The Prognosis-Predictive and Immunoregulatory Role of SUMOylation Related Genes: Potential Novel Targets in Prostate Cancer Treatment
Abstract
1. Introduction
2. Result
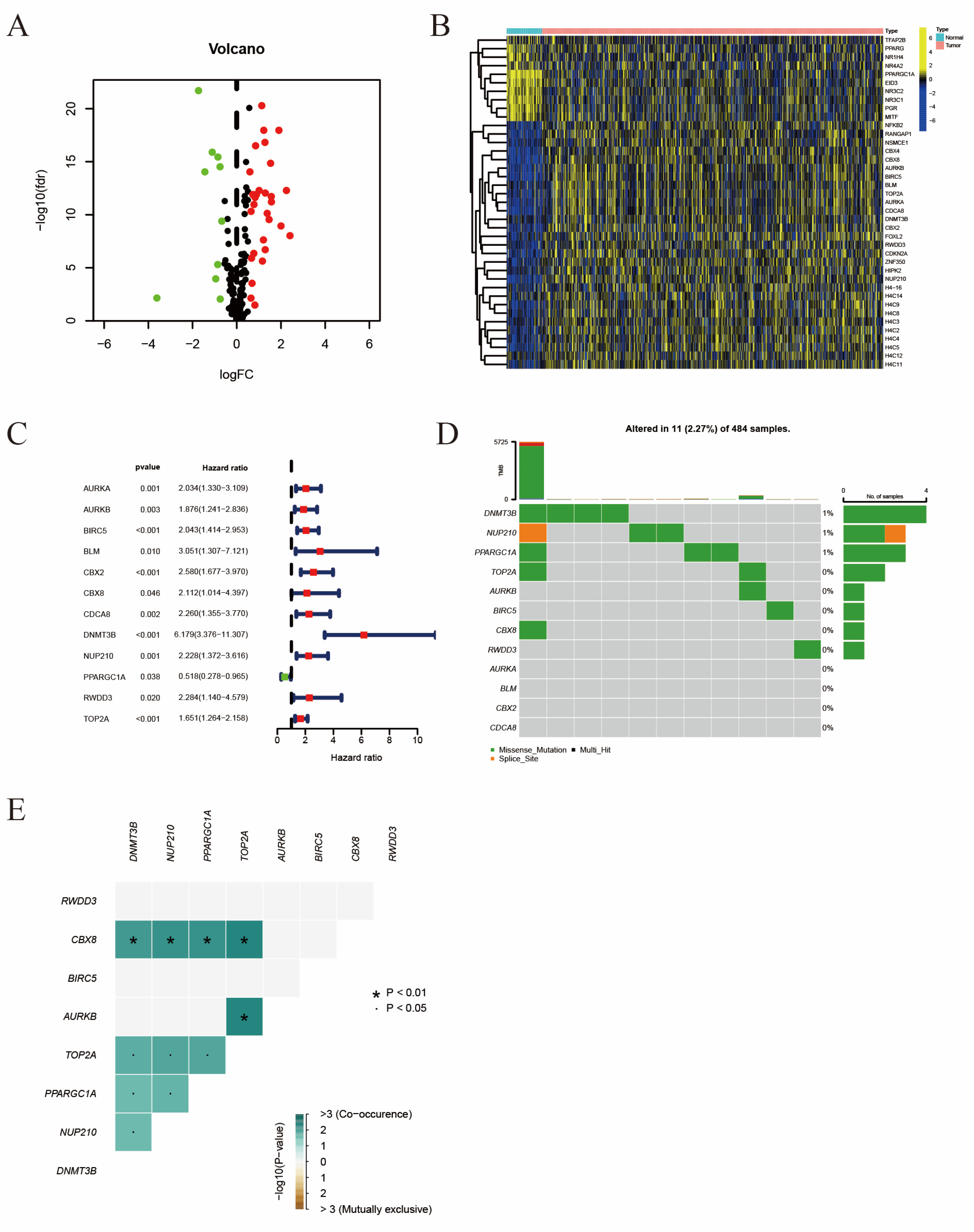
2.1. The Identification of Differentially Expressed Prognosis Related SRGs
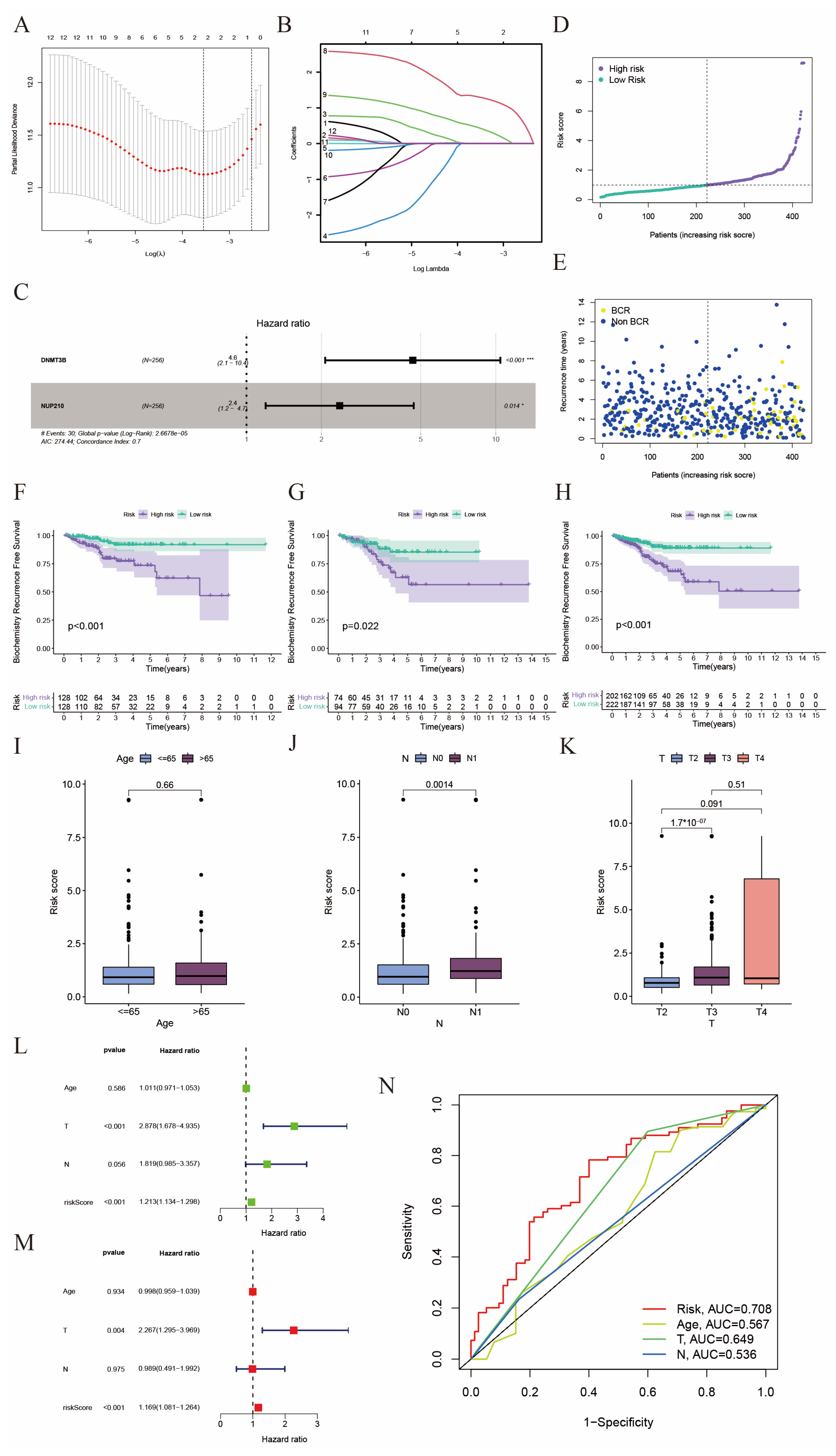
2.2. Construction of a Prognostic Model in the TCGA Cohort
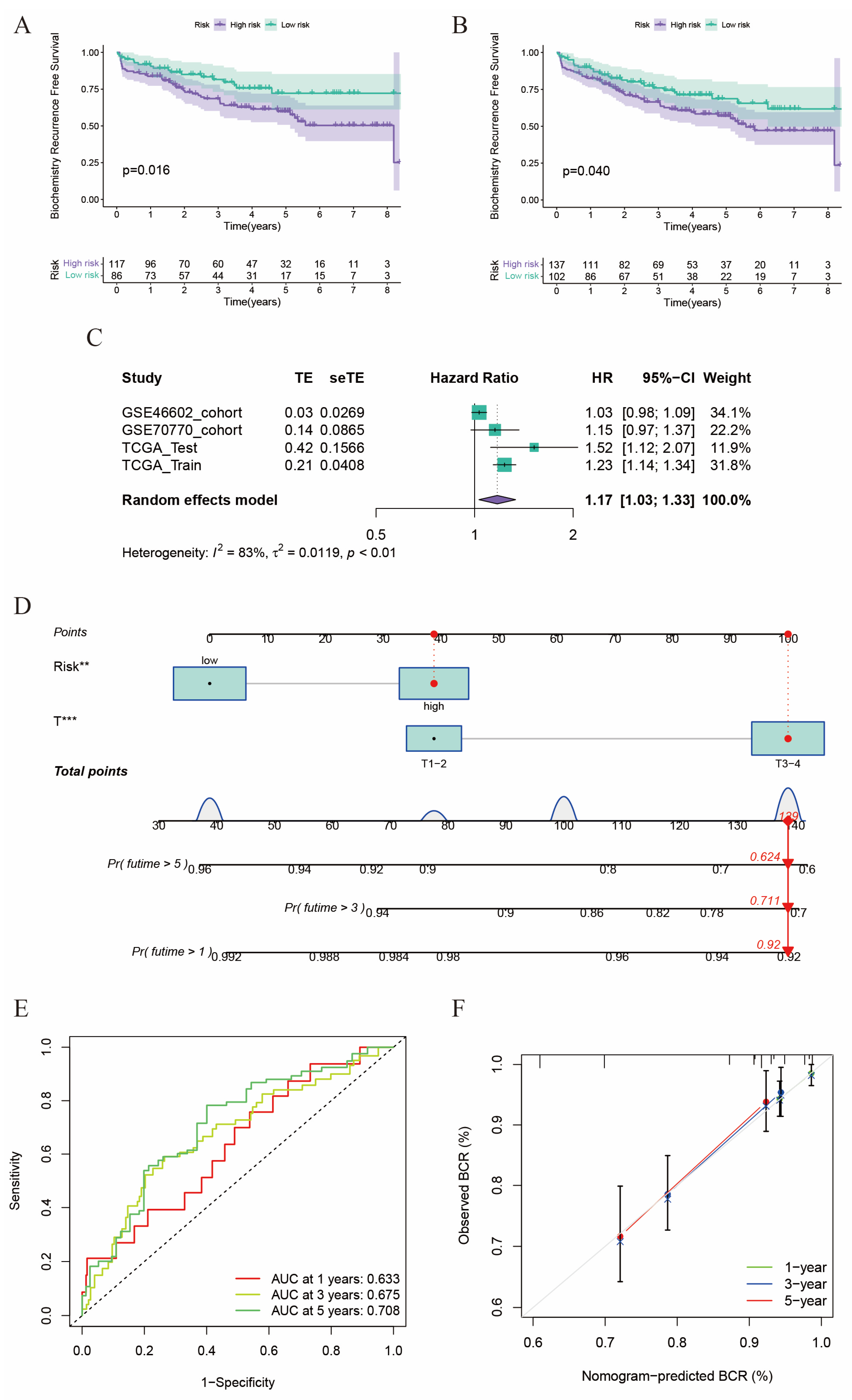
2.3. Validation of the Risk Score in GEO Cohorts
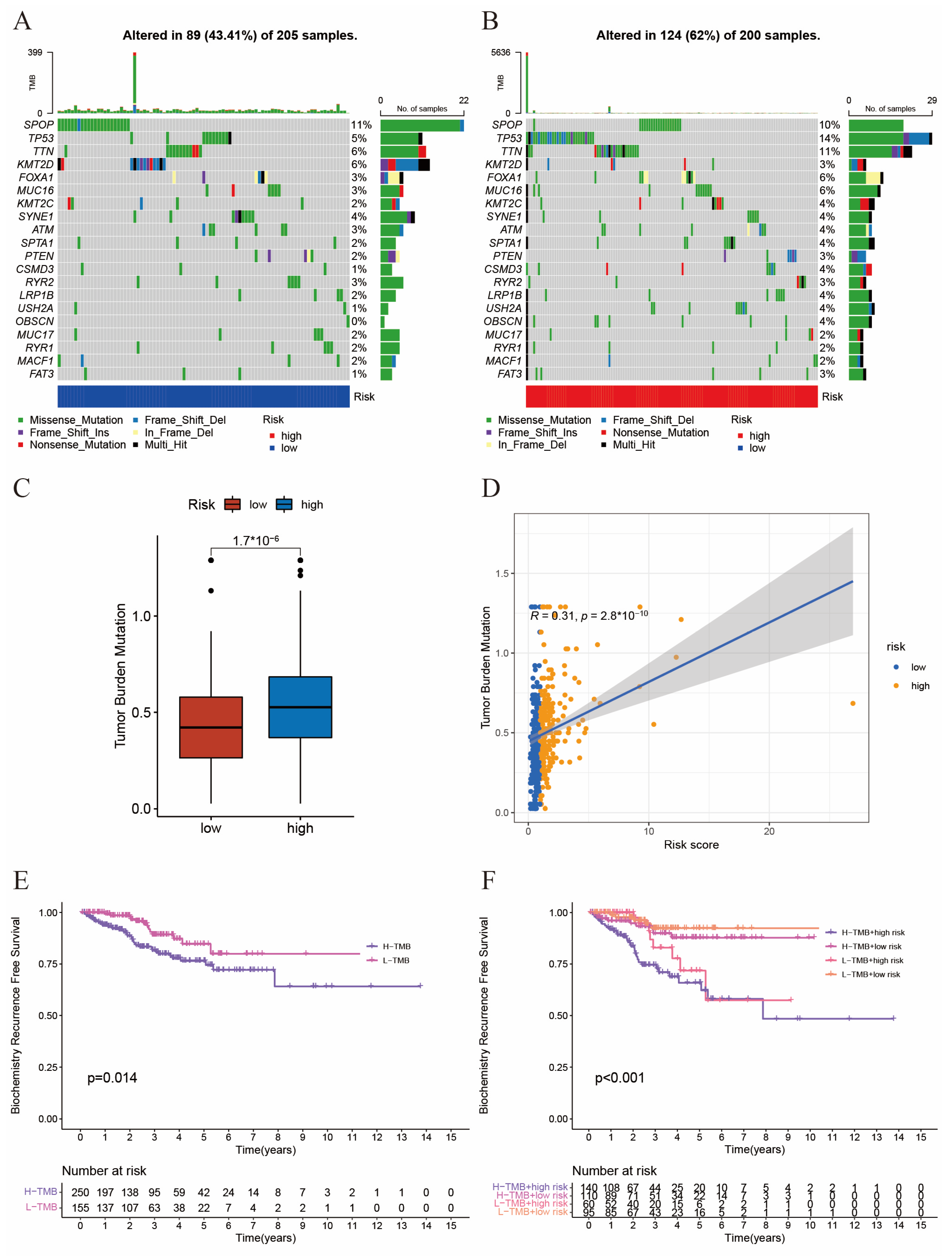
2.4. The Interaction between Tumor Mutation Burden and Risk Score
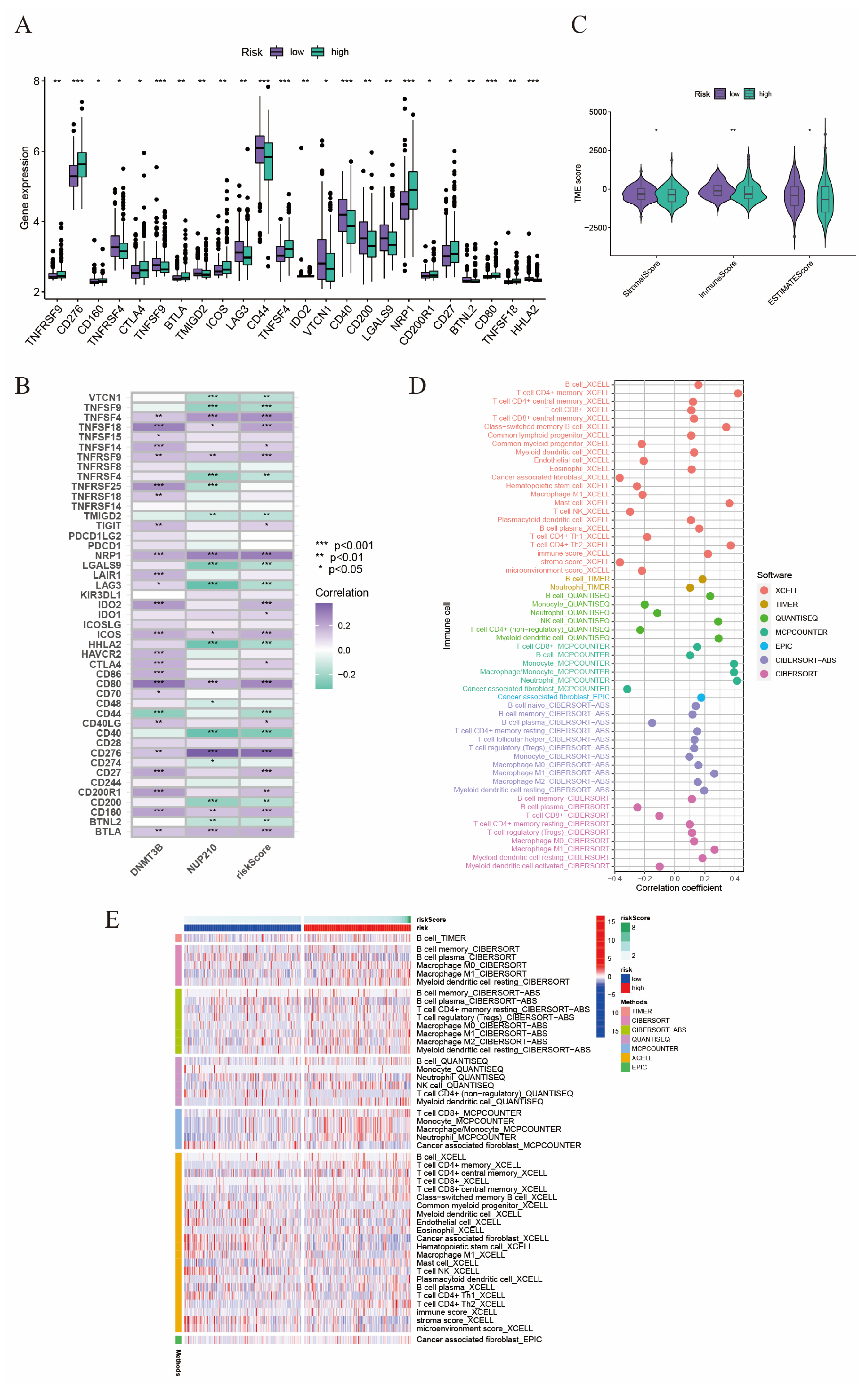
2.5. The Relationship between Tumor Microenvironment and Risk Score
2.6. Drug Sensitivity Analyses in Immunotherapy and Chemotherapy
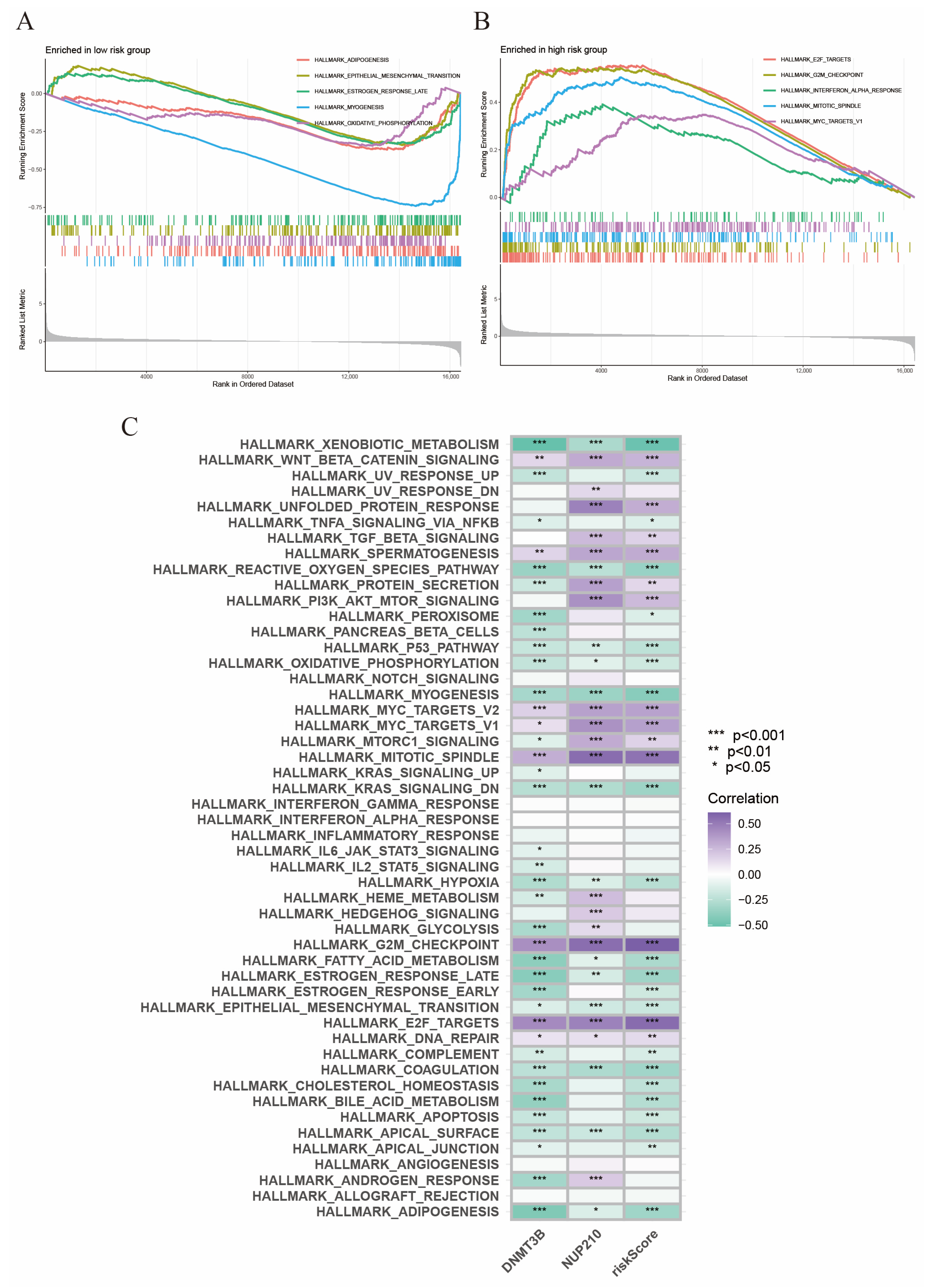
2.7. Gene Set Enrichment Analysis
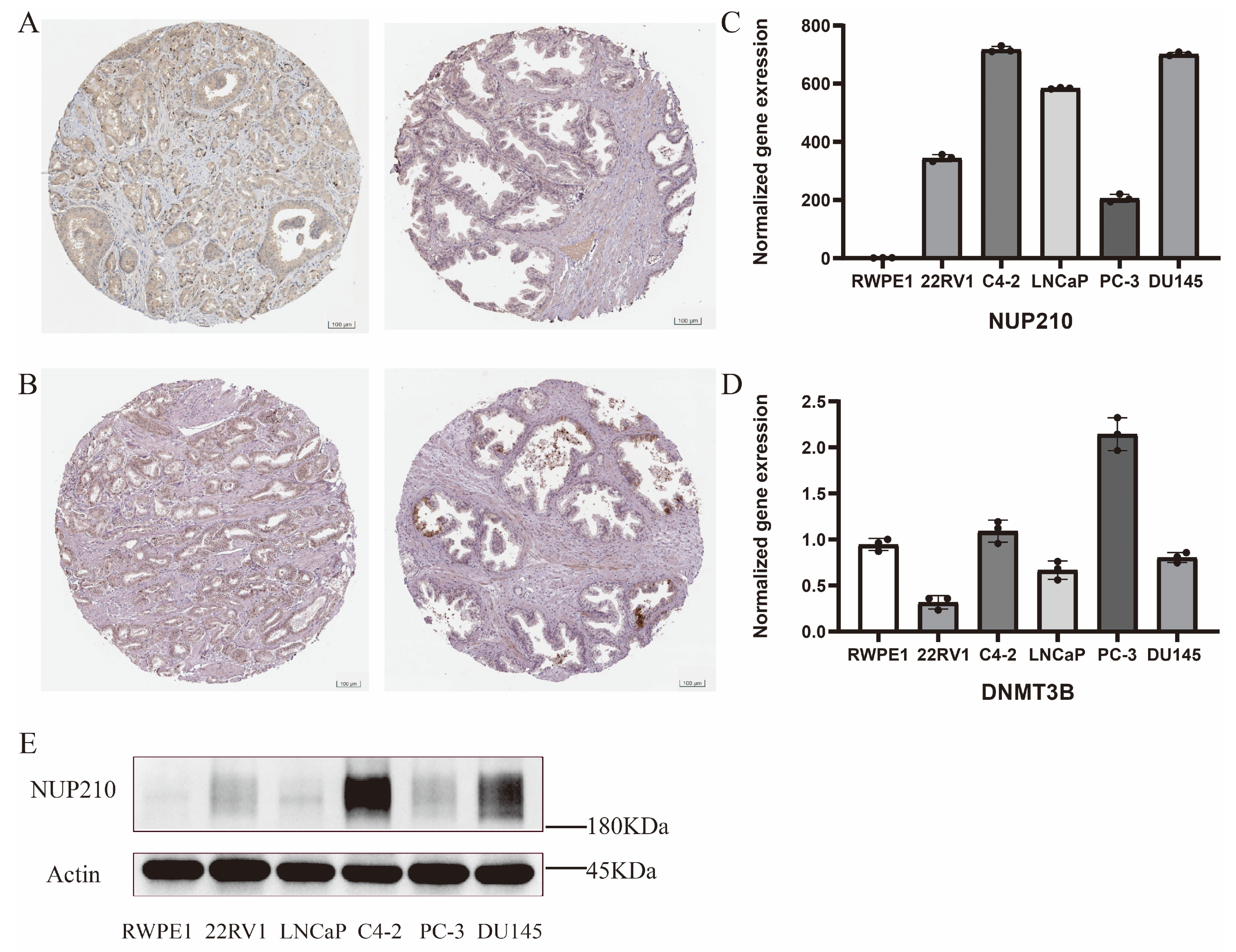
2.8. Experimental Verification of Two Vital SRGs
3. Discussion
4. Materials and Methods
4.1. Cell Culture and the Extraction of RNA and Protein
4.2. RT-qPCR, Western Blotting and Immunohistochemical Stain
4.3. Data Sources
4.4. Identification of Differentially Expressed SRGs
4.5. Construction and Validation of SUMOylation Risk Signature
