Abstract
Epigenetics is a rapidly developing science that has gained a lot of interest in recent years due to the correlation between characteristic epigenetic marks and cardiovascular diseases (CVDs). Epigenetic modifications contribute to a change in gene expression while maintaining the DNA sequence. The analysis of these modifications provides a thorough insight into the cardiovascular system from its development to its further functioning. Epigenetics is strongly influenced by environmental factors, including known cardiovascular risk factors such as smoking, obesity, and low physical activity. Similarly, conditions affecting the local microenvironment of cells, such as chronic inflammation, worsen the prognosis in cardiovascular diseases and additionally induce further epigenetic modifications leading to the consolidation of unfavorable cardiovascular changes. A deeper understanding of epigenetics may provide an answer to the continuing strong clinical impact of cardiovascular diseases by improving diagnostic capabilities, personalized medical approaches and the development of targeted therapeutic interventions. The aim of the study was to present selected epigenetic pathways, their significance in cardiovascular diseases, and their potential as a therapeutic target in specific medical conditions.
1. Introduction
Epigenetics involves a diverse set of reversible modifications to the genome that do not alter the deoxyribonucleic acid (DNA) sequence. Both the external environment and the internal microenvironment of cells and tissues influence epigenetic mechanisms that play a key role in the specification of cell fate and development of the organism [1,2]. Growing evidence suggests that epigenetic activity plays a fundamental role in the regulation of pathophysiological cellular processes [3,4,5]. The involvement of epigenetic mechanisms has been demonstrated at the level of protein and ribonucleic acid (RNA) gene expression and DNA replication in repair processes, cell differentiation and embryogenesis [6]. Epigenetic marks have traditionally been considered immutable, potentially transmissible to progeny, and underlying stable differentiation into different cell types that express their own distinctive patterns of gene expression [7]. It has now become clear that chromatin epigenetic markers are dynamically regulated in response to environmental cues [8,9,10]. This has led to a shift in the use of epigenetics to account for transient changes in chromatin in response to external stimuli that control gene expression [11,12,13,14,15]. Epigenetic pathways can be divided into three main categories: DNA methylation, histone modifications, and non-coding RNAs [16]. The first two are referred to as direct epigenetic mechanisms due to their involvement in chromatin remodeling, while non-coding RNAs are considered to be indirect due to modulation of gene expression mainly in the post-transcriptional phase.
1.1. DNA Methylation
DNA methylation is mainly associated with the inhibition of gene expression [17]. Active genes are located primarily in unmethylated sites. Cytosines contained in the DNA chain undergo covalent methylation in a reaction catalyzed by DNA methyltransferases (DNMT). High levels of 5-methylcytosine (5-mC) formed in this way are characteristic of the inactive form of chromatin-heterochromatin. The 5-mC molecule acts similarly to ordinary cytosine (C) by combining with guanine (G) in double-stranded DNA. However, when methylated cytosines are present at 5′–C–phosphate–G–3′ (CpG) sites in the promoter and enhancer regions, genes are often repressed [18,19]. CpG islands are regions in the genome with an increased content of CpG dinucleotides compared to the average value for the entire genome [20]. In humans, the presence of CpG islands within the promoter affects as much as 60–70% of genes. CpG islands located near housekeeping genes are usually not methylated. In the case of genes whose expression is characteristic only for specific tissues, methylation occurs selectively—CpG islands are not methylated only in the cells of these specific tissues; in others, they are methylated and not expressed [21].
More than every fifth transcription factor is inhibited from binding when the recognition sequence contains 5-mC. In addition, the presence of 5-mC in the promoter region can attract methyl-CpG-binding domain proteins that interact with nucleosome remodeling and histone deacetylase, leading to gene silencing. There are two types of DNA methylation—maintenance and de novo. Maintenance methylation is responsible for the methylation of the newly synthesized DNA strand due to replication at sites complementary to the methylated sites in the parent strand. This allows the methylation pattern to be inherited after cell division. DNMT-1 is involved in this reaction, responsible for maintaining epigenetic markers, which allows them to be reproduced with each cell division. The second type of methylation—de novo—uses DNMT-3A and DNMT-3B and consists of attaching methyl groups in completely new places, which changes the methylation pattern [22]. This allows it to maintain a specific pattern of methylated DNA sequences for various organs and tissues, which is passed on to the daughter cell. Thanks to this, it knows which genes should be expressed. Additionally, transcription from methylated CpG islands is strongly and hereditarily suppressed [22]. The reverse process—demethylations of 5-mC—catalyze ten-eleven translocation (TET) enzymes [23].
In eukaryotes, DNA is packaged into repeating units called nucleosomes by wrapping multimeric histone proteins. When nucleosomes are organized in tightly packed bundles (heterochromatin), transcription is inhibited by blocking the access of the transcriptional machinery. Conversely, when the chromatin is relaxed (euchromatin), the nucleosomes resemble balls on a string and this condition is associated with active transcription [24]. A large part of the modifications within histones relates to the change in the affinity of histones for DNA, which translates into the state of chromatin order. Although histone modifications occur throughout the sequence, the unstructured N-termini of histones, called histone tails, are particularly heavily modified [25]. Among the many modifications that occur, such as acetylation, methylation, ubiquitination, phosphorylation, sumoylation, ribosylation, and citrullination, the best described mechanism is histone acetylation. Histone acetylation involves the attachment of an acetyl group to a lysine (K) residue at the N-terminus of the core histone. These ends form protruding tails, and their acetylation reduces the affinity of histones for DNA, loosening the chromatin structure. In heterochromatin, histones are mostly non-acetylated. Histone acetyltransferases (HATs) are involved in acetylation. Similarly, histone methylation and demethylation are achieved by histone methyltransferases and histone demethylases, respectively [26]. Histone methylation can induce both activation and repression of transcription, depending on the number and location of the methyl groups [27,28]. Research on the diversity of histone modifications and the interactions resulting from them led to the formulation of the histone code hypothesis. According to the hypothesis, specific histone modification patterns determine the state of chromatin and gene activity in its area, which causes a specific biological effect. However, it is now known that the function of many histone-modifying enzymes extends independently and goes beyond their catalytic activity [29]. A full understanding of the exact mechanisms by which these histone tail changes affect DNA–histone interactions remains in the realm of research, though some specific examples have been developed in detail.
1.2. Indirect Epigenetic Mechanisms
A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. Particularly noteworthy in the context of epigenetic machinery are long ncRNAs (lncRNAs), and small RNAs, which include, among others: microRNAs (miRNAs), small interfering RNAs (siRNAs) and piwi-interacting RNAs (piRNAs). Approximately 60% of human protein-coding genes appear to be regulated by miRNAs [30]. MiRNA binds to messenger RNA (mRNA) that is complementary to it, leading to gene silencing and blocking the translation of mRNA into protein. As post-transcriptional repressors, they act on the 3′ untranslated region of mRNA to induce its degradation or translational repression. Each miRNA expressed in a cell can target about 100 to 200 messenger RNAs that it downregulates [31]. At the same time, about 50% of miRNA genes are associated with CpG islands [32], therefore, the expression of these genes is dependent on DNA methylation. Other miRNAs are regulated epigenetically by histone modifications or by combined DNA methylation and histone modification [32]. lncRNAs in the vast majority of cases do not encode any protein, there is evidence that some of them can carry information about micro- and polypeptides [33,34]. Specific lncRNAs show different subcellular locations—most of these molecules can be found in the nucleus, less in the cytoplasm or in both locations simultaneously. As a result, lncRNA molecules can efficiently regulate gene expression at various levels, from epigenetic DNA modifications and transcriptional changes to modulation of mRNA stability and translational or post-translational control [35]. In addition, lncRNAs can act as competitive endogenous RNAs (ceRNAs), thereby blocking miRNAs and regulating epigenetic mechanisms [36]. Figure 1 shows major epigenetic mechanisms involved in CVDs.
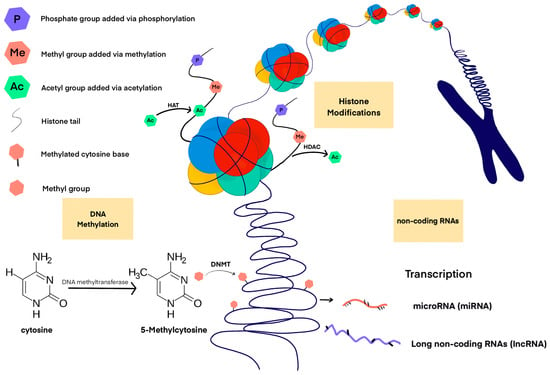
Figure 1.
Major epigenetic mechanisms involved in CVDs.
1.3. Epigenome-Wide Association Study
An epigenome-wide association study (EWAS) is a study that analyzes a set of measurable epigenetic characteristics such as DNA methylation across a genome. It is conducted in different patients to establish associations between epigenetic variation and a specific identifiable feature that can be considered within particular cardiovascular and cardiometabolic diseases [8]. An important meta-analysis of a genome-wide association study is a paper published in late 2022 in Nature, which covered n = 115,150 cases of all-cause heart failure and n = 1,550,331 controls of diverse genetic background [37]. The researchers identified 47 heart failure (HF) risk loci. Colocalization, gene expression profiling and Mendelian randomization have provided consistent evidence for a special role for branched-chain keto acid dehydrogenase E1 subunit alpha (BCKDHA) in the development of HF from all causes. In total, proteome-wide Mendelian randomization identified nine circulating HF-related proteins, and the strongest evidence of colocalization involved nine genes- DNAJC18, MTSS1, SQLE, BCKDHA, ABO, ALPK3, and PROM1. Another study demonstrated the utility of methyl-binding domain-capture sequencing to evaluate peripheral blood DNA methylation markers in a cohort of cardiac surgical patients with severe multi-vessel coronary artery disease and phenotypic extremes of heart failure [38]. DNA methylation patterns from peripheral blood allowed to distinguish patients with coronary artery disease with and without HF. The analysis of the methylation of CpG sites during the first diagnosis and follow-up enables the identification of new prognostic biomarkers and genes involved in the pathomechanism of CVDs. This approach was used in a paper from this year that shows significant upregulation of SOCS3, ITGAL, NFIC, NCOR2, and PGK1 mRNA (qRT-PCR) in peripheral blood mononuclear cells from pulmonary arterial hypertension (PAH) patients compared to healthy controls (p ≤ 0.05) [39]. This is an important study that shows new therapeutic and prognostic hopes in the care of patients with a poor prognosis. Further promising clinical implications, from two studies published in 2023, identify new hypermethylated or hypomethylated CpG and gene sites associated with anthracycline-induced cardiomyopathy [40,41]. Researchers indicate that the differential expression of epigenetic modifiers in early and late cardiotoxic heart failure reveals DNA methylation as a key regulator of cardiotoxicity during anthracycline treatment [41]. Since anthracycline-induced cardiomyopathy is still the leading cause of late morbidity in childhood cancer survivors [40], these epigenetic findings hold great potential for clinical applications. Further research in this area may contribute to reducing the cardiotoxicity of oncological treatment, which should become the subject of clinical trials. The variability of methyl tags of selected genes and miRNAs among HF patients is summarized in Table 1.

