Chromosomal Rearrangements and Satellite DNAs: Extensive Chromosome Reshuffling and the Evolution of Neo-Sex Chromosomes in the Genus Pyrrhulina (Teleostei; Characiformes)
Abstract
1. Introduction
2. Results
2.1. SatDNA Content of P. marilynae and P. semifasciata
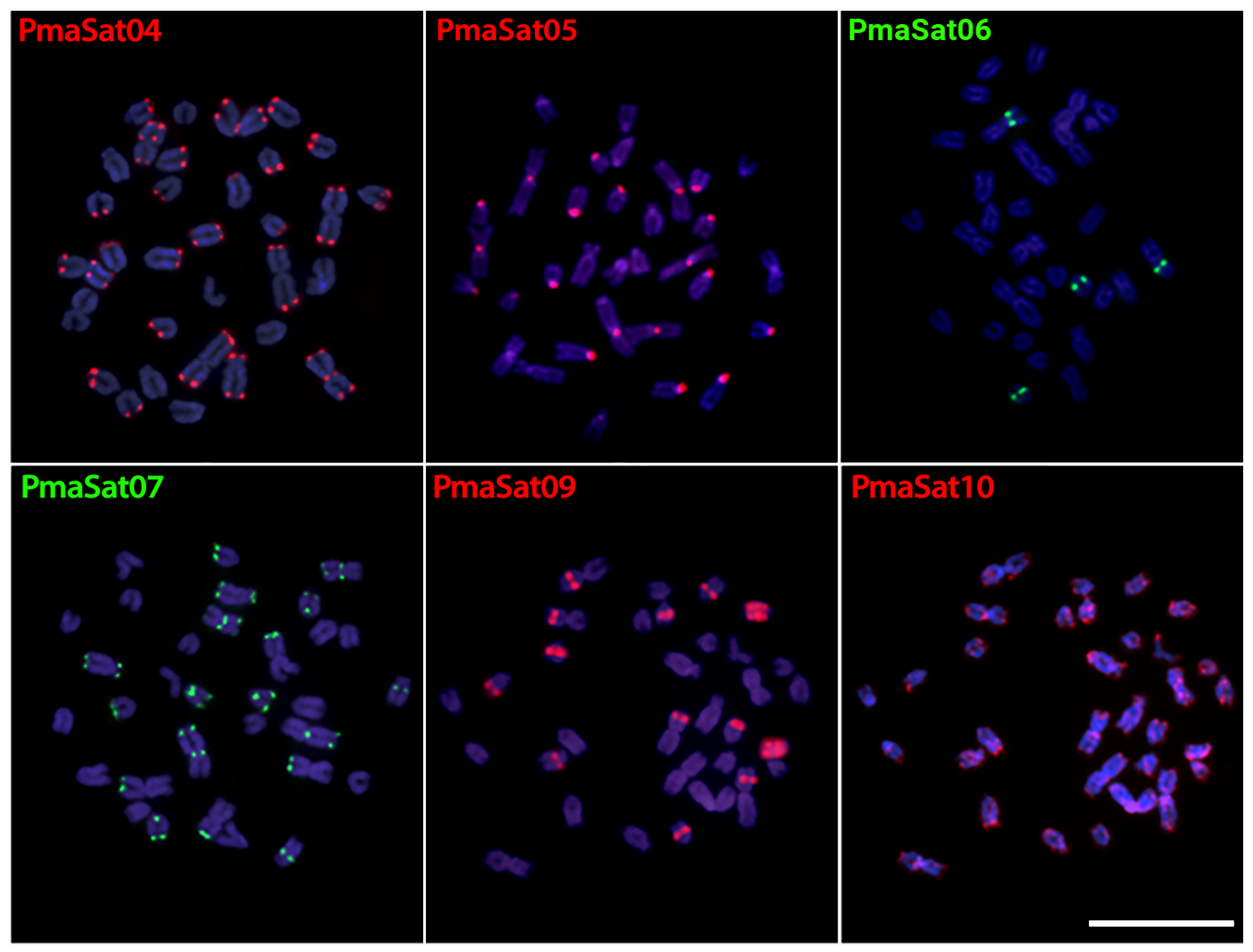
2.2. Chromosomal Distribution of PmaSatDNA in P. marilynae
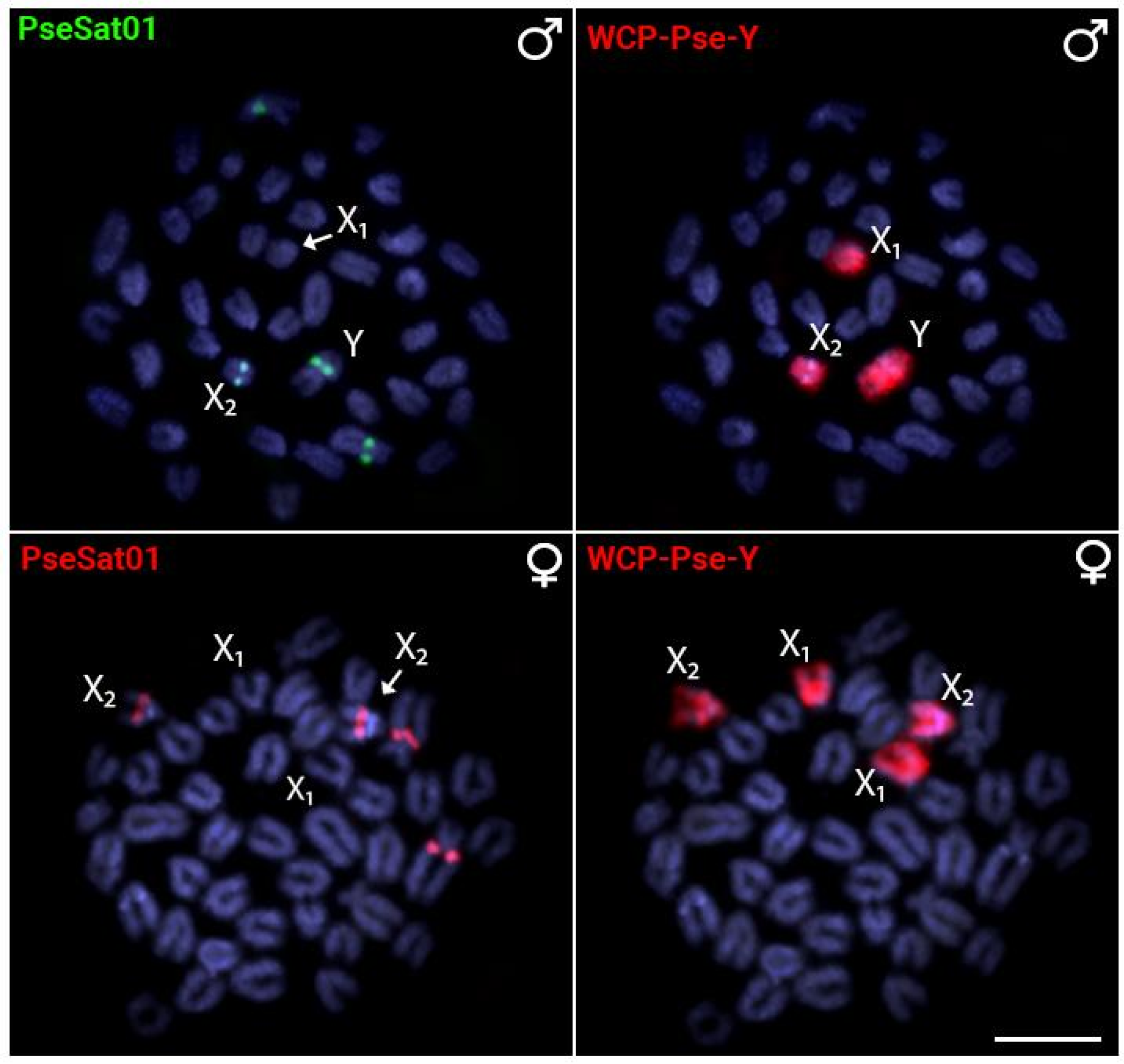
2.3. Chromosomal Distribution of PseSatDNA in P. semifasciata
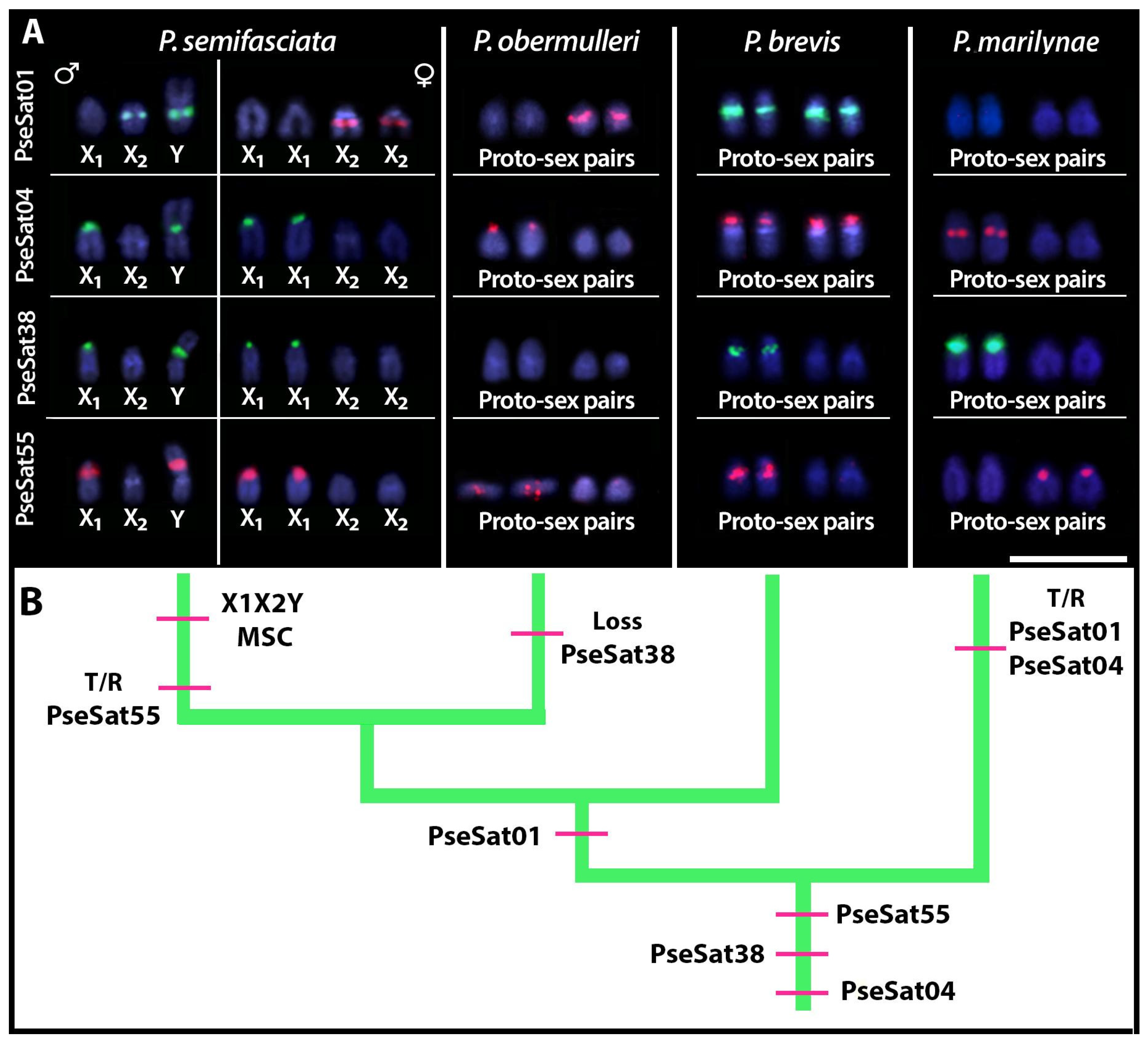
2.4. Chromosomal Distribution of PseSatDNA in Other Pyrrhulina Species
2.5. Minimum Spanning Trees: MSTs
3. Discussion
3.1. General Features P. marilynae and P. semifasciata Satellitomes
3.2. Satellite DNA Contribution in the Significant 2n Reduction Observed in P. marilynae
3.3. SatDNAs and the Evolution of Multiple Sex Chromosomes
4. Materials and Methods
4.1. Material, Mitotic Chromosomes and DNA Sequencing
4.2. Bioinformatic Analyses and satDNA Library
4.3. Primer Design and DNA Amplification via Polymerase Chain Reaction (PCR)
4.4. Fluorescence In Situ Hybridization (FISH)
4.5. Images and Confirmation of Results
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oliveira, R.R.; Feldberg, E.; dos Anjos, M.B.; Zuanon, J. Occurrence of multiple sexual chromosomes (XX/XY1Y2 and Z1Z1Z2Z2/Z1Z2W1W2) in catfishes of the genus Ancistrus (Siluriformes: Loricariidae) from the Amazon basin. Genetica 2009, 134, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Bateson, W. Heredity and variation in modern lights. In Darwin and Modern Science; Seward, A.C., Ed.; Cambridge University Press: Cambridge, UK, 1909; pp. 85–101. [Google Scholar]
- Dobzhansky, T. On the sterility of the interracial hybrids in Drosophila pseudoobscura. Proc. Natl. Acad. Sci. USA 1933, 19, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.J. Isolation mechanisms, evolution and temperature. Biol. Symp. 1942, 6, 71. Available online: https://www.google.de/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&ved=2ahUKEwiI1OuM3MyAAxXHhv0HHeVXDCAQFnoECBAQAQ&url=https%3A%2F%2Fwww.ucl.ac.uk%2Ftaxome%2Flit%2Fmuller%25201942%2520new%2520enh.pdf&usg=AOvVaw0Xf86o-hX4H9vA0QhXx295&opi=89978449 (accessed on 23 July 2023).
- Faria, R.; Navarro, A. Chromosomal speciation revisited: Rearranging theory with pieces of evidence. Trends in ecology and Evolution. Trends Ecol. Evol. 2010, 25, 660–669. [Google Scholar] [CrossRef]