4.6. Tumor Mutation Burden, TME Infiltration, and Drug Response
4.7. Gene Set Variation Analysis (GSVA) and Gene Set Enrichment Analysis (GSEA)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Culp, M.B.; Soerjomataram, I.; Efstathiou, J.A.; Bray, F.; Jemal, A. Recent Global Patterns in Prostate Cancer Incidence and Mortality Rates. Eur. Urol. 2020, 77, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Van den Broeck, T.; van den Bergh, R.C.N.; Arfi, N.; Gross, T.; Moris, L.; Briers, E.; Cumberbatch, M.; De Santis, M.; Tilki, D.; Fanti, S.; et al. Prognostic Value of Biochemical Recurrence Following Treatment with Curative Intent for Prostate Cancer: A Systematic Review. Eur. Urol. 2018, 75, 967–987. [Google Scholar] [CrossRef]
- Venne, A.S.; Kollipara, L.; Zahedi, R.P. The next level of complexity: Crosstalk of posttranslational modifications. Proteomics 2014, 14, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.J.; Feng, Y.H.; Gu, B.H.; Li, Y.M.; Chen, H. The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 2018, 52, 1081–1094. [Google Scholar] [CrossRef]
- Vertegaal, A.C.O. Signalling mechanisms and cellular functions of SUMO. Nat. Rev. Mol. Cell Biol. 2022, 23, 715–731. [Google Scholar] [CrossRef] [PubMed]