Table 1.
Methylation of selected protein-coding genes and miRNAs in HF patients. Tabulated data come from analyses that reached statistical significance of p < 0.05.
EWAS provides broad insights into the DNA methylation landscape of heart tissue among patients and confirms the role of DNA methylation in regulating genes involved in the progression of CVDs. The data collected with EWAS not only indicate new targets for potential epidrugs, but also provide the opportunity to conduct additional functional analysis of the role of identified methylation-sensitive genes in order to determine their exact mechanistic role in the development of CVDs. Further exploration of cell type-specific methylation patterns of the epigenetic machinery will provide valuable insights into the involvement of individual cells (cardiomyocytes, fibroblasts, endothelial cells, inflammatory cells) in the progression of CVDs that either drive or result from CVDs pathogenesis. This proposed analysis would indeed explain the differential cellular composition that can be seen in some CVD pathologies [42].
2. The Role of Epigenetics in Inflammation
Much evidence indicates that epigenetic mechanisms mediate the development of inflammation by modulating the expression of tumor necrosis factor alpha (TNF-α), interleukin 1 (IL-1), interleukin 2 (IL-2), interleukin 6 (IL-6), interleukin 8 (IL-8), interleukin 12 (IL-12), granulocyte-macrophage colony-stimulating factor (GM-CSF), tumor suppressor genes, oncogenes and autocrine and paracrine activation of the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) transcription factor [43,44,45,46,47,48,49,50]. It is believed that cardiac stress, e.g., during myocardial infarction (MI), leads to NF-κB-dependent stimulation of pro-inflammatory cytokine gene expression in resident heart cells [51,52,53]. The release of these mediators, as well as the molecular patterns associated with damage, induces the recruitment of inflammatory cells such as neutrophils and monocytes to the myocardium. This condition may have a beneficial dimension, e.g., by promoting the phagocytic removal of dead cells after an infarction [54,55]. On the other hand, persistent extravasation of inflammatory cells may lead to inflammation of the heart with its subsequent remodeling [56,57]. Inflammation-induced vascular change pathways result in both hypertrophic and hypertensive responses. Many of them are caused by the activation of the transcription factor NFκB, which ultimately leads to changes in gene transcription and the perpetuation of a hypertensive state [58,59].
Epigenetic regulation of both local and high-order chromatin structure is one of the mechanisms involved in the development of HF. Recently, early events of adaptive chromatin remodeling have been shown to precede pathological phenotypes and are reinforced in the failing heart [60]. Myocardial stress gene transcription responds through the dynamics of H3K27 acetylation enhancer enrichment and co-regulatory gene network [61]. Changes in gene expression in the course of HF correlated with differential enrichment of H3K27 histone acetylation in their respective proximal and distal interacting genomic enhancers encapsulated in these static chromatin structures [61]. Researchers indicate that a robust and intact CCCTC-binding factor loop is required to elicit a rapid and accurate stress response [61]. In order for transcription factors to activate their target genes, DNA and chromatin must be remodeled, mainly due to the aforementioned modifications within histones by histone acetyltransferase (HAT) and histone deacetylase (HDAC), which either open or close DNA strands to transcription factors. Normally, this balance is tightly controlled, but under conditions of stress and inflammation, activation of pro-inflammatory cytokines can result in increased HDAC activation and histone acetylation, which correlates with an increase in NFκB activity and a further increase in pro-inflammatory cytokine expression [62]. Due to the plasticity of epigenetic changes and their readiness to respond to the intervention of small molecule inhibitors, there is great potential for the development of new therapeutic agents that will serve as direct or complementary therapeutic compounds [63,64]. HDAC inhibitors have shown great potential in inhibiting the development of inflammatory diseases [65,66] and play a clear immunomodulatory role in the heart [67,68] reducing cardiac dysfunction associated with its aging [69]. In addition, HDAC inhibitors reduce myocardial fibrosis by multiple mechanisms including inhibition of proliferation and/or migration of cardiac fibroblasts and induction of genes that suppress extracellular matrix production by fibroblasts [65]. They also inhibit proinflammatory signals of fibrosis, differentiation of monocyte precursors into mature collagen-producing fibrocytes, and endothelial–mesenchymal transition (EndoMT), which defines the process of pathological dedifferentiation of vascular endothelial cells into matrix-producing mesenchymal cells [65].
Since inflammation has long been recognized as an important component of the pathogenesis of heart failure (HF) [58,59,70,71], the therapeutic efficacy of HDAC inhibitors has been investigated in animal models of HF. Numerous studies have shown that HDAC inhibitors are effective therapeutic agents that can halt and reverse the remodeling and dysfunction of the heart associated with the development of CVDs [72]. One study involved spontaneously hypertensive rats treated for 20 weeks with valproic acid (VPA), a weak HDAC inhibitor. Researchers observed reductions in the expression of IL-1β and TNF-α in the left ventricle, which showed a correlation with attenuated myocardial hypertrophy and fibrosis, and improved cardiac function [73]. Another pan-HDAC inhibitor, suberoylanilide hydroxamic acid (SAHA), approved by the Food and Drug Administration (FDA) for the treatment of cutaneous T-cell lymphoma, decreased plasma concentrations of more than 20 inflammatory cytokines including IL-1α, IL2, and TNF-α, inflammatory cell infiltration, interstitial collagen deposition, systolic hypertension and myocardial interstitial fibrosis in a model of hypertension-induced cardiac remodeling [74]. Another pan-HDAC inhibitor, trichostatin A (TSA), has shown the ability to inhibit NOS2 expression caused by various toll-like receptors (TLR) activators by lipopolysaccharides (LPS) or Escherichia coli, which are activators of TLR4 signaling, double-stranded RNA, which is an activator of TLR3 signaling, or peptidoglycan, which is an activator of TLR2 signaling. TSA led to increased acetylation of mitogen-activated protein kinase (MAPK) phosphatase-1, thereby promoting its ability to inhibit pro-inflammatory p38 kinase signaling. TSA also inhibited the expression of TNF-α, IL-6 and IL-1β in LPS-stimulated macrophages [75]. In addition, HDAC-mediated deacetylation of the regulatory factor interferon-7 has been shown to be required for effective DNA binding of this proinflammatory transcription factor [76]. Finally, the protein65 (p65) subunit of NF-κB can be deacetylated by HDAC3, promoting its nuclear shutdown by binding to nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (IκBα) [77].
Genome-wide association studies have revealed that genetic variants at the locus corresponding to HDAC-9 are associated with the risk of stroke, coronary artery disease and atherosclerosis due to varying levels of HDAC-9 expression [78,79,80]. HDAC-9 has been shown to play an essential role in cholesterol homeostasis and inflammation, as deletion of HDAC-9 in systemic cells and bone marrow resulted in upregulation of lipid homeostasis genes, downregulation of inflammatory genes and polarization of macrophages towards the M2 phenotype through increased accumulation total acetylated H3 and H3K9 on the ATP-binding cassette transporter (ABCA-1) and ATP-binding cassette subfamily G member 1 (ABCG-1) promoters in macrophages [80]. Researchers believe that targeted inhibition of HDAC9 may be a viable strategy to prevent atherosclerosis [81].
The presence of inflammatory activated macrophages in the aortic wall is a pathological feature of abdominal aortic aneurysm (AAA) development [82]. Thanks to the characterization of surgical samples and studies on animal models, the role of epigenetic modifications in monocytes and macrophages formed from them in the pathogenesis of aortic aneurysms has been demonstrated [7,83]. Global methylation levels in peripheral blood mononuclear cells are significantly altered in CpG islands in AAA patients compared to controls and positively correlated with increased aortic diameter [84]. Human AAA samples showed between five- and fifteen-fold higher levels of HDAC1, 2, 4, and 7 mRNA compared to healthy controls, while no differences were observed for HDAC3, 5, and 8 [85]. In mouse models of AAA, treatment with class I HDAC inhibitors (MS-275) or class IIa (MC-1568) reduced the incidence of AAA, the inflammatory phenotype of macrophages, and the concentration of pro-inflammatory mediators [85]. Another epigenetic therapeutic target for the treatment of AAA may be microRNA-33. MiR-33-deficient bone marrow transplantation reduced AAA formation in a mouse model by reducing inflammation by several mechanisms, including reducing matrix metalloproteinase-9 in macrophages and monocyte chemotactic protein-1 in vascular smooth muscle cells [86].
Other important molecules include the bromodomain and extra terminal (BET)-containing protein family. BET proteins act as an epigenetic reader that is a key positive regulator of NF-κB and transforming growth factor TGF-β dependent pro-inflammatory gene expression [87], acting as a critical co-activator in the process of cardiomyocyte hypertrophy [88]. Bromodomain-containing protein 4 (BRD4) binds acetyl-lysine via bromodomains and co-activates transcription, forming complexes that signal to RNA polymerase II and react directly with NF-κB through acetylated Lys310 of the p65 subunit. This interaction is required for NF-κB transactivation [89]. Treatment with the BET inhibitor thienodiazepine (JQ1) was shown to have a therapeutic effect during severe HF induced by prolonged pressure overload as well as massive anterior MI in a mouse model, and early intervention with JQ1 at the very onset of pressure overload in mice prevented the development of cardiac hypertrophy and left ventricular (LV) systolic dysfunction. JQ1 attenuated many of the hallmarks of HF progression in vivo, including cardiomegaly, pulmonary edema, LV systolic dysfunction, LV cavity dilation, LV wall thickening, and LV fibrosis [88]. Because BRD4is involved in the maintenance and progression of cancer cells in a wide range of malignancies [90]. BRD inhibitors, including JQ1 derivatives, are the subject of human therapy studies in early clinical phases [91,92]. Unlike several cancer drugs that cause cardiotoxicity, researchers indicate that BET bromodomain inhibitors, such as HDACs, may be a privileged class of cancer drugs that have cardioprotective properties due to the regulation of pro-inflammatory genes [88].