- Šíchová, J.; Voleníková, A.; Dincă, V.; Nguyen, P.; Sahara, K.; Marec, F. Dynamic karyotype evolution and unique sex determination systems in Leptidea wood white butterflies. BMC Evol. Biol. 2015, 15, 89. [Google Scholar] [CrossRef] [PubMed]
- Vieira, C.P.; Coelho, P.A.; Vieira, J. Inferences on the evolutionary history of the Drosophila americana polymorphic X/4 fusion from patterns of polymorphism at the X-linked paralytic and elav genes. Genetics 2003, 164, 1459–1469. [Google Scholar] [CrossRef] [PubMed]
- Biémont, C.; Vieira, C. Junk DNA as an evolutionary force. Nature 2006, 443, 521–524. [Google Scholar] [CrossRef]
- Khosraviani, N.; Ostrowski, L.A.; Mekhail, K. Roles for non-coding RNAs in spatial genome organization. Front. Cell Dev. Biol. 2019, 7, 336. [Google Scholar] [CrossRef]
- Šatović-Vukšić, E.; Plohl, M. Satellite DNAs—From localized to highly dispersed genome components. Genes 2023, 14, 742. [Google Scholar] [CrossRef]
- Montiel, E.E.; Panzera, F.; Palomeque, T.; Lorite, P.; Pita, S. Satellitome analysis of Rhodnius prolixus, one of the main Chagas disease vector species. Int. J. Mol. Sci. 2021, 22, 6052. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Hu, K.B.; Clark, A.G. Three recent sex chromosome-to-autosome fusions in a Drosophila virilis strain with high satellite DNA content. Genetics 2023, 224, iyad062. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, A.B.S.M.; Milani, D.; Palacios-Gimenez, O.M.; Ruiz-Ruano, F.J.; Cabral-de-Mello, D.C. High dynamism for neo-sex chromosomes: Satellite DNAs reveal complex evolution in a grasshopper. Heredity 2020, 125, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Bardella, V.B.; Milani, D.; Cabral-de-Mello, D.C. Analysis of Holhymenia histrio genome provides insight into the satDNA evolution in an insect with holocentric chromosomes. Chromosome Res. 2020, 28, 369–380. [Google Scholar] [CrossRef]
- Vozdova, M.; Kubickova, S.; Martínková, N.; Galindo, D.J.; Bernegossi, A.M.; Cernohorska, H.; Kadlcikova, D.; Musilová, P.; Duarte, J.M.; Rubes, J. Satellite DNA in neotropical deer species. Genes 2021, 12, 123. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Zrzavá, M.; Kubíčková, S.; Rendón, P.; Marec, F. The role of satellite DNAs in genome architecture and sex chromosome evolution in Crambidae moths. Front. Genet. 2021, 12, 661417. [Google Scholar] [CrossRef]
