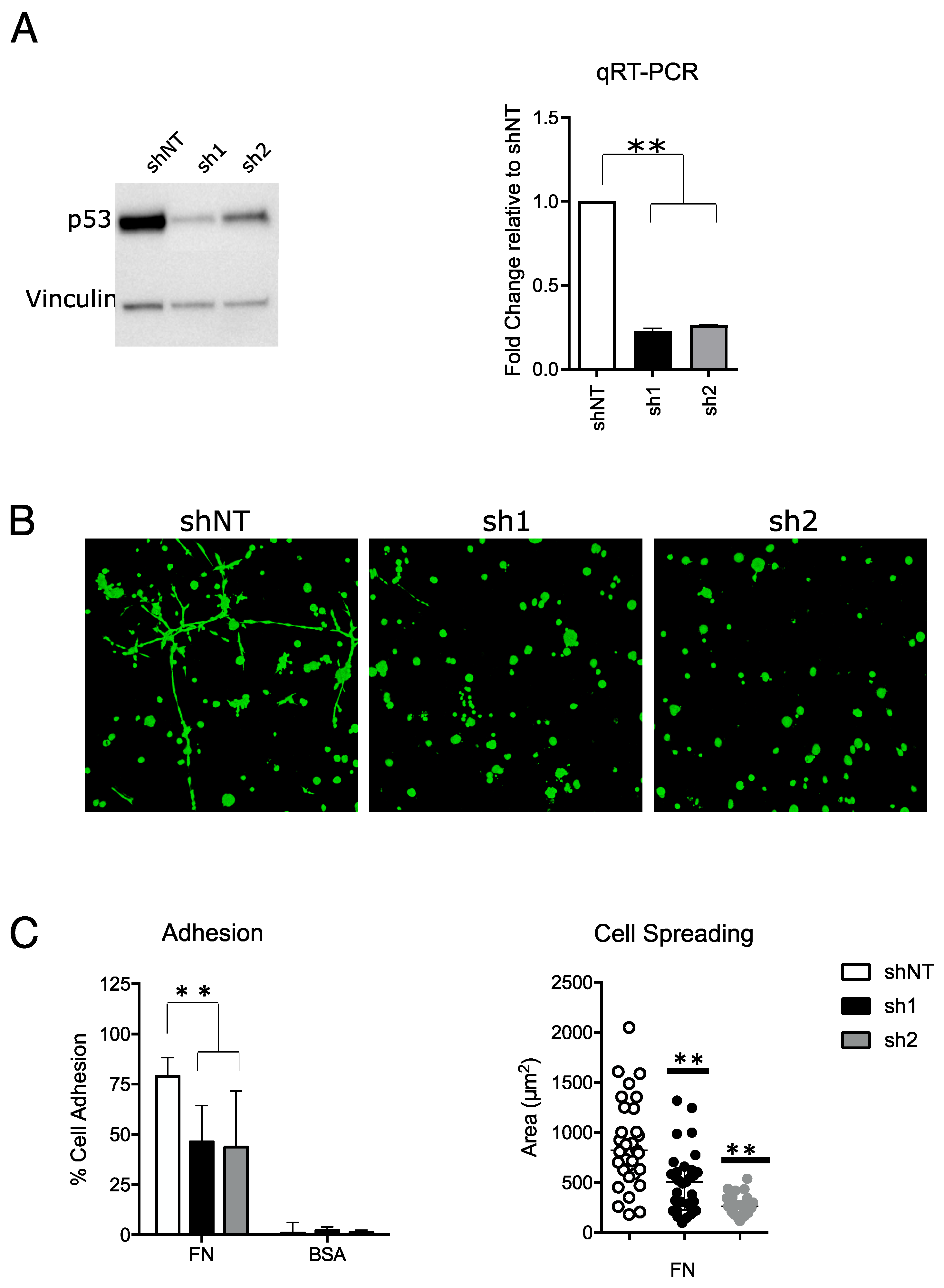
2.1. Mutant p53 Confers a Pro-Invasive Phenotype in MDA-MB-231 Cell Lines
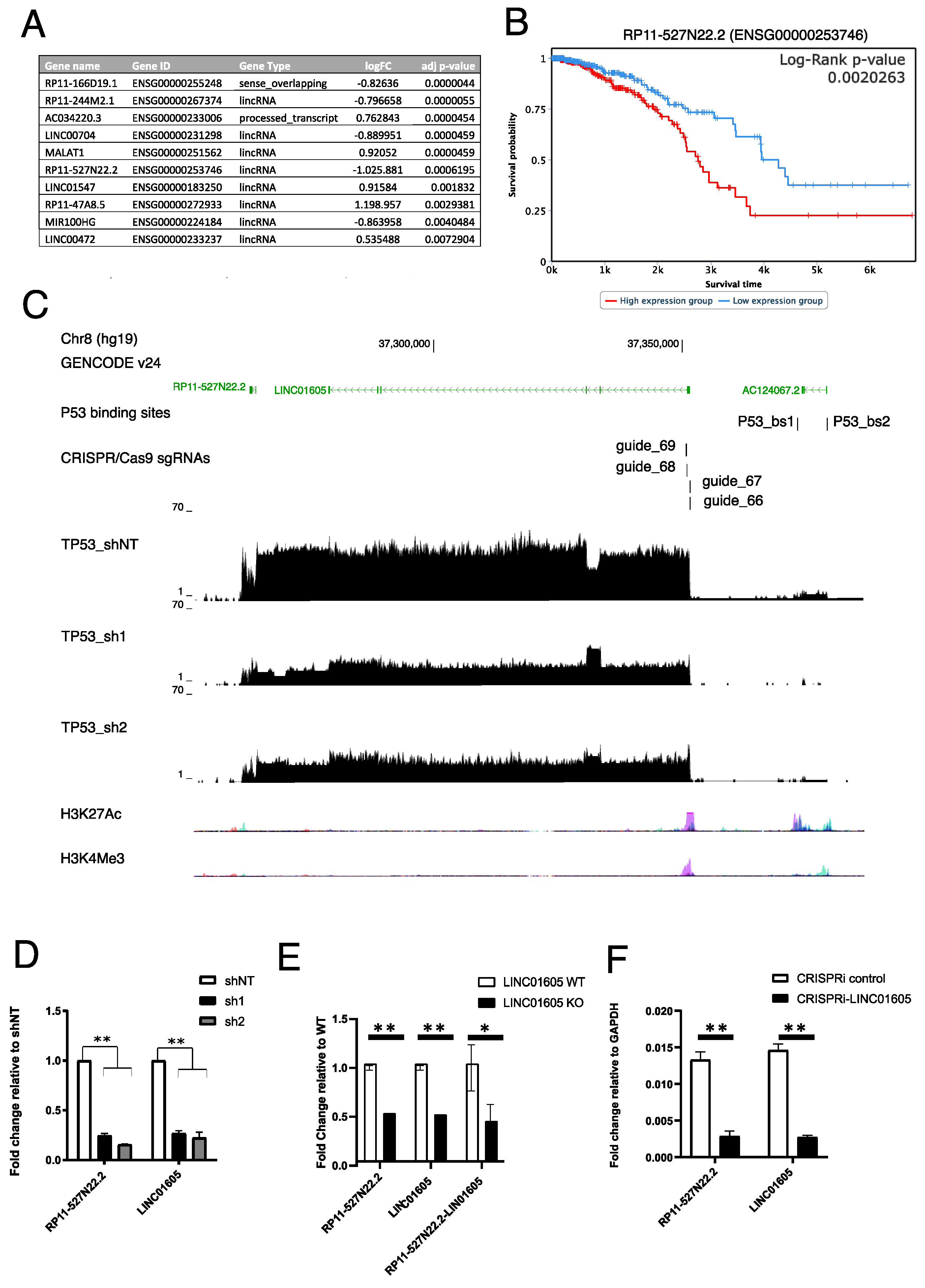
2.2. RP11-527N22.2 and LINC01605 Represent a Unique lncRNA Transcript Regulated by mut_p53 in Breast and Ovarian Cancer Cell Lines
2.3. Identification of a Putative mut_p53-Dependent DNA Regulatory Element near LINC01605 Transcription Start Site
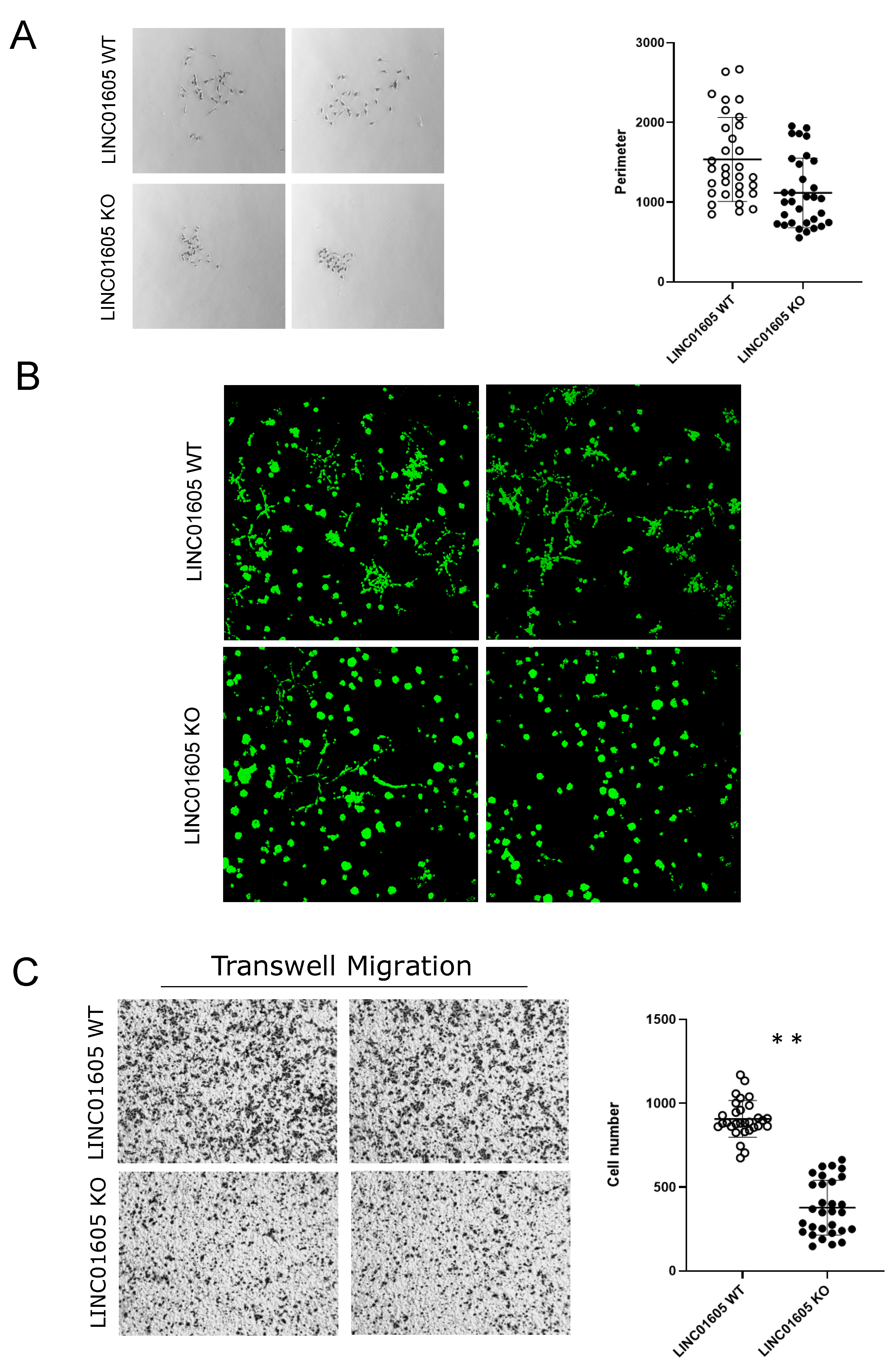
2.4. LINC01605 Regulates Breast Cancer Cell Migration
2.5. Pathway Analysis Reveals Resemblance between the Effect of mut_TP53 and LINC01605 in MDA-MB-231
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Cell Cultures and Lentiviral Transduction
4.2. CRISPR-Cas9 System
4.3. ChIP Assay
4.4. RNA Extraction, cDNA Synthesis and qRT-PCR
4.5. Luciferase Reporter Assay
4.6. Gene Expression and ChIP-Seq Analysis
4.7. Adhesion Assay, Colony Assay, Matrigel and Motility Experiments
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Stein, Y.; Rotter, V.; Aloni-Grinstein, R. Gain-of-function mutant p53: All the roads lead to tumorigenesis. Int. J. Mol. Sci. 2019, 20, 6197. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.J.; Vousden, K.H. P53 mutations in cancer. Nat. Cell Biol. 2013, 15, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019, 26, 199–212. [Google Scholar] [CrossRef]
- Vousden, K.H.; Prives, C. Blinded by the Light: The Growing Complexity of p53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef]
- McCann, J.J.; Vasilevskaya, I.A.; McNair, C.; Gallagher, P.; Neupane, N.P.; de Leeuw, R.; Shafi, A.A.; Dylgjeri, E.; Mandigo, A.C.; Schiewer, M.J.; et al. Mutant p53 elicits context-dependent pro-tumorigenic phenotypes. Oncogene 2021, 41, 444–458. [Google Scholar] [CrossRef]