Another FDA-approved targeting molecule for BDR4 is apabetalone (APA)—it affects several characteristics of the atherosclerotic process, such as lipid metabolism, oxidative stress and vasculitis. Adding APA therapy to patients receiving high-dose statins led to an increase in high-density lipoprotein cholesterol (HDL-C) by 15.4% and a decrease in high-sensitivity C-reactive protein (hsCRP) by 18.4% after 24 weeks of treatment, which was a better result than the placebo group. Improvement in the lipid profile was associated with fewer cardiovascular events in patients treated with APA than with placebo [93]. In pooled phase 2 data analyses, APA reduced the rate of cardiovascular events by approximately 60%. However, in the BETOnMACE Phase III study, APA reduced the risk of major adverse cardiac events (MACE) by 50% in the chronic kidney disease subpopulation, indicating a beneficial effect along the kidney–heart axis [94,95]. Certainly, further clinical trials are needed to better investigate the safety and efficacy of APA in the treatment of CVDs. An overview of studies on the use of HDCAC inhibitors in CVDs is summarized in Table 2. An overview of clinical trials of both new epidrug molecules and already commonly used drugs due to their additional epigenetic mechanisms is summarized in Table 3.

Table 2.
An overview of studies on the use of HDCAC inhibitors in CVDs.

Table 3.
Clinical trials on the use of drugs in cardiovascular diseases.
Many drugs using epigenetic mechanisms of gene silencing have long been used in the treatment of humans, mainly for oncological causes. These include DNMT inhibitors, such as 5-azacytidine and 5-aza-2′-deoxycytidine, or HDAC inhibitors, such as vorinostat and romidepsin. Numerous more modern drugs from these groups (belinostat, panobinostat, etinostat, SAHA, givinostat, guadecitabine) are currently used as part of standard oncological treatment and clinical trials [109]. Non-oncology therapies include, but are not limited to, rheumatic diseases (systemic onset juvenile idiopathic arthritis—givinostat) or neurological diseases (amyotrophic lateral sclerosis—sodium phenylbutyrate–taurursodiol) [110,111,112]. Drugs commonly used in the treatment of CVDs, such as statins, sodium–glucose cotransporter-2 inhibitors (SGLT2i), hydralazine or metformin, in which the pleiotropic effect has been observed for a long time, have recently been identified as drugs showing an epigenetic mechanism [113]. Metformin, by activating AMPK, consequently leads to the modification of histones and promotes an increase or decrease in the expression of several genes. Metformin also indirectly increases HAT1 activity as observed in a mouse embryonic fibroblast model [114]. Metformin treatment also reduced the activity of two important co-activators of a large number of genes involved in inflammation and gluconeogenesis, HAT p300 and CREB-binding protein [114,115]. Both phenomena are strongly represented in patients with HF [113]. Moreover, long-term exposure to metformin was associated with a protective effect on the incidence of newly diagnosed symptomatic heart failure with preserved ejection fraction (HFpEF), diastolic dysfunction and LV hypertrophy in patients with type 2 diabetes and hypertension, which may be beneficial in delaying the progression of HFpEF [116]. Hydralazine, a vasodilator, exerts epigenetic effects by reducing the expression of DNMT1. In addition, this drug can modulate calcium homeostasis in cardiomyocytes by reducing methylation of the SERCA2a promoter, while enhancing the protein and activity of SERCA2a [117]. The epigenetic effects of this drug are also linked to specific gene expression by methylation of CpG islands in gene promoters [113]. Going forward, dapagliflozin (SGLT2i) modulates miRNAs involved in the pathophysiology of HF, such as miR199a-3p and miR30e-5p, which are involved in the regulation of PPARδ levels of mitochondrial fatty acid oxidation [113,118].
Statins are drugs that regulate epigenetic mechanisms at all levels—DNA methylation, histone modification and via non-coding RNAs [113]. They normalize subtelomeric DNA methylation in type 2 diabetes patients [119] and induce epigenetic changes by modulating Sirt1 transcription, resulting in regulation of inflammatory and apoptotic mechanisms. Statins increase miRNA-221/222 levels, the downregulation of which is associated with coronary artery disease [120] while upregulated miR-22 levels are associated with myocardial hypertrophy [121]. Statin therapy, by restoring KLF4-miR-483 expression and inhibiting EndoMT, may benefit patients with acute Kawasaki disease [122]. However, a pilot study showed that statins differentially modulate microRNA expression in the peripheral cells of patients with hyperlipidemia, which may be related to the variable response to these drugs [123].
3. DNA Methylation and Its Importance in the Regulation of the Cardiovascular System
The best-known epigenetic modification of DNA is 5-cytosine methylation, which is essential for proper gene expression, transposon silencing, alternative splicing and genome stability [124]. Cardiovascular risk factors such as smoking, low dietary folic acid, and elevated plasma homocysteine induce an abnormal DNA methylation pattern that may determine an individual’s predisposition to develop CVDs [8,125]. The methylation status of hundreds of CpG sites has been used as an epigenetic clock to measure the biological age that determines the rate of human aging [126,127]. In 832 participants without CVDs, researchers determined the difference between chronological and biological age using DNA methylation analysis and the Havroth formula. There were 153 cases of CVDs during the 10-year follow-up. For each year of increased biological age, the researchers observed a 4% increase in CVD risk. (HR = 1.033, 95% CI 1.004–1.063, p = 0.024) [128]. Since abnormal DNA methylation patterns occur before macrovascular damage [129], their identification may improve individual cardiovascular risk stratification. A 2023 screening study analyzed various methylated genes using a methylated DNA immunoprecipitation chip (MeDIP-chip). The study revealed that hypomethylation within the vascular endothelial growth factor B (VEGF-B), placental growth factor (PLGF), phospholipase C beta1 (PLCB1) and fatty acid transporter 4 (FATP4) genes could represent new biomarkers for CVDs. In addition, the VEGF-receptor signaling pathway regulated by DNA methylation may play a role in the pathogenesis of cardiovascular diseases in diabetes [130]. Moreover, global DNA hypomethylation is observed in atherosclerotic lesions both in humans and in animal models [131,132], whereas the promoter regions of atherosclerotic protective genes such as estrogen receptor β, ABCA1 and Krüppel-like factor 4 (KLF4) are often hypermethylated in atherosclerosis [133]. To date, the mechanisms underlying changes in DNA methylation in cardiovascular aging and CVDs remain under investigation. Several observations documented so far relate, for example, to the correlation between increased DNMT1 in macrophages and decreased PPAR-γ and increased pro-inflammatory cytokines in apolipoprotein E (ApoE)-deficient mice fed an atherogenic diet as well as in patients with atherosclerosis [134]. Another study describes hypomethylated CpG in the endothelial nitric oxide synthase (eNOS) promoter of eNOS-expressing endothelial cells, in stark contrast to dense DNA methylation in eNOS-negative cells (e.g., vascular smooth muscle cells) [135]. DNMT1 has been shown to be a critical regulator that negatively regulates arterial stiffening by maintaining the vascular contractility phenotype of smooth muscle cells [136], and mitochondrial DNA hypermethylation in vascular smooth muscle cells impairs cell contractility [137]. Methylome profiling allows a thorough insight into the pathomechanism of individual diseases, reports from 2023 reveal the key electrophysiology and immune dysregulation in hypertrophic cardiomyopathy, which brings new therapeutic possibilities [138]. DNA methylation also has an important aspect in radiation-induced cardiovascular disease (RICVD), which is an important problem in thoracic radiotherapy with complex pathophysiology. Current studies describe a systemic radiation fingerprint at the level of DNA methylation, explaining the possible relationship of DNA methylation to the pathophysiology of RICVD [139]. Figure 2 illustrates a cause-and-effect diagram between the drivers of proinflammatory epigenetic changes that contribute to the development of CVDs.
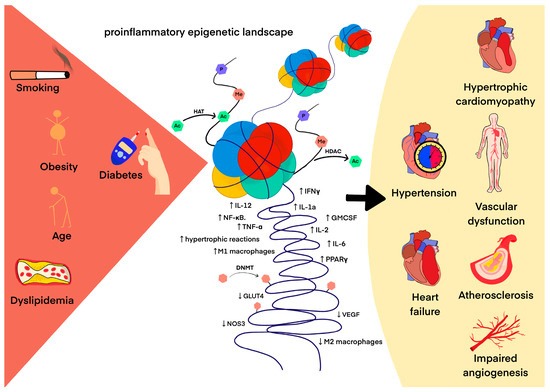
Figure 2.
The pro-inflammatory epigenetic landscape as a result of adverse environmental interactions in the development of CVDs. The arrow indicates cause and effect.
4. The Role of Tet Enzymes in Active DNA Demethylation and Its Relationship with the Differentiation and Function of Cardiovascular Cells
Members of the methylcytosine dioxygenase family, Tet enzymes regulate transcription by playing a major role in DNA demethylation by catalyzing the initial hydroxylation of 5-mC to 5-hmC. All three related human proteins, Tet1, Tet2 and Tet3, possess 5-mC oxidase activity but differ in domain architecture and tissue specificity of their expression levels [140]. The basic characteristics are summarized in Table 4.

Table 4.
The basic characteristics of Tet enzymes.