- Cabral-de-Melo, D.C.; Mora, P.; Rico-Porras, J.M.; Ferretti, A.B.S.M.; Palomeque, T.; Lorite, P. The spread of satellite DNAs in euchromatin and insights into the multiple sex chromosome evolution in Hemiptera revealed by repeatome analysis of the bug Oxycarenus hyalinipennis. Insect Mol. Biol. 2023. online version of record before inclusion in an issue. [Google Scholar] [CrossRef]
- Gržan, T.; Dombi, M.; Despot-Slade, E.; Veseljak, D.; Volarić, M.; Meštrović, N.; Plohl, M.; Mravinac, B. The Low-Copy-Number Satellite DNAs of the Model Beetle Tribolium castaneum. Genes 2023, 14, 999. [Google Scholar] [CrossRef]
- Peona, V.; Kutschera, V.E.; Blom, M.P.; Irestedt, M.; Suh, A. Satellite DNA evolution in Corvoidea inferred from short and long reads. Mol. Ecol. 2023, 32, 1288–1305. [Google Scholar] [CrossRef]
- Henikoff, S.; Ahmad, K.; Malik, H.S. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 2001, 293, 1098–1102. [Google Scholar] [CrossRef]
- O’Neill, R.J.; Eldridge, M.D.B.; Metcalfe, C.J. Centromere dynamics and chromosome evolution in marsupials. J. Hered. 2004, 95, 375–381. [Google Scholar] [CrossRef]
- Plohl, M.; Meštrović, N.; Mravinac, B. Satellite DNA evolution. Repetitive DNA 2012, 7, 126–152. [Google Scholar] [CrossRef]
- Weissensteiner, M.H.; Suh, A. Repetitive DNA: The dark matter of avian genomics. In Avian Genomics in Ecology and Evolution: From the Lab into the Wild; Kraus, R., Ed.; Springer: Cham, Switzerland, 2019; pp. 93–150. [Google Scholar] [CrossRef]
- Shatskikh, A.S.; Kotov, A.A.; Adashev, V.E.; Bazlev, S.S.; Olenina, L.V. Functional significance of satellite DNAs: Insights from Drosophila. Front. Cell Dev. Biol. 2020, 8, 312. [Google Scholar] [CrossRef] [PubMed]
- Fricke, R.; Eschmeyer, W.N.; van der Laan, R. Eschmeyer’s Catalog of Fishes: Genera, Species, References. 2023. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 12 July 2023).
- Netto-Ferreira, A.L.; Marinho, M.M.F. New species of Pyrrhulina (Ostariophysi: Characiformes: Lebiasinidae) from the Brazilian shield, with comments on a putative monophyletic group of species in the genus. Zootaxa 2013, 3664, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Moraes, R.L.R.; Bertollo, L.A.C.; Marinho, M.M.F.; Yano, C.F.; Hatanaka, T.; Barby, F.F.; Troy, W.P.; Cioffi, M.B. Evolutionary relationships and cytotaxonomy considerations in the genus Pyrrhulina (Characiformes, Lebiasinidae). Zebrafish 2017, 14, 536–546. [Google Scholar] [CrossRef]
- Moraes, R.L.R.; Sember, A.; Bertollo, L.A.C.; Oliveira, E.A.; Ráb, P.; Hatanaka, T.; Marinho, M.M.F.; Liehr, T.; Al-Rikabi, A.B.H.; Feldberg, E.; et al. Comparative cytogenetics and neo-Y formation in sall-sized fish species of the genus Pyrrhulina (Characiformes, Lebiasinidae). Front. Genet. 2019, 10, 678. [Google Scholar] [CrossRef]