- Khadiullina, R.; Mirgayazova, R.; Davletshin, D.; Khusainova, E.; Chasov, V.; Bulatov, E. Assessment of Thermal Stability of Mutant p53 Proteins via Differential Scanning Fluorimetry. Life 2022, 13, 31. [Google Scholar] [CrossRef]
- Walerych, D.; Lisek, K.; del Sal, G. Multi-omics reveals global effects of mutant p53 gain-of-function. Cell Cycle 2016, 15, 3009–3010. [Google Scholar] [CrossRef][Green Version]
- Walerych, D.; Lisek, K.; del Sal, G. Mutant p53: One, No One, and One Hundred Thousand. Front. Oncol. 2015, 5, 289. [Google Scholar] [CrossRef]
- Walerych, D.; Napoli, M.; Collavin, L.; Del Sal, G. The rebel angel: Mutant p53 as the driving oncogene in breast cancer. Carcinogenesis 2012, 33, 2007–2017. [Google Scholar] [CrossRef]
- Kim, T.; Veronese, A.; Pichiorri, F.; Lee, T.J.; Jeon, Y.-J.; Volinia, S.; Pineau, P.; Marchio, A.; Palatini, J.; Suh, S.-S.; et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 2011, 208, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Karaayvaz, M.; Jia, N.; Kaneuchi, M.; Hamada, J.; Watari, H.; Sudo, S.; Ju, J.; Sakuragi, N. Mutant p53 gain-of-function induces epithelial–mesenchymal transition through modulation of the miR-130b–ZEB1 axis. Oncogene 2013, 32, 3286–3295. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, L.; Chen, L.L. The diversity of long noncoding RNAs and their generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Lagarde, J. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat. Genet. 2017, 49, 1731–1740. [Google Scholar] [CrossRef]
- Derrien, T. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Fang, S. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2020, 22, 96–118. [Google Scholar] [CrossRef]
- Ali, T.; Grote, P. Beyond the RNA-dependent function of LncRNA genes. ELife 2020, 9, e60583. [Google Scholar] [CrossRef]