The 5-hmC and 5-mC profiles mapped in embryonic, neonatal, adult, and hypertrophic mouse and human cardiomyocytes indicate dynamic modulation of DNA methylation and hydroxymethylation during development and heart disease [141]. Deposition of 5-hmC on the gene body is strongly positively correlated with gene expression and identifies heart-specific genes. Disruption of these processes contributes to the phenotypic expression of CHDs such as tetralogy of Fallot (TOF) or ventricular septal defect (VSD) [142]. Genome-wide methylation analysis shows that Tet knockout (Tet-KD) causes hypermethylation of the promoter of genes encoding WNT inhibitors, leading to hyperactivated WNT signaling and defects in cardiac mesoderm patterning. Human embryonic stem cells in which all three Tet genes were inactivated had defective structural gene specifications for cardiac progenitor cells and formed into cardiomyocytes with altered mesodermal patterning. Among other factors, this was due to the inability to maintain the Tet-dependent hypomethylation state and the expression of the NKX2-5 gene [141]. In another study, Tet2/3 double-KD mice display an embryonic lethal ventricular non-compaction cardiomyopathy [143]. Recent studies of DNA methylation in biopsies of human heart tissue from patients with chronic heart failure (CHF) of various etiologies have shown clear epigenomic patterns in important DNA elements of the heart genome in end-stage HF [142,144]. Interestingly, DNA methylation patterns in CVDs share some common features with developmental changes. Methylation patterns of cardiomyocytes isolated from failing mouse hearts partly resemble those of newborn mice [145]. While DNA methylation in failing cardiomyocytes returned to the fetal methylation pattern, the observed changes were less pronounced during physiological myocyte aging. Research is currently lacking to determine whether human cardiomyocyte DNA methylome exhibits similar characteristics during development and disease. The 5-hmC landscape is also shifting towards a neonatal distribution pattern in pathologically hypertrophied hearts. The RNA-seq data revealed that all three Tet genes were highly expressed in the fetal cardiomyocyte genome and downregulated in adult cells, and undergoing some changes during hypertrophy induction [146]. The most strongly expressed member was Tet2, positively correlated with the expression of the absolute 5-hmC content. These findings led the researchers to hypothesize that Tet2 plays the largest role in the fetal cardiomyocyte genome. Researchers showed that Tet2 regulates the expression of key cardiac genes, such as the Myh7 fetal cardiac α-myosin heavy chain gene, by depositing 5-hmC on the gene body and within enhancers. Tet2-KD resulted in significant deregulation of a large number of genes including those related to the cell cycle, cardiac development and contraction of the heart muscle. The researchers emphasize that Myh7 and the myosin light chain gene Myl4—which encodes the basic structural elements of the embryonic heart muscle—were profoundly affected by Tet2-KD, going through six and three down adjustments, respectively. Although Tet2-KD significantly reduced the endogenous expression of Tet2 (Western blotting confirmed a 70% reduction of the Tet2 protein), the researchers found no effect of Tet2-KD on the global level of 5-hmC, which they explain by compensation by Tet3, whose level was significantly increased after Tet2-KD. This relationship emphasizes the high importance of the Tet family in the processes of differentiation of cells of the cardiovascular system. It has recently been discovered that dysmetabolic conditions such as diabetes mellitus can alter DNA demethylation in conjunction with extranuclear Tet relocation, which is dependent on AMPK activity [147]. It is currently unknown whether extranuclear Tet compartmentalization occurs in the heart of diabetic patients and whether this phenomenon plays a role in this context. In contrast, a study from 2022 with n = 150 patients with AMI [148] showed a significant increase in Tet-2 levels, positive correlation of Tet2 with infarct size, cTNT levels and Gensini score (all, p < 0.001). The researchers included the representation of people with CV-risk factors, including diabetes, in the control and study groups, noting the lowest Tet2 levels in the group with the lowest CV risk. In the same year, facilitated expression of Tet2 by c-MYC binding to the miR-29a-3p promoter was shown to have a therapeutic effect on ventricular remodeling in MI rats [149]. An increase in Tet2 improved cardiac function, reduced infarct size, myocardial apoptotic death, reduced oxidative stress, inflammatory response, and normalized collagen deposition. There are also preliminary reports on the use of Tet1 overexpression in controlling arrhythmias dependent on the secretory activity of cancer cells [150]. Researchers have linked gastrointestinal cancer secretions to DNA methylation of potassium voltage-gated channel subfamily D member 3 (KCND3) ion channel genes as a result of activation of transforming growth factor β/phosphoinositide 3-kinase (TGF-β/PI3K) signaling. Tet1 overexpression normalized the methylation state of the CpG islands of the promoters of the KCND3 channel genes in human cardiomyocytes, which protected the ion currents from the influence of tumor secretory activity [150]. Making changes in Tet expression certainly has a wide therapeutic potential and is currently an attractive subject of numerous scientific studies.
5. Conclusions
Epigenetics provides a thorough insight into the cardiovascular system, from its development to its continued functioning. The evidence cited suggests that epigenetics is a rapidly evolving science that may provide solutions to the continuing strong clinical impact of CVDs. This is possible by targeting pharmaceutical interventions to reprogram the pro-inflammatory epigenetic landscape associated with cardiometabolic and vascular diseases and to improve individual risk stratification.
Author Contributions
Conceptualization—A.W., Ł.W., M.K. and G.G.; writing—original draft preparation—A.J., A.W., Ł.W., M.K., J.O. and J.H.; writing—review and editing Ł.W. and G.G.; visualization A.J., A.W., M.K., J.O. and J.H; supervision M.K. and G.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Raghubeer, S. The influence of epigenetics and inflammation on cardiometabolic risks. Semin. Cell Dev. Biol. 2023. [Google Scholar] [CrossRef]
- Keshawarz, A.; Joehanes, R.; Ma, J.; Lee, G.Y.; Costeira, R.; Tsai, P.C.; Masachs, O.M.; Bell, J.T.; Wilson, R.; Thorand, B.; et al. Dietary and supplemental intake of vitamins C and E is associated with altered DNA methylation in an epigenome-wide association study meta-analysis. Epigenetics 2023, 18, 2211361. [Google Scholar] [CrossRef]
- Benak, D.; Kolar, F.; Zhang, L.; Devaux, Y.; Hlavackova, M. RNA modification m6Am: The role in cardiac biology. Epigenetics 2023, 18, 2218771. [Google Scholar] [CrossRef]
- Zhan, M.; Song, H.; Tian, D.; Wen, Q.; Shi, X.; Wang, Y.; Mao, X.; Wang, J. Molecular features, biological behaviors and clinical implications of m5C RNA methylation modification regulators in gastrointestinal cancers. Cancer Biol. Ther. 2023, 24, 2223382. [Google Scholar] [CrossRef]
- Becker, L.; Montes-Mojarro, I.A.; Layland, S.L.; Nsair, A.; Fend, F.; Marzi, J.; Schenke-Layland, K. Exploring the Relationship between Epigenetic DNA Methylation and Cardiac Fibrosis through Raman Microspectroscopy. Am. J. Physiol. Cell Physiol. 2023, 325, C332–C343. [Google Scholar] [CrossRef]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef]
- Davis, F.M.; Gallagher, K.A. Epigenetic Mechanisms in Monocytes/Macrophages Regulate Inflammation in Cardiometabolic and Vascular Disease. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 623–634. [Google Scholar] [CrossRef]
- Carreras-Gallo, N.; Dwaraka, V.B.; Cáceres, A.; Smith, R.; Mendez, T.L.; Went, H.; Gonzalez, J.R. Impact of tobacco, alcohol, and marijuana on genome-wide DNA methylation and its relationship with hypertension. Epigenetics 2023, 18, 2214392. [Google Scholar] [CrossRef]
- Stephenson, M.; Bollepalli, S.; Cazaly, E.; Salvatore, J.E.; Barr, P.; Rose, R.J.; Dick, D.; Kaprio, J.; Ollikainen, M. Associations of Alcohol Consumption with Epigenome-Wide DNA Methylation and Epigenetic Age Acceleration: Individual-Level and Co-twin Comparison Analyses. Alcohol. Clin. Exp. Res. 2021, 45, 318–328. [Google Scholar] [CrossRef]
- Dugué, P.A.; Wilson, R.; Lehne, B.; Jayasekara, H.; Wang, X.; Jung, C.H.; Joo, J.E.; Makalic, E.; Schmidt, D.F.; Baglietto, L.; et al. Alcohol consumption is associated with widespread changes in blood DNA methylation: Analysis of cross-sectional and longitudinal data. Addict. Biol. 2021, 26, e12855. [Google Scholar] [CrossRef]