- Moraes, R.L.R.; Sassi, F.D.M.C.; Bertollo, L.A.C.; Marinho, M.M.F.; Viana, P.F.; Feldberg, E.; Sales-Oliveira, V.C.; Deon, G.A.; Al-Rikabi, B.H.; Liehr, T.; et al. Tracking the evolutionary trends among small-size fishes of the genus Pyrrhulina (Characiforme, Lebiasinidae): New insights from a molecular cytogenetic perspective. Front. Genet. 2021, 12, 769984. [Google Scholar] [CrossRef]
- dos Santos, L.L.; Benone, N.L.; Brasil, L.S.; Pires, T.H.; Begot, T.O.; Dantas, D.D.F.; de Assis Montag, L.F. The use of taxonomic families as biological surrogates of the diversity of the Amazonian stream fish. Ecol. Ind. 2022, 141, 109094. [Google Scholar] [CrossRef]
- Sassi, F.M.C.; Oliveira, E.; Bertollo, L.A.C.; Nirchio, M.; Hatanaka, T.; Marinho, M.M.F.; Moreira-Filho, O.; Aroutiounian, R.; Liehr, T.; Al-Rikabi, H.B.H.; et al. Chromosomal evolution and evolutionary relationships of Lebiasina species (Characiformes, Lebiasinidae). Int. J. Mol. Sci. 2019, 20, 2944. [Google Scholar] [CrossRef]
- Benzaquem, D.C.; Oliveira, C.; da Silva Batista, J.; Zuanon, J.; Porto, J.I.R. DNA Barcoding in Pencilfishes (Lebiasinidae: Nannostomus) Reveals Cryptic Diversity across the Brazilian Amazon. PLoS ONE 2015, 10, e0112217. [Google Scholar] [CrossRef]
- Toma, G.A.; Moraes, R.L.R.; Sassi, F.M.C.; Bertollo, L.A.C.; Oliveira, E.A.; Rab, P.; Sember, A.; Liehr, T.; Hatanaka, T.; Viana, P.F.; et al. Cytogenetics of the small-sized fish, Copeina Guttata (Characiformes, Lebiasinidae): Novel insights into the karyotype differentiation of the family. PLoS ONE 2019, 14, e0226746. [Google Scholar] [CrossRef] [PubMed]
- Sember, A.; de Oliveira, E.A.; Ráb, P.; Bertollo, L.A.C.; Freitas, N.L.; Viana, P.F.; Yano, C.F.; Hatanaka, T.; Marinho, M.M.F.; Moraes, R.L.R.; et al. Centric fusions behind the karyotype evolution of neotropical Nannostomus pencilfishes (Characiforme, Lebiasinidae): First insights from a molecular cytogenetic perspective. Genes 2020, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, C.; Zhu, Y.; Li, Y.; Wei, Y.; Ruan, H. Mitochondrial genomes of four American characins and phylogenetic relationships within the family Characidae (Teleostei: Characiformes). Gene 2020, 762, 145041. [Google Scholar] [CrossRef]
- Froese, R.; Pauly, D. Fish Base: World Wide Web Electronic Publication. 2023. Available online: www.fishbase.org (accessed on 22 July 2023).