- Lin, T.; Hou, P.-F.; Meng, S.; Chen, F.; Jiang, T.; Li, M.-L.; Shi, M.-L.; Liu, J.-J.; Zheng, J.-N.; Bai, J. Emerging Roles of p53 Related lncRNAs in Cancer Progression: A Systematic Review. Int. J. Biol. Sci. 2019, 15, 1287–1298. [Google Scholar] [CrossRef]
- Vousden, K.H.; Lane, D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 275–283. [Google Scholar] [CrossRef]
- Hall, J.R.; Messenger, Z.J.; Tam, H.W.; Phillips, S.L.; Recio, L.; Smart, R.C. Long noncoding RNA lincRNA-p21 is the major mediator of UVB-induced and p53-dependent apoptosis in keratinocytes. Cell Death Dis. 2015, 6, e1700. [Google Scholar] [CrossRef] [PubMed]
- Tano, K.; Onoguchi-Mizutani, R.; Yeasmin, F.; Uchiumi, F.; Suzuki, Y.; Yada, T.; Akimitsu, N. Identification of minimal p53 promoter region regulated by MALAT1 in human lung adenocarcinoma cells. Front. Genet. 2018, 9, 208. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Lan, J.; Xu, L.; Feng, X.; Liao, H.; Xie, K.; Wu, H.; Zeng, Y. Long noncoding RNA TLNC1 promotes the growth and metastasis of liver cancer via inhibition of p53 signalling. Mol. Cancer. 2022, 21, 105. [Google Scholar] [CrossRef] [PubMed]
- Verduci, L.; Ferraiuolo, M.; Sacconi, A.; Ganci, F.; Vitale, J.; Colombo, T.; Paci, P.; Strano, S.; Macino, G.; Rajewsky, N.; et al. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD tran-scription-competent complex. Genome Biol. 2017, 18, 237. [Google Scholar] [CrossRef]
- Nandwani, A.; Rathore, S.; Datta, M. LncRNAs in cancer: Regulatory and therapeutic implications. Cancer Lett. 2021, 501, 162–171. [Google Scholar] [CrossRef]
- Wang, W.; He, X.; Wang, Y.; Liu, H.; Zhang, F.; Wu, Z.; Mo, S.; Chen, D. LINC01605 promotes aerobic glycolysis through lactate dehydrogenase A in triple-negative breast cancer. Cancer Sci. 2022, 113, 2484–2495. [Google Scholar] [CrossRef]