- Owerbach, D.; McKay, E.M.; Yeh, E.T.; Gabbay, K.H.; Bohren, K.M. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 2005, 337, 517–520. [Google Scholar] [CrossRef]
- Wang, Y.; Dasso, M. SUMOylation and deSUMOylation at a glance. J. Cell Sci. 2009, 122, 4249–4252. [Google Scholar] [CrossRef]
- Ochaba, J.; Monteys, A.M.; O’rourke, J.G.; Reidling, J.C.; Steffan, J.S.; Davidson, B.L.; Thompson, L.M. PIAS1 Regulates Mutant Huntingtin Accumulation and Huntington’s Disease-Associated Phenotypes In Vivo. Neuron 2016, 90, 507–520. [Google Scholar] [CrossRef]
- Gupta, M.K.; Gulick, J.; Liu, R.; Wang, X.; Molkentin, J.D.; Robbins, J. Sumo E2 enzyme UBC9 is required for efficient protein quality control in cardiomyocytes. Circ. Res. 2014, 115, 721–729. [Google Scholar] [CrossRef]
- Du, L.; Li, Y.-J.; Fakih, M.; Wiatrek, R.L.; Duldulao, M.; Chen, Z.; Chu, P.; Garcia-Aguilar, J.; Chen, Y. Role of SUMO activating enzyme in cancer stem cell maintenance and self-renewal. Nat. Commun. 2016, 7, 12326. [Google Scholar] [CrossRef] [PubMed]