- Ferreira, P.H.N.; Souza, F.H.S.; Moraes, R.L.R.M.; Perez, M.F.; Sassi, F.M.C.; Viana, P.F.; Feldberg, E.; Ezaz, T.; Liehr, T.; Bertollo, L.A.C.; et al. The genetic differentiation of Pyrrhulina (Teleostei, Characiformes) species is likely influenced by both geographical distribution and chromosomal rearrangements. Front. Genet. 2022, 13, 869073. [Google Scholar] [CrossRef]
- Silva, D.M.D.A.; Utsunomia, R.; Ruiz-Ruano, F.J.; Daniel, S.N.; Porto-Foresti, F.; Hashimoto, D.T.; Oliveira, C.; Camacho, J.P.M.; Foresti, F. High-throughput analysis unveils a highly shared satellite DNA library among three species of fish genus Astyanax. Sci. Rep. 2017, 7, 12726. [Google Scholar] [CrossRef]
- dos Santos, R.Z.; Calegari, R.M.; Silva, D.M.Z.D.A.; Ruiz-Ruano, F.J.; Melo, S.; Oliveira, C.; Foresti, F.; Uliano-Silva, M.; Porto-Foresti, F.; Utisunomia, R. A long-term conserved satellite DNA that remains unexpanded in several genomes of Characiformes fish is actively transcribed. Genome Biol. Evol. 2021, 13, evab002. [Google Scholar] [CrossRef] [PubMed]
- Goes, C.A.G.; Santos, R.Z.; Aguiar, W.R.C.; Alves, D.C.V.; Silva, D.M.Z.A.; Foresti, F.; Oliveira, C.; Utsunomia, R.; Porto-Foresti, F. Revealing the satellite DNA history in Psalidodon and Astyanax characid fish by comparative satellitomics. Front. Genet. 2023, 13, 884072. [Google Scholar] [CrossRef]
- Lisachov, A.; Rumyantsev, A.; Prokopov, D.; Ferguson-Smith, M.; Trifonov, V. Conservation of major satellite DNAs in snake heterochromatin. Animals 2023, 13, 334. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Singchat, W.; Jehangir, M.; Suntronpong, A.; Panthum, T.; Malaivijitnond, S.; Srikulnath, K. Dark Matter of Primate Genomes: Satellite DNA Repeats and Their Evolutionary Dynamics. Cells 2020, 9, 2714. [Google Scholar] [CrossRef]
- Guzmán-Markevich, K.; Roco, A.S.; Ruiz-García, A.; Bullejos, M. Cytogenetic analysis in the toad species Bufo spinosus, Bufotes viridis and Epidalea calamita (Anura, Bufonidae) from the mediterranean area. Genes 2022, 13, 1475. [Google Scholar] [CrossRef]
- Fry, K.; Salser, W. Nucleotide sequences of HS-α satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 1977, 12, 1069–1084. [Google Scholar] [CrossRef] [PubMed]
- Camacho, J.P.M.; Cabrero, J.; López-León, M.D.; Martín-Pecina, M.; Perfectti, F.; Garrido-Ramos, M.A.; Ruiz-Ruano, F.J. Satellitome comparison of two oedipodine grasshoppers highlights the contingent nature of satellite DNA evolution. BMC Biol. 2022, 20, 36. [Google Scholar] [CrossRef]
- Podgornaya, O.I. Nuclear organization by satellite DNA, SAF-A/hnRNPU and matrix attachment regions. Semin. Cell Dev. Biol. 2022, 128, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Thakur, J.; Packiaraj, J.; Henikoff, S. Sequence, chromatin and evolution of satellite DNA. Int. J. Mol. Sci. 2021, 22, 4309. [Google Scholar] [CrossRef]
- Tunjić-Cvitanić, M.; Pasantes, J.J.; García-Souto, D.; Cvtanic, T.; Plohl, M.; Šatović-Vukšić, E. Satellitome analysis of the pacific oyster Crassostrea gigas reveals new pattern of satellite DNA organization, highly scattered across the genome. Int. J. Mol. Sci. 2021, 22, 6798. [Google Scholar] [CrossRef]
- Pereira, J.A.; Milani, D.; Ferretti, A.B.S.; Bardella, V.B.; Cabral-de-Mello, D.C.; Lopes, D.M. The extensive amplification of heterochromatin in Melipona bees revealed by high throughput genomic and chromosomal analysis. Chromosoma 2021, 130, 251–262. [Google Scholar] [CrossRef]
- Valeri, M.P.; Dias, G.B.; do Espírito Santo, A.A.; Moreira, C.N.; Yonenaga-Yassuda, Y.; Sommer, I.B.; Kuhn, G.C.; Svartman, M. First description of a satellite DNA in manatees’ centromeric regions. Front. Genet. 2021, 12, 694866. [Google Scholar] [CrossRef]
- Crepaldi, C.; Martí, E.; Gonçalves, E.M.; Martí, D.A.; Parise-Maltempi, P.P. Genomic differences between the sexes in a fish species seen through satellite DNAs. Front. Genet. 2021, 12, 728670. [Google Scholar] [CrossRef]
- Serrano-Freitas, E.A.; Silva, A.M.Z.A.; Ruiz-Ruano, F.J.; Utsunomia, R.; Araya-Jaime, C.; Oliveira, C.; Camacho, J.P.M.; Foresti, F. Satellite DNA content of B chromosomes in the characid fish Characidium gomesi supports their origin from sex chromosomes. Mol. Genet. Genom. 2020, 295, 195–207. [Google Scholar] [CrossRef]