- Sonego, M.; Schiappacassi, M.; Lovisa, S.; Dall’Acqua, A.; Bagnoli, M.; Lovat, F.; Libra, M.; D’Andrea, S.; Canzonieri, V.; Militello, L.; et al. Stathmin regulates mutant p53 stability and transcriptional activity in ovarian cancer. EMBO Mol. Med. 2013, 5, 707–722. [Google Scholar] [CrossRef]
- Walerych, D.; Lisek, K.; Sommaggio, R.; Piazza, S.; Ciani, Y.; Dalla, E.; Rajkowska, K.; Gaweda-Walerych, K.; Ingallina, E.; Tonelli, C.; et al. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat. Cell Biol. 2016, 18, 897–909. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Y.; Wang, L.; Xu, P.-C.; Huang, F.-J.; Jian, X.; Wei, Z.-C.; Chen, Y.-Q. LINC01605 promotes the proliferation of laryngeal squamous cell carcinoma through targeting miR-493-3p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10379–10386. [Google Scholar] [CrossRef]
- Yue, M.; Liu, T.; Yan, G.; Luo, X.; Wang, L. LINC01605, regulated by the EP300-SMYD2 complex, potentiates the binding between METTL3 and SPTBN2 in colorectal cancer. Cancer Cell Int. 2021, 21, 504. [Google Scholar] [CrossRef]
- Forrest, M.E.; Saiakhova, A.; Beard, L.; Buchner, D.A.; Scacheri, P.C.; LaFramboise, T.; Markowitz, S.; Khalil, A.M. Colon Cancer-Upregulated Long Non-Coding RNA lincDUSP Regulates Cell Cycle Genes and Potentiates Resistance to Apoptosis. Sci. Rep. 2018, 8, 7324. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jiang, Y.; Guan, D.; Li, J.; Yin, H.; Pan, Y.; Xie, D.; Chen, Y. Critical Roles of p53 in Epithelial-Mesenchymal Transition and Metastasis of Hepatocellular Carcinoma Cells. PLoS ONE 2013, 8, e72846. [Google Scholar] [CrossRef]
- Sorrentino, G.; Ruggeri, N.; Specchia, V.; Cordenonsi, M.; Mano, M.; Dupont, S.; Manfrin, A.; Ingallina, E.; Sommaggio, R.; Piazza, S.; et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat. Cell Biol. 2014, 16, 357–366. [Google Scholar] [CrossRef]