- Seeler, J.-S.; Dejean, A. SUMO and the robustness of cancer. Nat. Rev. Cancer 2017, 17, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Eifler, K.; Vertegaal, A.C. SUMOylation-Mediated Regulation of Cell Cycle Progression and Cancer. Trends Biochem. Sci. 2015, 40, 779–793. [Google Scholar] [CrossRef]
- Moschos, S.J.; Jukic, D.M.; Athanassiou, C.; Bhargava, R.; Dacic, S.; Wang, X.; Kuan, S.-F.; Fayewicz, S.L.; Galambos, C.; Acquafondata, M.; et al. Expression analysis of Ubc9, the single small ubiquitin-like modifier (SUMO) E2 conjugating enzyme, in normal and malignant tissues. Hum. Pathol. 2010, 41, 1286–1298. [Google Scholar] [CrossRef] [PubMed]
- McDoniels-Silvers, A.L.; Nimri, C.F.; Stoner, G.D.; Lubet, R.A.; You, M. Differential gene expression in human lung adenocarcinomas and squamous cell carcinomas. Clin. Cancer Res. 2002, 8, 1127–1138. [Google Scholar]
- Tomasi, M.L.; Tomasi, I.; Ramani, K.; Pascale, R.M.; Xu, J.; Giordano, P.; Mato, J.M.; Lu, S.C. S-adenosyl methionine regulates ubiquitin-conjugating enzyme 9 protein expression and sumoylation in murine liver and human cancers. Hepatology 2012, 56, 982–993. [Google Scholar] [CrossRef]