- Garrido-Ramos, M.A. Satellite DNA: An evolving topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef]
- Paço, A.; Chaves, R.; Vieira-da-Silva, A.; Adega, F. The involvement of repetitive sequences in the remodelling of karyotypes: The Phodopus genomes (Rodentia, Cricetidae). Micron 2013, 46, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Vieira-da-Silva, A.; Louzada, S.; Adega, F.; Chaves, R. A high-resolution comparative chromosome map of Cricetus cricetus and Peromyscus eremicus reveals the involvement of constitutive heterochromatin in breakpoint regions. Cytogenet. Genome Res. 2015, 145, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Gatto, K.P.; Mattos, J.V.; Seger, K.R.; Lourenço, L.B. Sex chromosome differentiation in the frog genus Pseudis involves satellite DNA and chromosome rearrangements. Front. Genet. 2018, 9, 301. [Google Scholar] [CrossRef] [PubMed]
- Escudeiro, A.; Ferreira, D.; Mendes-da-Silva, A.; Heslop-Harrison, J.S.; Adega, F.; Chaves, R. Bovine satellite DNAs–a history of the evolution of complexity and its impact in the Bovidae family. Eur. Zool. J. 2019, 86, 20–37. [Google Scholar] [CrossRef]
- de Lima, L.G.; Ruiz-Ruano, F.J. In-depth satellitome analyses of 37 Drosophila species illuminate repetitive DNA evolution in the Drosophila genus. Genome Biol. Evol. 2022, 14, evac064. [Google Scholar] [CrossRef]
- Slamovits, C.H.; Cook, J.A.; Lessa, E.P.; Rossi, S.M. Recurrent amplifications and deletions of satellite DNA accompanied chromosomal diversification in South American tuco-tucos (genus Ctenomys, Rodentia: Octodontidae): A phylogenetic approach. Mol. Biol. Evol. 2001, 18, 1708–1709. [Google Scholar] [CrossRef]
- Ugarković, Ð.; Plohl, M. Variation in satellite DNA profiles—Causes and effects. EMBO J. 2002, 18, 1708–1719. [Google Scholar] [CrossRef]
- Kopecna, O.; Kubickova, S.; Cernohorska, H.; Cabelova, K.; Vahala, J.; Martinkova, N.; Rubes, J. Tribe-specific satellite DNA in non-domestic Bovidae. Chromosome Res. 2014, 22, 277–291. [Google Scholar] [CrossRef]
- Vozdova, M.; Kubickova, S.; Cernohorska, H.; Fröhlich, J.; Vodicka, R.; Rubes, J. Comparative study of the bush dog (Speothos venaticus) karyotype and analysis of satellite DNA sequences and their chromosome distribution in six species of Canidae. Cytogenet. Genome Res. 2019, 159, 88–96. [Google Scholar] [CrossRef]
- Franchini, P.; Irisarri, I.; Fudickar, A.; Schmidt, A.; Meyer, A.; Wikelski, M.; Partecke, J. Animal tracking meets migration genomics: Transcriptomic analysis of a partially migratory bird species. Mol. Ecol. 2017, 26, 3204–3216. [Google Scholar] [CrossRef]
- Franchini, P.; Kautt, A.F.; Nater, A.; Antonini, G.; Castiglia, R.; Meyer, A.; Solano, E. Reconstructing the evolutionary history of chromosomal races on islands: A genome-wide analysis of natural house mouse populations. Mol. Biol. Evol. 2020, 37, 2825–2837. [Google Scholar] [CrossRef] [PubMed]
- Vara, C.; Paytuví-Gallart, A.; Cuartero, Y.; Álvarez-González, L.; Marín-Gual, L.; Garcia, F.; Florit-Sabater, B.; Capilla, L.; Sanchéz-Guillén, R.A.; Sarrate, Z.; et al. The impact of chromosomal fusions on 3D genome folding and recombination in the germ line. Nat. Commun. 2021, 12, 2981. [Google Scholar] [CrossRef] [PubMed]
- Naish, M.; Alonge, M.; Wlodzimierz, P.; Tock, A.J.; Abramson, B.W.; Schmücker, A.; Mandáková, T.; Jamge, B.; Lambing, C.; Kuo, P.; et al. The genetic and epigenetic landscape of the Arabidopsis centromeres. Science 2021, 374, eabi7489. [Google Scholar] [CrossRef]
- Yang, T.J.; Yu, Y.; Chang, S.B.; Jong, H.; Oh, C.S.; Ahn, S.N.; Fang, E.; Wing, R.A. Toward closing rice telomere gaps: Mapping and sequence characterization of rice subtelomere regions. Theor. Appl. Genet. 2005, 111, 467–478. [Google Scholar] [CrossRef]
- Kipling, D.; Ackford, H.E.; Taylor, B.A.; Cooke, H.J. Mouse minor satellite DNA genetically maps to the centromere and is physically linked to the proximal telomere. Genomics 1991, 11, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Glugoski, L.; Nogaroto, V.; Deon, G.A.; Azambuja, M.; Moreira-Filho, O.; Vicari, M.R. Enriched tandem repeats in chromosomal fusion points of Rineloricaria latirostris (Boulenger, 1900) (Siluriformes: Loricariidae). Genome 2022, 65, 479–489. [Google Scholar] [CrossRef]