- Parrales, A.; Iwakuma, T. p53 as a Regulator of Lipid Metabolism in Cancer. Int. J. Mol. Sci. 2016, 17, 2074. [Google Scholar] [CrossRef]
- Kabadi, A.M.; Ousterout, D.G.; Hilton, I.B.; Gersbach, C.A. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014, 42, e147. [Google Scholar] [CrossRef]

| mutTP53-silenced | LINC01605-KO | |
|---|---|---|
| HALLMARK_OXIDATIVE_PHOSPHOYLATION | enriched | enriched |
| HALLMARK_ADIPOGENESIS | enriched | enriched |
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | enriched | enriched |
| HALLMARK_DNA_REPAIR | enriched | enriched |
| HALLMARK_XENOBIOTIC_METABOLISM | enriched | enriched |
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | enriched | enriched |
| HALLMARK_PROTEIN_SECRETION | enriched | enriched |
| HALLMARK_UV_RESPONSE_UP | enriched | enriched |
| HALLMARK_FATTY_ACID_METABOLISM | enriched | enriched |
| HALLMARK_PEROXISOME | enriched | enriched |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | enriched | enriched |
| HALLMARK_MYOGENESIS | enriched | enriched |
| HALLMARK_MTORC1_SIGNALING | enriched | enriched |
| HALLMARK_HYPOXIA | enriched | |
| HALLMARK_GLYCOLYSIS | enriched | |
| HALLMARK_P53_PATHWAY | enriched | |
| HALLMARK_APOPTOSIS | enriched | |
| HALLMARK_ESTROGEN_RESPONSE_LATE | enriched | |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | enriched | |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | enriched | |
| HALLMARK_E2F_TARGETS | enriched | |
| HALLMARK_HEME_METABOLISM | enriched | |
| HALLMARK_IL2_STAT5_SIGNALING | enriched | |
| HALLMARK_COAGULATION | enriched | |
| HALLMARK_COMPLEMENT | enriched | |
| HALLMARK_ANDROGEN_RESPONSE | enriched | |
| HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY | enriched | |
| HALLMARK_UV_RESPONSE_DN | enriched | |