- Lee, M.H.; Cho, E.R.; Lim, J.E.; Jee, S.H. Association between serum persistent organic pollutants and DNA methylation in Korean adults. Environ. Res. 2017, 158, 333–341. [Google Scholar] [CrossRef]
- Goodman, S.; Chappell, G.; Guyton, K.Z.; Pogribny, I.P.; Rusyn, I. Epigenetic alterations induced by genotoxic occupational and environmental human chemical carcinogens: An update of a systematic literature review. Mutat. Res. Rev. Mutat. Res. 2022, 789, 108408. [Google Scholar] [CrossRef]
- Lind, P.M.; Salihovic, S.; Lind, L. High plasma organochlorine pesticide levels are related to increased biological age as calculated by DNA methylation analysis. Environ. Int. 2018, 113, 109–113. [Google Scholar] [CrossRef]
- Xu, Y.; Lindh, C.H.; Fletcher, T.; Jakobsson, K.; Engström, K. Perfluoroalkyl substances influence DNA methylation in school-age children highly exposed through drinking water contaminated from firefighting foam: A cohort study in Ronneby, Sweden. Environ. Epigenet. 2022, 8, dvac004. [Google Scholar] [CrossRef]
- Si, J.; Chen, L.; Yu, C.; Guo, Y.; Sun, D.; Pang, Y.; Millwood, I.Y.; Walters, R.G.; Yang, L.; Chen, Y.; et al. Healthy lifestyle, DNA methylation age acceleration, and incident risk of coronary heart disease. Clin. Epigenetics 2023, 15, 52. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Zemach, A.; McDaniel, I.E.; Silva, P.; Zilberman, D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Kumar, S.; Chinnusamy, V.; Mohapatra, T. Epigenetics of Modified DNA Bases: 5-Methylcytosine and Beyond. Front. Genet. 2018, 9, 640. [Google Scholar] [CrossRef]
- Miranda, T.B.; Jones, P.A. DNA methylation: The nuts and bolts of repression. J. Cell Physiol. 2007, 213, 384–390. [Google Scholar] [CrossRef]
- Illingworth, R.S.; Gruenewald-Schneider, U.; Webb, S.; Kerr, A.R.; James, K.D.; Turner, D.J.; Smith, C.; Harrison, D.J.; Andrews, R.; Bird, A.P. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010, 6, e1001134. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Bestor, T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005, 74, 481–514. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhao, B.S.; He, C. TET family proteins: Oxidation activity, interacting molecules, and functions in diseases. Chem. Rev. 2015, 115, 2225–2239. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Pilotto, S.; Speranzini, V.; Tortorici, M.; Durand, D.; Fish, A.; Valente, S.; Forneris, F.; Mai, A.; Sixma, T.K.; Vachette, P.; et al. Interplay among nucleosomal DNA, histone tails, and corepressor CoREST underlies LSD1-mediated H3 demethylation. Proc. Natl. Acad. Sci. USA 2015, 112, 2752–2757. [Google Scholar] [CrossRef]
- Dimitrova, E.; Turberfield, A.H.; Klose, R.J. Histone demethylases in chromatin biology and beyond. EMBO Rep. 2015, 16, 1620–1639. [Google Scholar] [CrossRef] [PubMed]
- Assi, R.; Cherifi, C.; Cornelis, F.M.F.; Zhou, Q.; Storms, L.; Pazmino, S.; Coutinho de Almeida, R.; Meulenbelt, I.; Lories, R.J.; Monteagudo, S. Inhibition of KDM7A/B histone demethylases restores H3K79 methylation and protects against osteoarthritis. Ann. Rheum. Dis. 2023, 82, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ge, M.X.; Li, Y.H.; Li, J.L.; Yu, Q.; Xiao, F.H.; Ao, H.S.; Yang, L.Q.; Li, J.; He, Y.; et al. Longevity-Associated Transcription Factor ATF7 Promotes Healthspan by Suppressing Cellular Senescence and Systematic Inflammation. Aging Dis. 2023, 14, 1374–1389. [Google Scholar] [CrossRef]
- Morgan, M.A.J.; Shilatifard, A. Epigenetic moonlighting: Catalytic-independent functions of histone modifiers in regulating transcription. Sci. Adv. 2023, 9, eadg6593. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yao, H.; Lin, S.; Zhu, X.; Shen, Z.; Lu, G.; Poon, W.S.; Xie, D.; Lin, M.C.; Kung, H.F. Transcriptional and epigenetic regulation of human microRNAs. Cancer Lett. 2013, 331, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A.; Pasut, A.; Matsumoto, M.; Yamashita, R.; Fung, J.; Monteleone, E.; Saghatelian, A.; Nakayama, K.I.; Clohessy, J.G.; Pandolfi, P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017, 541, 228–232. [Google Scholar] [CrossRef]
- Deng, L.; Han, X.; Wang, Z.; Nie, X.; Bian, J. The Landscape of Noncoding RNA in Pulmonary Hypertension. Biomolecules 2022, 12, 796. [Google Scholar] [CrossRef]
- Sen, R.; Ghosal, S.; Das, S.; Balti, S.; Chakrabarti, J. Competing endogenous RNA: The key to posttranscriptional regulation. Sci. World J. 2014, 2014, 896206. [Google Scholar] [CrossRef]
- Levin, M.G.; Tsao, N.L.; Singhal, P.; Liu, C.; Vy, H.M.T.; Paranjpe, I.; Backman, J.D.; Bellomo, T.R.; Bone, W.P.; Biddinger, K.J.; et al. Genome-wide association and multi-trait analyses characterize the common genetic architecture of heart failure. Nat. Commun. 2022, 13, 6914. [Google Scholar] [CrossRef]
- Bain, C.R.; Ziemann, M.; Kaspi, A.; Khan, A.W.; Taylor, R.; Trahair, H.; Khurana, I.; Kaipananickal, H.; Wallace, S.; El-Osta, A.; et al. DNA methylation patterns from peripheral blood separate coronary artery disease patients with and without heart failure. ESC Heart Fail. 2020, 7, 2468–2478. [Google Scholar] [CrossRef]
- Benincasa, G.; Maron, B.A.; Affinito, O.; D’Alto, M.; Franzese, M.; Argiento, P.; Schiano, C.; Romeo, E.; Bontempo, P.; Golino, P.; et al. Association Between Circulating CD4+ T Cell Methylation Signatures of Network-Oriented SOCS3 Gene and Hemodynamics in Patients Suffering Pulmonary Arterial Hypertension. J. Cardiovasc. Transl. Res. 2023, 16, 17–30. [Google Scholar] [CrossRef]
- Singh, P.; Zhou, L.; Shah, D.A.; Cejas, R.B.; Crossman, D.K.; Jouni, M.; Magdy, T.; Wang, X.; Sharafeldin, N.; Hageman, L.; et al. Identification of novel hypermethylated or hypomethylated CpG sites and genes associated with anthracycline-induced cardiomyopathy. Sci. Rep. 2023, 13, 12683. [Google Scholar] [CrossRef]
- Robinson, E.L.; Ameri, P.; Delrue, L.; Vanderheyden, M.; Bartunek, J.; Altieri, P.; Heymans, S.; Heggermont, W.A. Differential expression of epigenetic modifiers in early and late cardiotoxic heart failure reveals DNA methylation as a key regulator of cardiotoxicity. Front. Cardiovasc. Med. 2023, 10, 884174. [Google Scholar] [CrossRef] [PubMed]
- Glezeva, N.; Moran, B.; Collier, P.; Moravec, C.S.; Phelan, D.; Donnellan, E.; Russell-Hallinan, A.; O’Connor, D.P.; Gallagher, W.M.; Gallagher, J.; et al. Targeted DNA Methylation Profiling of Human Cardiac Tissue Reveals Novel Epigenetic Traits and Gene Deregulation Across Different Heart Failure Patient Subtypes. Circ. Heart Fail. 2019, 12, e005765. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wu, S.; Guo, W.; Liang, S.; Li, X.; Yang, X. Epigenetic regulation of pro-inflammatory cytokine genes in lipopolysaccharide -stimulated peripheral blood mononuclear cells from broilers. Immunobiology 2017, 222, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Liu, Y.; Ren, X.; Gao, K.; Li, Y.; Li, S.; Yao, J.; Yang, X. Changes in DNA Methylation and Chromatin Structure of Pro-inflammatory Cytokines Stimulated by LPS in Broiler Peripheral Blood Mononuclear Cells. Poult. Sci. 2016, 95, 1636–1645. [Google Scholar] [CrossRef]
- Stylianou, E. Epigenetics of chronic inflammatory diseases. J. Inflamm. Res. 2018, 12, 1–14. [Google Scholar] [CrossRef]
- Smith, J.A.; Zhao, W.; Wang, X.; Ratliff, S.M.; Mukherjee, B.; Kardia, S.L.R.; Liu, Y.; Roux, A.V.D.; Needham, B.L. Neighborhood characteristics influence DNA methylation of genes involved in stress response and inflammation: The Multi-Ethnic Study of Atherosclerosis. Epigenetics 2017, 12, 662–673. [Google Scholar] [CrossRef]
- Perkins, D.J.; Patel, M.C.; Blanco, J.C.; Vogel, S.N. Epigenetic Mechanisms Governing Innate Inflammatory Responses. J. Interferon Cytokine Res. 2016, 36, 454–461. [Google Scholar] [CrossRef]
- Xu, S.; Kamato, D.; Little, P.J.; Nakagawa, S.; Pelisek, J.; Jin, Z.G. Targeting epigenetics and non-coding RNAs in atherosclerosis: From mechanisms to therapeutics. Pharmacol. Ther. 2019, 196, 15–43. [Google Scholar] [CrossRef]
- Mo, Z.; Li, Q.; Cai, L.; Zhan, M.; Xu, Q. The effect of DNA methylation on the miRNA expression pattern in lipopolysaccharide-induced inflammatory responses in human dental pulp cells. Mol. Immunol. 2019, 111, 11–18. [Google Scholar] [CrossRef]
- Tan, S.Y.X.; Zhang, J.; Tee, W.W. Epigenetic Regulation of Inflammatory Signaling and Inflammation-Induced Cancer. Front. Cell Dev. Biol. 2022, 10, 931493. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. The immune system and cardiac repair. Pharmacol. Res. 2008, 58, 88–111. [Google Scholar] [CrossRef] [PubMed]