- Xiao, J.; Sun, F.; Wang, Y.-N.; Liu, B.; Zhou, P.; Wang, F.-X.; Zhou, H.-F.; Ge, Y.; Yue, T.-T.; Luo, J.-H.; et al. UBC9 deficiency enhances immunostimulatory macrophage activation and subsequent antitumor T cell response in prostate cancer. J. Clin. Investig. 2023, 133, e158352. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.-D.; Sun, J.-X.; Xun, Y.; Xiao, J.; Liu, C.-Q.; Xu, J.-Z.; An, Y.; Xu, M.-Y.; Liu, Z.; Wang, S.-G.; et al. SUMOylation Pattern Predicts Prognosis and Indicates Tumor Microenvironment Infiltration Characterization in Bladder Cancer. Front. Immunol. 2022, 13, 864156. [Google Scholar] [CrossRef]
- Kalantari, E.; Saadi, F.H.; Asgari, M.; Shariftabrizi, A.; Roudi, R.; Madjd, Z. Increased Expression of ALDH1A1 in Prostate Cancer is Correlated With Tumor Aggressiveness: A Tissue Microarray Study of Iranian Patients. Appl. Immunohistochem. Mol. Morphol. 2017, 25, 592–598. [Google Scholar] [CrossRef]
- Foroozan, M.; Roudi, R.; Abolhasani, M.; Gheytanchi, E.; Mehrazma, M. Clinical significance of endothelial cell marker CD34 and mast cell marker CD117 in prostate adenocarcinoma. Pathol. Res. Pract. 2017, 213, 612–618. [Google Scholar] [CrossRef]
- Sternsdorf, T.; Jensen, K.; Will, H. Evidence for covalent modification of the nuclear dot–associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 1997, 139, 1621–1634. [Google Scholar] [CrossRef]