| HALLMARK_PI3K_AKT_MTOR_SIGNALING | enriched | |
| HALLMARK_TGF_BETA_SIGNALING | enriched | |
| HALLMARK_BILE_ACID_METABOLISM | enriched | |
| HALLMARK_ANGIOGENESIS | enriched | |
| HALLMARK_PANCREAS_BETA_CELLS | enriched | |
| HALLMARK_MYC_TARGETS_V1 | enriched | |
| HALLMARK_MYC_TARGETS_V2 | enriched | |
| HALLMARK_APICAL_JUNCTION | enriched | |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | enriched |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coan, M.; Toso, M.; Cesaratto, L.; Rigo, I.; Borgna, S.; Dalla Pietà, A.; Zandonà, L.; Iuri, L.; Zucchetto, A.; Piazza, C.; et al. LINC01605 Is a Novel Target of Mutant p53 in Breast and Ovarian Cancer Cell Lines. Int. J. Mol. Sci. 2023, 24, 13736. https://doi.org/10.3390/ijms241813736
Coan M, Toso M, Cesaratto L, Rigo I, Borgna S, Dalla Pietà A, Zandonà L, Iuri L, Zucchetto A, Piazza C, et al. LINC01605 Is a Novel Target of Mutant p53 in Breast and Ovarian Cancer Cell Lines. International Journal of Molecular Sciences. 2023; 24(18):13736. https://doi.org/10.3390/ijms241813736
Chicago/Turabian StyleCoan, Michela, Martina Toso, Laura Cesaratto, Ilenia Rigo, Silvia Borgna, Anna Dalla Pietà, Luigi Zandonà, Lorenzo Iuri, Antonella Zucchetto, Carla Piazza, and et al. 2023. "LINC01605 Is a Novel Target of Mutant p53 in Breast and Ovarian Cancer Cell Lines" International Journal of Molecular Sciences 24, no. 18: 13736. https://doi.org/10.3390/ijms241813736
APA StyleCoan, M., Toso, M., Cesaratto, L., Rigo, I., Borgna, S., Dalla Pietà, A., Zandonà, L., Iuri, L., Zucchetto, A., Piazza, C., Baldassarre, G., Spizzo, R., & Nicoloso, M. S. (2023). LINC01605 Is a Novel Target of Mutant p53 in Breast and Ovarian Cancer Cell Lines. International Journal of Molecular Sciences, 24(18), 13736. https://doi.org/10.3390/ijms241813736