- Lakshminarayanan, V.; Lewallen, M.; Frangogiannis, N.G.; Evans, A.J.; Wedin, K.E.; Michael, L.H.; Entman, M.L. Reactive oxygen intermediates induce monocyte chemotactic protein-1 in vascular endothelium after brief ischemia. Am. J. Pathol. 2001, 159, 1301–1311. [Google Scholar] [CrossRef]
- Lu, L.; Chen, S.S.; Zhang, J.Q.; Ramires, F.J.; Sun, Y. Activation of nuclear factor-kappaB and its proinflammatory mediator cascade in the infarcted rat heart. Biochem. Biophys. Res. Commun. 2004, 321, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G.; Smith, C.W.; Entman, M.L. The inflammatory response in myocardial infarction. Cardiovasc. Res. 2002, 53, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Nahrendorf, M.; Swirski, F.K.; Aikawa, E.; Stangenberg, L.; Wurdinger, T.; Figueiredo, J.L.; Libby, P.; Weissleder, R.; Pittet, M.J. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J. Exp. Med. 2007, 204, 3037–3047. [Google Scholar] [CrossRef]
- Mann, D.L. Innate immunity and the failing heart: The cytokine hypothesis revisited. Circ. Res. 2015, 116, 1254–1268. [Google Scholar] [CrossRef]
- Hartupee, J.; Mann, D.L. Role of inflammatory cells in fibroblast activation. J. Mol. Cell Cardiol. 2016, 93, 143–148. [Google Scholar] [CrossRef]
- Zhang, Y.; Bauersachs, J.; Langer, H.F. Immune mechanisms in heart failure. Eur. J. Heart Fail. 2017, 19, 1379–1389. [Google Scholar] [CrossRef]
- Adamo, L.; Rocha-Resende, C.; Prabhu, S.D.; Mann, D.L. Reappraising the role of inflammation in heart failure. Nat. Rev. Cardiol. 2020, 17, 269–285. [Google Scholar] [CrossRef]
- Chapski, D.J.; Cabaj, M.; Morselli, M.; Mason, R.J.; Soehalim, E.; Ren, S.; Pellegrini, M.; Wang, Y.; Vondriska, T.M.; Rosa-Garrido, M. Early adaptive chromatin remodeling events precede pathologic phenotypes and are reinforced in the failing heart. J. Mol. Cell Cardiol. 2021, 160, 73–86. [Google Scholar] [CrossRef]
- Lee, D.P.; Tan, W.L.W.; Anene-Nzelu, C.G.; Lee, C.J.M.; Li, P.Y.; Luu, T.D.A.; Chan, C.X.; Tiang, Z.; Ng, S.L.; Huang, X.; et al. Robust CTCF-Based Chromatin Architecture Underpins Epigenetic Changes in the Heart Failure Stress-Gene Response. Circulation 2019, 139, 1937–1956. [Google Scholar] [CrossRef] [PubMed]
- Keslacy, S.; Tliba, O.; Baidouri, H.; Amrani, Y. Inhibition of tumor necrosis factor-alpha-inducible inflammatory genes by interferon-gamma is associated with altered nuclear factor-kappaB transactivation and enhanced histone deacetylase activity. Mol. Pharmacol. 2007, 71, 609–618. [Google Scholar] [CrossRef]
- Napoli, C.; Bontempo, P.; Palmieri, V.; Coscioni, E.; Maiello, C.; Donatelli, F.; Benincasa, G. Epigenetic Therapies for Heart Failure: Current Insights and Future Potential. Vasc Health Risk Manag. 2021, 17, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Wołowiec, Ł.; Mędlewska, M.; Osiak, J.; Wołowiec, A.; Grześk, E.; Jaśniak, A.; Grześk, G. MicroRNA and lncRNA as the Future of Pulmonary Arterial Hypertension Treatment. Int. J. Mol. Sci. 2023, 24, 9735. [Google Scholar] [CrossRef] [PubMed]
- Schuetze, K.B.; McKinsey, T.A.; Long, C.S. Targeting cardiac fibroblasts to treat fibrosis of the heart: Focus on HDACs. J. Mol. Cell Cardiol. 2014, 70, 100–107. [Google Scholar] [CrossRef]
- Habibian, J.; Ferguson, B.S. The Crosstalk between Acetylation and Phosphorylation: Emerging New Roles for HDAC Inhibitors in the Heart. Int. J. Mol. Sci. 2018, 20, 102. [Google Scholar] [CrossRef]
- McKinsey, T.A. Targeting inflammation in heart failure with histone deacetylase inhibitors. Mol. Med. 2011, 17, 434–441. [Google Scholar] [CrossRef]
- Wright, L.H.; Menick, D.R. A class of their own: Exploring the nondeacetylase roles of class IIa HDACs in cardiovascular disease. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, H199–H206. [Google Scholar] [CrossRef]
- Ferguson, B.S.; McKinsey, T.A. Non-sirtuin histone deacetylases in the control of cardiac aging. J. Mol. Cell Cardiol. 2015, 83, 14–20. [Google Scholar] [CrossRef]
- Briasoulis, A.; Androulakis, E.; Christophides, T.; Tousoulis, D. The role of inflammation and cell death in the pathogenesis, progression and treatment of heart failure. Heart Fail. Rev. 2016, 21, 169–176. [Google Scholar] [CrossRef]
- Dutka, M.; Bobiński, R.; Ulman-Włodarz, I.; Hajduga, M.; Bujok, J.; Pająk, C.; Ćwiertnia, M. Various aspects of inflammation in heart failure. Heart Fail. Rev. 2020, 25, 537–548. [Google Scholar] [CrossRef]
- Jin, G.; Wang, K.; Zhao, Y.; Yuan, S.; He, Z.; Zhang, J. Targeting histone deacetylases for heart diseases. Bioorg Chem. 2023, 138, 106601. [Google Scholar] [CrossRef]
- Cardinale, J.P.; Sriramula, S.; Pariaut, R.; Guggilam, A.; Mariappan, N.; Elks, C.M.; Francis, J. HDAC inhibition attenuates inflammatory, hypertrophic, and hypertensive responses in spontaneously hypertensive rats. Hypertension 2010, 56, 437–444. [Google Scholar] [CrossRef]
- Iyer, A.; Fenning, A.; Lim, J.; Le, G.T.; Reid, R.C.; Halili, M.A.; Fairlie, D.P.; Brown, L. Antifibrotic activity of an inhibitor of histone deacetylases in DOCA-salt hypertensive rats. Br. J. Pharmacol. 2010, 159, 1408–1417. [Google Scholar] [CrossRef]
- Cao, W.; Bao, C.; Padalko, E.; Lowenstein, C.J. Acetylation of mitogen-activated protein kinase phosphatase-1 inhibits Toll-like receptor signaling. J. Exp. Med. 2008, 205, 1491–1503. [Google Scholar] [CrossRef] [PubMed]
- Caillaud, A.; Prakash, A.; Smith, E.; Masumi, A.; Hovanessian, A.G.; Levy, D.E.; Marié, I. Acetylation of interferon regulatory factor-7 by p300/CREB-binding protein (CBP)-associated factor (PCAF) impairs its DNA binding. J. Biol. Chem. 2002, 277, 49417–49421. [Google Scholar] [CrossRef] [PubMed]
- Giridharan, S.; Srinivasan, M. Mechanisms of NF-κB p65 and strategies for therapeutic manipulation. J. Inflamm. Res. 2018, 11, 407–419. [Google Scholar] [CrossRef]
- Dichgans, M.; Malik, R.; König, I.R.; Rosand, J.; Clarke, R.; Gretarsdottir, S.; Thorleifsson, G.; Mitchell, B.D.; Assimes, T.L.; Levi, C.; et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: A genome-wide analysis of common variants. Stroke 2014, 45, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Malik, R.; Chauhan, G.; Traylor, M.; Sargurupremraj, M.; Okada, Y.; Mishra, A.; Rutten-Jacobs, L.; Giese, A.K.; van der Laan, S.W.; Gretarsdottir, S.; et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018, 50, 524–537. [Google Scholar] [CrossRef]
- Cao, Q.; Rong, S.; Repa, J.J.; St Clair, R.; Parks, J.S.; Mishra, N. Histone deacetylase 9 represses cholesterol efflux and alternatively activated macrophages in atherosclerosis development. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1871–1879. [Google Scholar] [CrossRef]
- Azghandi, S.; Prell, C.; van der Laan, S.W.; Schneider, M.; Malik, R.; Berer, K.; Gerdes, N.; Pasterkamp, G.; Weber, C.; Haffner, C.; et al. Deficiency of the stroke relevant HDAC9 gene attenuates atherosclerosis in accord with allele-specific effects at 7p21.1. Stroke 2015, 46, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Körfer, D.; Erhart, P.; Dihlmann, S.; Hakimi, M.; Böckler, D.; Peters, A.S. Histopathological Characterization of Abdominal Aortic Aneurysms from Patients with Multiple Aneurysms Compared to Patients with a Single Abdominal Aortic Aneurysm. Biomedicines 2023, 11, 1311. [Google Scholar] [CrossRef] [PubMed]
- Mangum, K.; Gallagher, K.; Davis, F.M. The Role of Epigenetic Modifications in Abdominal Aortic Aneurysm Pathogenesis. Biomolecules 2022, 12, 172. [Google Scholar] [CrossRef] [PubMed]
- Toghill, B.J.; Saratzis, A.; Freeman, P.J.; Sylvius, N.; UKAGS Collaborators; Bown, M.J. SMYD2 promoter DNA methylation is associated with abdominal aortic aneurysm (AAA) and SMYD2 expression in vascular smooth muscle cells. Clin. Epigenetics 2018, 10, 29. [Google Scholar] [CrossRef]
- Galán, M.; Varona, S.; Orriols, M.; Rodríguez, J.A.; Aguiló, S.; Dilmé, J.; Camacho, M.; Martínez-González, J.; Rodriguez, C. Induction of histone deacetylases (HDACs) in human abdominal aortic aneurysm: Therapeutic potential of HDAC inhibitors. Dis. Model. Mech. 2016, 9, 541–552. [Google Scholar] [CrossRef]
- Nakao, T.; Horie, T.; Baba, O.; Nishiga, M.; Nishino, T.; Izuhara, M.; Kuwabara, Y.; Nishi, H.; Usami, S.; Nakazeki, F.; et al. Genetic Ablation of MicroRNA-33 Attenuates Inflammation and Abdominal Aortic Aneurysm Formation via Several Anti-Inflammatory Pathways. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 2161–2170. [Google Scholar] [CrossRef]
- Yatchang, M.F.; Mathew, B.; Srivastava, R.K.; Khan, J.; Muzaffar, S.; Zhang, S.; Wu, M.; Zhai, L.; Ruiz, P.; Agarwal, A.; et al. Development of BRD4 inhibitors as anti-inflammatory agents and antidotes for arsenicals. Bioorg Med. Chem. Lett. 2022, 64, 128696. [Google Scholar] [CrossRef]