- Glugoski, L.; Deon, G.A.; Nogaroto, V.; Moreira-Filho, O.; Vicari, M.R. Robertsonian fusion site in Rineloricaria pentamaculata (Siluriformes: Loricariidae): Involvement of 5S rDNA and satellite sequences. Cytogenet. Genome Res. 2023. Online ahead of print. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Lee, H.; Darby, C.A.; Schatz, M.C. Piercing the dark matter: Bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet. 2018, 19, 329–346. [Google Scholar] [CrossRef]
- Louzada, S.; Lopes, M.; Ferreira, D.; Adega, F.; Escudeiro, A.; Gama-Carvalho, M.; Chaves, R. Decoding the role of satellite DNA in genome architecture and plasticity—An evolutionary and clinical affair. Genes 2020, 11, 72. [Google Scholar] [CrossRef]
- Montiel, E.E.; Mora, P.; Rico-Porras, J.M.; Palomeque, T.; Lorite, P. Satellitome of the red palm weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae), the most diverse among insects. Front. Ecol. Evol. 2022, 10, 826808. [Google Scholar] [CrossRef]
- Sember, A.; Nguyen, P.; Perez, M.F.; Altmanová, M.; Rab, P.; Cioffi, M.B. Multiple sex chromosomes in teleost fishes from a cytogenetic perspective: State of the art and future challenges. Phil. Trans. R. Soc. B 2021, 376, 20200098. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Dias, G.B.; De Lima, L.G.; Kuhn, G.C.S.; Ramos, E.; Martins, C.; Cabral-de-Mello, D.C. High-throughput analysis of the satellitome revealed enormous diversity of satellite DNAs in the neo-Y chromosome of the cricket Eneoptera surinamensis. Sci. Rep. 2017, 7, 6422. [Google Scholar] [CrossRef] [PubMed]
- da Silva, M.J.; Gazoni, T.; Haddad, C.F.B.; Parise-Maltempi, P.P. Analysis in Proceratophrys boiei genome illuminates the satellite DNA content in a frog from the Brazilian atlantic forest. Front. Genet. 2023, 14, 1101397. [Google Scholar] [CrossRef] [PubMed]
- Bertollo, L.A.; Fontes, M.S.; Fenocchio, A.S.; Cano, J. The X 1 X 2 Y sex chromosome system in the fish Hoplias malabaricus. I. G-, C-and chromosome replication banding. Chromosome Res. 1997, 5, 493–499. [Google Scholar] [CrossRef]
- Bertollo, L.A.C.; Oliveira, C.; Molina, W.F.; Margarido, V.P.; Fontes, M.S.; Pastori, M.C.; Falcão, J.N.; Fenocchio, A.S. Chromosome evolution in the erythrinid fish, Erythrinus erythrinus (Teleostei: Characiformes). Heredity 2004, 93, 228–233. [Google Scholar] [CrossRef][Green Version]
- Cioffi, M.B.; Kejnovský, E.; Marquioni, V.; Poltronieri, J.; Molina, W.F.; Diniz, D.; Bertollo, L.A.C. The key role of repeated DNAs in sex chromosome evolution in two fish species with ZW sex chromosome system. Mol. Cytogenet. 2012, 5, 28. [Google Scholar] [CrossRef]
- Feliciello, I.; Akrap, I.; Ugarković, Đ. Satellite DNA modulates gene expression in the beetle Tribolium castaneum after heat stress. PLoS Genet. 2015, 11, e100566. [Google Scholar] [CrossRef]
- Kuhn, G.C.; Küttler, H.; Moreira-Filho, O.; Heslop-Harrison, J.S. The 1.688 repetitive DNA of Drosophila: Concerted evolution at different genomic scales and association with genes. Mol. Biol. Evol. 2012, 29, 7–11. [Google Scholar] [CrossRef]
- Plohl, M.; Luchetti, A.; Meštrović, N.; Mantovani, B. Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero) chromatin. Gene 2008, 409, 72–78. [Google Scholar] [CrossRef]
- Prakhongcheep, O.; Thapana, W.; Suntronpong, A.; Singchat, W.; Pattanatanang, K.; Phatcharakullawarawat, R.; Muangmai, N.; Peyachoknagul, K.; Matsubara, K.; Ezaz, T.; et al. Lack of satellite DNA species-specific homogenization and relationship to chromosomal rearrangements in monitor lizards (Varanidae, Squamata). BMC Evol. Biol. 2017, 17, 193. [Google Scholar] [CrossRef]
- Feliciello, I.; Pezer, Ž.; Kordiš, D.; Bruvo Mađarić, B.; Ugarković, Đ. Evolutionary history of alpha satellite DNA repeats dispersed within human genome euchromatin. Genome Biol. Evol. 2020, 12, 2125–2138. [Google Scholar] [CrossRef]
- Zattera, M.L.; Bruschi, D.P. Transposable elements as a source of novel repetitive DNA in the eukaryote genome. Cells 2022, 11, 3373. [Google Scholar] [CrossRef] [PubMed]