- Tatham, M.H.; Geoffroy, M.C.; Shen, L.; Plechanovova, A.; Hattersley, N.; Jaffray, E.G.; Hay, R.T. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008, 10, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Lallemand-Breitenbach, V.; Jeanne, M.; Benhenda, S.; Nasr, R.; Lei, M.; Peres, L.; de Thé, H. Arsenic degrades PML or PML–RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 2008, 10, 547–555. [Google Scholar] [CrossRef]
- Guo, C.; Henley, J.M. Wrestling with stress: Roles of protein SUMOylation and deSUMOylation in cell stress response. IUBMB Life 2014, 66, 71–77. [Google Scholar] [CrossRef]
- Saitoh, H.; Hinchey, J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000, 275, 6252–6258. [Google Scholar] [CrossRef] [PubMed]
- Bossis, G.; Melchior, F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell 2006, 21, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, J. Dissecting multiple roles of SUMOylation in prostate cancer. Cancer Lett. 2021, 521, 88–97. [Google Scholar] [CrossRef]
- Ashikari, D.; Takayama, K.; Tanaka, T.; Suzuki, Y.; Obinata, D.; Fujimura, T.; Urano, T.; Takahashi, S.; Inoue, S. Androgen induces G3BP2 and SUMO-mediated p53 nuclear export in prostate cancer. Oncogene 2017, 36, 6272–6281. [Google Scholar] [CrossRef]
- Lin, W.; Luo, J.; Sun, Y.; Lin, C.; Li, G.; Niu, Y.; Chang, C. ASC-J9® suppresses prostate cancer cell invasion via altering the sumoylation-phosphorylation of STAT3. Cancer Lett. 2018, 425, 21–30. [Google Scholar] [CrossRef]
- Poukka, H.; Karvonen, U.; Jänne, O.A.; Palvimo, J.J. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl. Acad. Sci. USA 2000, 97, 14145–14150. [Google Scholar] [CrossRef]
- Mukherjee, S.; Cruz-Rodríguez, O.; Bolton, E.; Iñiguez-Lluhí, J.A. The in vivo role of androgen receptor SUMOylation as revealed by androgen insensitivity syndrome and prostate cancer mutations targeting the proline/glycine residues of synergy control motifs. J. Biol. Chem. 2012, 287, 31195–31206. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Ishii, K.; Mirosevich, J.; Kuwajima, S.; Oppenheimer, S.R.; Roberts, R.L.; Jiang, M.; Yu, X.; Shappell, S.B.; Caprioli, R.M.; et al. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development 2005, 132, 3431–3443. [Google Scholar] [CrossRef] [PubMed]