- Duan, Q.; McMahon, S.; Anand, P.; Shah, H.; Thomas, S.; Salunga, H.T.; Huang, Y.; Zhang, R.; Sahadevan, A.; Lemieux, M.E.; et al. BET bromodomain inhibition suppresses innate inflammatory and profibrotic transcriptional networks in heart failure. Sci. Transl. Med. 2017, 9, eaah5084. [Google Scholar] [CrossRef]
- Huang, B.; Yang, X.D.; Zhou, M.M.; Ozato, K.; Chen, L.F. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol. Cell Biol. 2009, 29, 1375–1387. [Google Scholar] [CrossRef]
- Shu, S.; Lin, C.Y.; He, H.H.; Witwicki, R.M.; Tabassum, D.P.; Roberts, J.M.; Janiszewska, M.; Huh, S.J.; Liang, Y.; Ryan, J.; et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 2016, 529, 413–417. [Google Scholar] [CrossRef]
- Von Schaper, E. Roche bets on bromodomains. Nat. Biotechnol. 2016, 34, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Berthon, C.; Raffoux, E.; Thomas, X.; Vey, N.; Gomez-Roca, C.; Yee, K.; Taussig, D.C.; Rezai, K.; Roumier, C.; Herait, P.; et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: A dose-escalation, phase 1 study. Lancet Haematol. 2016, 3, e186–e195. [Google Scholar] [CrossRef]
- Nicholls, S.J.; Schwartz, G.G.; Buhr, K.A.; Ginsberg, H.N.; Johansson, J.O.; Kalantar-Zadeh, K.; Kulikowski, E.; Toth, P.P.; Wong, N.; Sweeney, M. Apabetalone and hospitalization for heart failure in patients following an acute coronary syndrome: A prespecified analysis of the BETonMACE study. Cardiovasc. Diabetol. 2021, 20, 13. [Google Scholar] [CrossRef] [PubMed]
- Gilham, D.; Wasiak, S.; Rakai, B.D.; Fu, L.; Tsujikawa, L.M.; Sarsons, C.D.; Carestia, A.; Lebioda, K.; Johansson, J.O.; Sweeney, M.; et al. Apabetalone Downregulates Fibrotic, Inflammatory and Calcific Processes in Renal Mesangial Cells and Patients with Renal Impairment. Biomedicines 2023, 11, 1663. [Google Scholar] [CrossRef]
- Ray, K.K.; Nicholls, S.J.; Buhr, K.A.; Ginsberg, H.N.; Johansson, J.O.; Kalantar-Zadeh, K.; Kulikowski, E.; Toth, P.P.; Wong, N.; Sweeney, M.; et al. Effect of Apabetalone Added to Standard Therapy on Major Adverse Cardiovascular Events in Patients with Recent Acute Coronary Syndrome and Type 2 Diabetes: A Randomized Clinical Trial. JAMA 2020, 323, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Tong, Q.; Zhang, Z.; Wang, S.; Zheng, Y.; Liu, Q.; Qian, L.B.; Chen, S.Y.; Sun, J.; Cai, L. Inhibition of HDAC3 prevents diabetic cardiomyopathy in OVE26 mice via epigenetic regulation of DUSP5-ERK1/2 pathway. Clin. Sci. 2017, 131, 1841–1857. [Google Scholar] [CrossRef] [PubMed]
- Seok, Y.M.; Lee, H.A.; Park, K.M.; Hwangbo, M.H.; Kim, I.K. Lysine deacetylase inhibition attenuates hypertension and is accompanied by acetylation of mineralocorticoid receptor instead of histone acetylation in spontaneously hypertensive rats. Naunyn Schmiedebergs Arch. Pharmacol. 2016, 389, 799–808. [Google Scholar] [CrossRef]
- Xie, M.; Kong, Y.; Tan, W.; May, H.; Battiprolu, P.K.; Pedrozo, Z.; Wang, Z.V.; Morales, C.; Luo, X.; Cho, G.; et al. deacetylase inhibition blunts ischemia/reperfusion injury by inducing cardiomyocyte autophagy. Circulation 2014, 129, 1139–1151. [Google Scholar] [CrossRef]
- Demos-Davies, K.M.; Ferguson, B.S.; Cavasin, M.A.; Mahaffey, J.H.; Williams, S.M.; Spiltoir, J.I.; Schuetze, K.B.; Horn, T.R.; Chen, B.; Ferrara, C.; et al. HDAC6 contributes to pathological responses of heart and skeletal muscle to chronic angiotensin-II signaling. Am. J. Physiol. Heart Circ. Physiol. 2014, 307, H252–H258. [Google Scholar] [CrossRef]
- Jeong, M.Y.; Lin, Y.H.; Wennersten, S.A.; Demos-Davies, K.M.; Cavasin, M.A.; Mahaffey, J.H.; Monzani, V.; Saripalli, C.; Mascagni, P.; Reece, T.B.; et al. Histone deacetylase activity governs diastolic dysfunction through a nongenomic mechanism. Sci. Transl. Med. 2018, 10, eaao0144. [Google Scholar] [CrossRef]
- Raal, F.J.; Kallend, D.; Ray, K.K.; Turner, T.; Koenig, W.; Wright, R.S.; Wijngaard, P.L.J.; Curcio, D.; Jaros, M.J.; Leiter, L.A.; et al. Inclisiran for the Treatment of Heterozygous Familial Hypercholesterolemia. N. Engl. J. Med. 2020, 382, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
- Täubel, J.; Hauke, W.; Rump, S.; Viereck, J.; Batkai, S.; Poetzsch, J.; Rode, L.; Weigt, H.; Genschel, C.; Lorch, U.; et al. Novel antisense therapy targeting microRNA-132 in patients with heart failure: Results of a first-in-human Phase 1b randomized, double-blind, placebo-controlled study. Eur. Heart J. 2021, 42, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, S.J.; Gordon, A.; Johannson, J.; Ballantyne, C.M.; Barter, P.J.; Brewer, H.B.; Kastelein, J.J.; Wong, N.C.; Borgman, M.R.; Nissen, S.E. ApoA-I induction as a potential cardioprotective strategy: Rationale for the SUSTAIN and ASSURE studies. Cardiovasc Drugs Ther. 2012, 26, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Zamani, P.; Akers, S.; Soto-Calderon, H.; Beraun, M.; Koppula, M.R.; Varakantam, S.; Rawat, D.; Shiva-Kumar, P.; Haines, P.G.; Chittams, J.; et al. Isosorbide Dinitrate, With or Without Hydralazine, Does Not Reduce Wave Reflections, Left Ventricular Hypertrophy, or Myocardial Fibrosis in Patients With Heart Failure With Preserved Ejection Fraction. J. Am. Heart Assoc. 2017, 6, e004262. [Google Scholar] [CrossRef]
- Mulkareddy, V.; Simon, M.A. Metformin in Pulmonary Hypertension in Left Heart Disease. Front. Med. 2020, 19, 425. [Google Scholar] [CrossRef]
- Peikert, A.; Martinez, F.A.; Vaduganathan, M.; Claggett, B.L.; Kulac, I.J.; Desai, A.S.; Jhund, P.S.; de Boer, R.A.; DeMets, D.; Hernandez, A.F.; et al. Efficacy and Safety of Dapagliflozin in Heart Failure With Mildly Reduced or Preserved Ejection Fraction According to Age: The DELIVER Trial. Circ. Heart Fail. 2022, 15, e010080. [Google Scholar] [CrossRef]
- Marfella, R.; D’Onofrio, N.; Trotta, M.C.; Sardu, C.; Scisciola, L.; Amarelli, C.; Balestrieri, M.L.; Grimaldi, V.; Mansueto, G.; Esposito, S.; et al. Sodium/glucose cotransporter 2 (SGLT2) inhibitors improve cardiac function by reducing JunD expression in human diabetic hearts. Metabolism 2022, 127, 154936. [Google Scholar] [CrossRef]
- Kjekshus, J.; Apetrei, E.; Barrios, V.; Böhm, M.; Cleland, J.G.; Cornel, J.H.; Dunselman, P.; Fonseca, C.; Goudev, A.; Grande, P.; et al. Rosuvastatin in older patients with systolic heart failure. N. Engl. J. Med. 2007, 357, 2248–2261. [Google Scholar] [CrossRef]
- Yoon, S.; Eom, G.H. HDAC and HDAC Inhibitor: From Cancer to Cardiovascular Diseases. Chonnam Med. J. 2016, 52, 1–11. [Google Scholar] [CrossRef]
- Sahafnejad, Z.; Ramazi, S.; Allahverdi, A. An Update of Epigenetic Drugs for the Treatment of Cancers and Brain Diseases: A Comprehensive Review. Genes. 2023, 14, 873. [Google Scholar] [CrossRef]
- Paganoni, S.; Macklin, E.A.; Hendrix, S.; Berry, J.D.; Elliott, M.A.; Maiser, S.; Karam, C.; Caress, J.B.; Owegi, M.A.; Quick, A.; et al. Trial of Sodium Phenylbutyrate-Taurursodiol for Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2020, 383, 919–930. [Google Scholar] [CrossRef]
- Vojinovic, J.; Damjanov, N. HDAC inhibition in rheumatoid arthritis and juvenile idiopathic arthritis. Mol. Med. 2011, 17, 397–403. [Google Scholar] [CrossRef]
- Gorica, E.; Mohammed, S.A.; Ambrosini, S.; Calderone, V.; Costantino, S.; Paneni, F. Epi-Drugs in Heart Failure. Front. Cardiovasc. Med. 2022, 9, 923014. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiu, J.; Wang, X.; Zhang, Y.; Xia, M. AMP-activated protein kinase suppresses endothelial cell inflammation through phosphorylation of transcriptional coactivator p300. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2897–2908. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Sabet, A.; Djedjos, S.; Miller, R.; Sun, X.; Hussain, M.A.; Radovick, S.; Wondisford, F.E. Metformin and insulin suppress hepatic gluconeogenesis through phosphorylation of CREB binding protein. Cell 2009, 137, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Yin, Z.F.; Zhang, J.F.; Wang, C.Q. Association between long-term prescription of metformin and the progression of heart failure with preserved ejection fraction in patients with type 2 diabetes mellitus and hypertension. Int. J. Cardiol. 2020, 306, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.H.; Cheng, C.C.; Chen, Y.C.; Chung, C.C.; Lee, T.I.; Chen, S.A.; Chen, Y.J. Hydralazine-induced promoter demethylation enhances sarcoplasmic reticulum Ca2+ -ATPase and calcium homeostasis in cardiac myocytes. Lab Invest. 2011, 91, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- el Azzouzi, H.; Leptidis, S.; Dirkx, E.; Hoeks, J.; van Bree, B.; Brand, K.; McClellan, E.A.; Poels, E.; Sluimer, J.C.; van den Hoogenhof, M.M.; et al. The hypoxia-inducible microRNA cluster miR-199a∼214 targets myocardial PPARδ and impairs mitochondrial fatty acid oxidation. Cell Metab. 2013, 18, 341–354. [Google Scholar] [CrossRef]