- Scalvenzi, T.; Pollet, N. Insights on genome size evolution from a miniature inverted repeat transposon driving a satellite DNA. Mol. Phylogenet. Evol. 2014, 81, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Guarracino, A.; Buonaiuto, S.; de Lima, L.G.; Potapova, T.; Rhie, A.; Koren, S.; Rubinstein, B.; Fischer, C.; Consortium, H.P.R.; Gerton, J.L.; et al. Recombination between heterologous human acrocentric chromosomes. Nature 2023, 617, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Yazdi, H.P.; Olito, C.; Kawakami, T.; Unneberg, P.; Schalk, M.F.; Cloete, S.W.P.; Hansson, B.; Cornwallis, C.K. The evolutionary maintenance of ancient recombining sex chromosomes in the ostrich. PLoS Genet. 2023, 19, e1010801. [Google Scholar] [CrossRef] [PubMed]
- Kratochvíl, L.; Stöck, M.; Rovatsos, M.; Bullejos, M.; Herpin, A.; Jeffries, D.L.; Peichel, C.L.; Perrin, N.; Valenzuela, N.; Pokorná, M.J. Expanding the classical paradigm: What we have learnt from vertebrates about sex chromosome Evolution. Phil. Trans. R. Soc. B 2021, 376, 20200097. [Google Scholar] [CrossRef]
- Bertollo, L.A.C. Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae). Braz. J. Genet. 1978, 1, 103–120. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef]
- Schmieder, R.; Eswards, R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef]
- Smith, C.J.; Castanon, O.; Said, K.; Volf, V.; Khoshakhlagh, P.; Hornik, A.; Ferreira, R.; Wu, C.T.; Guell, M.; Garg, S.; et al. Enabling large-scale genome editing at repetitive elements by reducing DNA nicking. Nucleic Acids Res. 2020, 48, 5183–5195. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, M.; Sousa, A.; Ramirez, M.; Francisco, A.P.; Carriço, J.A.; Vaz, C. PHYLOViZ 2.0: Providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 2017, 33, 128–129. [Google Scholar] [CrossRef] [PubMed]
- Pinkel, D.; Straume, T.; Gray, J.W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA 1986, 83, 2934–2938. [Google Scholar] [CrossRef] [PubMed]
- Sassi, F.M.C.; Toma, G.A.; Cioffi, M.B. FISH-in fish chromosomes. In Cytogenetics and Molecular Cytogenetics, 1st ed.; Liehr, T., Ed.; CRC Press: Boca Raton, FL, USA, 2022; pp. 281–297. [Google Scholar] [CrossRef]



| Species | Locality | N | Voucher |
|---|---|---|---|
| P. brevis | Adolfo Ducke Reserve- Igarapé Barro Branco, Manaus–AM (2°56′04.6″ S 59°58′10.6″ W) | 04♂; 07♀ | MZUSP 123077 |
| P. marilynae | Ipiranga do Norte–MT (11°36′02″ S 55°56′27″ W) | 13♂; 04♀ | UFPB 12080 |
| P. obermulleri | Tefé-AM (3°25′50.7″ S 64°44′54.8″ W) | 06♂; 04♀ | UFPB 12079 |
| P. semifasciata | Adolfo Ducke Reserve- Igarapé Barro Branco, Manaus–AM (2°56′04.6″ S 59°58′10.6″ W) | 07♀; 12♂ | MZUSP 123080 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Moraes, R.L.R.; de Menezes Cavalcante Sassi, F.; Vidal, J.A.D.; Goes, C.A.G.; dos Santos, R.Z.; Stornioli, J.H.F.; Porto-Foresti, F.; Liehr, T.; Utsunomia, R.; de Bello Cioffi, M. Chromosomal Rearrangements and Satellite DNAs: Extensive Chromosome Reshuffling and the Evolution of Neo-Sex Chromosomes in the Genus Pyrrhulina (Teleostei; Characiformes). Int. J. Mol. Sci. 2023, 24, 13654. https://doi.org/10.3390/ijms241713654
de Moraes RLR, de Menezes Cavalcante Sassi F, Vidal JAD, Goes CAG, dos Santos RZ, Stornioli JHF, Porto-Foresti F, Liehr T, Utsunomia R, de Bello Cioffi M. Chromosomal Rearrangements and Satellite DNAs: Extensive Chromosome Reshuffling and the Evolution of Neo-Sex Chromosomes in the Genus Pyrrhulina (Teleostei; Characiformes). International Journal of Molecular Sciences. 2023; 24(17):13654. https://doi.org/10.3390/ijms241713654
Chicago/Turabian Stylede Moraes, Renata Luiza Rosa, Francisco de Menezes Cavalcante Sassi, Jhon Alex Dziechciarz Vidal, Caio Augusto Gomes Goes, Rodrigo Zeni dos Santos, José Henrique Forte Stornioli, Fábio Porto-Foresti, Thomas Liehr, Ricardo Utsunomia, and Marcelo de Bello Cioffi. 2023. "Chromosomal Rearrangements and Satellite DNAs: Extensive Chromosome Reshuffling and the Evolution of Neo-Sex Chromosomes in the Genus Pyrrhulina (Teleostei; Characiformes)" International Journal of Molecular Sciences 24, no. 17: 13654. https://doi.org/10.3390/ijms241713654
APA Stylede Moraes, R. L. R., de Menezes Cavalcante Sassi, F., Vidal, J. A. D., Goes, C. A. G., dos Santos, R. Z., Stornioli, J. H. F., Porto-Foresti, F., Liehr, T., Utsunomia, R., & de Bello Cioffi, M. (2023). Chromosomal Rearrangements and Satellite DNAs: Extensive Chromosome Reshuffling and the Evolution of Neo-Sex Chromosomes in the Genus Pyrrhulina (Teleostei; Characiformes). International Journal of Molecular Sciences, 24(17), 13654. https://doi.org/10.3390/ijms241713654