- Makino, N.; Maeda, T.; Abe, N. Short telomere subtelomeric hypomethylation is associated with telomere attrition in elderly diabetic patients 1. Can. J. Physiol. Pharmacol. 2019, 97, 335–339. [Google Scholar] [CrossRef]
- Minami, Y.; Satoh, M.; Maesawa, C.; Takahashi, Y.; Tabuchi, T.; Itoh, T.; Nakamura, M. Effect of atorvastatin on microRNA 221/222 expression in endothelial progenitor cells obtained from patients with coronary artery disease. Eur. J. Clin. Investig. 2009, 39, 359–367. [Google Scholar] [CrossRef]
- Tu, Y.; Wan, L.; Bu, L.; Zhao, D.; Dong, D.; Huang, T.; Cheng, Z.; Shen, B. MicroRNA-22 downregulation by atorvastatin in a mouse model of cardiac hypertrophy: A new mechanism for antihypertrophic intervention. Cell Physiol. Biochem. 2013, 31, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Chen, Z.; Martin, M.; Zhang, J.; Sangwung, P.; Woo, B.; Tremoulet, A.H.; Shimizu, C.; Jain, M.K.; Burns, J.C.; et al. miR-483 Targeting of CTGF Suppresses Endothelial-to-Mesenchymal Transition: Therapeutic Implications in Kawasaki Disease. Circ. Res. 2017, 120, 354–365. [Google Scholar] [CrossRef]
- Zambrano, T.; Hirata, R.D.C.; Hirata, M.H.; Cerda, Á.; Salazar, L.A. Statins differentially modulate microRNAs expression in peripheral cells of hyperlipidemic subjects: A pilot study. Eur. J. Pharm. Sci. 2018, 117, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Bogan, S.N.; Strader, M.E.; Hofmann, G.E. Associations between DNA methylation and gene regulation depend on chromatin accessibility during transgenerational plasticity. BMC Biol. 2023, 21, 149. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.C.E.; Mens, M.M.J.; Kühnel, B.; van Meurs, J.B.J.; Uitterlinden, A.G.; Peters, A.; Prokisch, H.; Herder, C.; Grallert, H.; Kunze, S.; et al. Smoking-related changes in DNA methylation and gene expression are associated with cardio-metabolic traits. Clin. Epigenetics 2020, 12, 157. [Google Scholar] [CrossRef]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115, Erratum in Genome Biol. 2015, 16, 96. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sanlés, A.; Sayols-Baixeras, S.; Curcio, S.; Subirana, I.; Marrugat, J.; Elosua, R. DNA Methylation and Age-Independent Cardiovascular Risk, an Epigenome-Wide Approach: The REGICOR Study (REgistre GIroní del COR). Arterioscler. Thromb. Vasc. Biol. 2018, 38, 645–652. [Google Scholar] [CrossRef]
- Lind, L.; Ingelsson, E.; Sundström, J.; Siegbahn, A.; Lampa, E. Methylation-based estimated biological age and cardiovascular disease. Eur. J. Clin. Investig. 2018, 48, 12872. [Google Scholar] [CrossRef]
- Lund, G.; Andersson, L.; Lauria, M.; Lindholm, M.; Fraga, M.F.; Villar-Garea, A.; Ballestar, E.; Esteller, M.; Zaina, S. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J. Biol. Chem. 2004, 279, 29147–29154. [Google Scholar] [CrossRef]
- Hu, S.; Chen, L.; Zeng, T.; Wang, W.; Yan, Y.; Qiu, K.; Xie, Y.; Liao, Y. DNA methylation profiling reveals novel pathway implicated in cardiovascular diseases of diabetes. Front. Endocrinol. 2023, 14, 1108126. [Google Scholar] [CrossRef]
- Zhang, W.; Song, M.; Qu, J.; Liu, G.H. Epigenetic Modifications in Cardiovascular Aging and Diseases. Circ. Res. 2018, 123, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.P.; Mishra, B.H.; Raitoharju, E.; Mononen, N.; Viikari, J.; Juonala, M.; Hutri-Kähönen, N.; Kähönen, M.; Raitakari, O.T.; Lehtimäki, T. Gene Set Based Integrated Methylome and Transcriptome Analysis Reveals Potential Molecular Mechanisms Linking Cigarette Smoking and Related Diseases. OMICS 2023, 27, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, Y.S.; Wang, K.C.; Chien, S. Epigenetic Mechanism in Regulation of Endothelial Function by Disturbed Flow: Induction of DNA Hypermethylation by DNMT1. Cell Mol. Bioeng. 2014, 7, 218–224. [Google Scholar] [CrossRef]
- Yu, J.; Qiu, Y.; Yang, J.; Bian, S.; Chen, G.; Deng, M.; Kang, H.; Huang, L. DNMT1-PPARγ pathway in macrophages regulates chronic inflammation and atherosclerosis development in mice. Sci. Rep. 2016, 6, 30053. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.; Fish, J.E.; D’Abreo, C.; Lin, S.; Robb, G.B.; Teichert, A.M.; Karantzoulis-Fegaras, F.; Keightley, A.; Steer, B.M.; Marsden, P.A. The cell-specific expression of endothelial nitric-oxide synthase: A role for DNA methylation. J. Biol. Chem. 2004, 279, 35087–35100. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.A.; Zhang, T.; Wang, J.; Zhao, F.; Zhang, Y.P.; Yao, W.J.; Hur, S.S.; Yeh, Y.T.; Pang, W.; Zheng, L.S.; et al. Matrix stiffness determines the phenotype of vascular smooth muscle cell in vitro and in vivo: Role of DNA methyltransferase 1. Biomaterials 2018, 155, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.F.; Zhu, J.J.; Yu Tian, X.; Liu, H.; Zhang, T.; Zhang, Y.P.; Xie, S.A.; Zheng, M.; Kong, W.; Yao, W.J.; et al. Hypermethylation of mitochondrial DNA in vascular smooth muscle cells impairs cell contractility. Cell Death Dis. 2020, 11, 35. [Google Scholar] [CrossRef]
- Li, X.; Fan, H.; Song, X.; Song, B.; Liu, W.; Dong, R.; Zhang, H.; Guo, S.; Liang, H.; Schrodi, S.J.; et al. DNA methylome and transcriptome profiling reveal key electrophysiology and immune dysregulation in hypertrophic cardiomyopathy. Epigenetics 2023, 18, 2195307. [Google Scholar] [CrossRef]
- Sallam, M.; Mysara, M.; Benotmane, M.A.; Crijns, A.P.G.; Spoor, D.; Van Nieuwerburgh, F.; Deforce, D.; Baatout, S.; Guns, P.J.; Aerts, A.; et al. DNA Methylation Alterations in Fractionally Irradiated Rats and Breast Cancer Patients Receiving Radiotherapy. Int. J. Mol. Sci. 2022, 23, 16214. [Google Scholar] [CrossRef]
- Jin, S.G.; Zhang, Z.M.; Dunwell, T.L.; Harter, M.R.; Wu, X.; Johnson, J.; Li, Z.; Liu, J.; Szabó, P.E.; Lu, Q.; et al. Tet3 Reads 5-Carboxylcytosine through Its CXXC Domain and Is a Potential Guardian against Neurodegeneration. Cell Rep. 2016, 14, 493–505. [Google Scholar] [CrossRef]
- Lan, Y.; Banks, K.M.; Pan, H.; Verma, N.; Dixon, G.R.; Zhou, T.; Ding, B.; Elemento, O.; Chen, S.; Huangfu, D.; et al. Stage-specific regulation of DNA methylation by TET enzymes during human cardiac differentiation. Cell Rep. 2021, 37, 110095. [Google Scholar] [CrossRef]
- Grunert, M.; Dorn, C.; Cui, H.; Dunkel, I.; Schulz, K.; Schoenhals, S.; Sun, W.; Berger, F.; Chen, W.; Sperling, S.R. Comparative DNA methylation and gene expression analysis identifies novel genes for structural congenital heart diseases. Cardiovasc Res. 2016, 112, 464–477. [Google Scholar] [CrossRef]
- Fang, S.; Li, J.; Xiao, Y.; Lee, M.; Guo, L.; Han, W.; Li, T.; Hill, M.C.; Hong, T.; Mo, W.; et al. Tet inactivation disrupts YY1 binding and long-range chromatin interactions during embryonic heart development. Nat. Commun. 2019, 10, 4297. [Google Scholar] [CrossRef]
- Movassagh, M.; Choy, M.K.; Knowles, D.A.; Cordeddu, L.; Haider, S.; Down, T.; Siggens, L.; Vujic, A.; Simeoni, I.; Penkett, C.; et al. Distinct epigenomic features in end-stage failing human hearts. Circulation 2011, 124, 2411–2422. [Google Scholar] [CrossRef] [PubMed]
- Gilsbach, R.; Preissl, S.; Grüning, B.A.; Schnick, T.; Burger, L.; Benes, V.; Würch, A.; Bönisch, U.; Günther, S.; Backofen, R.; et al. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun. 2014, 5, 5288. [Google Scholar] [CrossRef] [PubMed]
- Greco, C.M.; Kunderfranco, P.; Rubino, M.; Larcher, V.; Carullo, P.; Anselmo, A.; Kurz, K.; Carell, T.; Angius, A.; Latronico, M.V.; et al. DNA hydroxymethylation controls cardiomyocyte gene expression in development and hypertrophy. Nat. Commun. 2016, 7, 12418. [Google Scholar] [CrossRef]
- Wu, D.; Hu, D.; Chen, H.; Shi, G.; Fetahu, I.S.; Wu, F.; Rabidou, K.; Fang, R.; Tan, L.; Xu, S.; et al. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature 2018, 559, 637–641. [Google Scholar] [CrossRef]
- Wang, Q.; Fu, S.; Li, S.; Huang, X.; Wang, H. Correlation of TET-2 Levels with Disease Evaluation in AMI Patients. Contrast Media Mol. Imaging 2022, 2022, 9983071. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Cheng, S.; Wang, S.; Li, W.; Liu, J. C-MYC ameliorates ventricular remodeling of myocardial infarction rats via binding to the promoter of microRNA-29a-3p to facilitate TET2 expression. Int. J. Cardiol. 2022, 357, 105–112. [Google Scholar] [CrossRef]
- Zhong, R.; Zhang, F.; Yang, Z.; Li, Y.; Xu, Q.; Lan, H.; Lang, S.; Cyganek, L.; Burgermeister, E.; El-Battrawy, I.; et al. Regulation of Ion Channel Function in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Cancer Cell Secretion Through DNA Methylation. Front. Cardiovasc. Med. 2022, 9, 839104. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).