- Sutinen, P.; Rahkama, V.; Rytinki, M.; Palvimo, J.J. Nuclear mobility and activity of FOXA1 with androgen receptor are regulated by SUMOylation. Mol. Endocrinol. 2014, 28, 1719–1728. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Swatkoski, S.; Holly, A.; Lee, L.C.; Giroux, V.; Lee, C.-S.; Hsu, D.; Smith, J.L.; Yuen, G.; Yue, J.; et al. Oncogenesis driven by the Ras/Raf pathway requires the SUMO E2 ligase Ubc9. Proc. Natl. Acad. Sci. USA 2015, 112, E1724–E1733. [Google Scholar] [CrossRef] [PubMed]
- Kessler, J.D.; Kahle, K.T.; Sun, T.; Meerbrey, K.L.; Schlabach, M.R.; Schmitt, E.M.; Skinner, S.O.; Xu, Q.; Li, M.Z.; Hartman, Z.C.; et al. A SUMOylation-dependent transcriptional subprogram is required for myc-driven tumorigenesis. Science 2012, 335, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Dünnebier, T.; Bermejo, J.L.; Haas, S.; Fischer, H.-P.; Pierl, C.B.; Justenhoven, C.; Brauch, H.; Baisch, C.; Gilbert, M.; Harth, V.; et al. Polymorphisms in the UBC9 and PIAS3 genes of the SUMO-conjugating system and breast cancer risk. Breast Cancer Res. Treat. 2009, 121, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Dünnebier, T.; Bermejo, J.L.; Haas, S.; Fischer, H.-P.; Pierl, C.B.; Justenhoven, C.; Brauch, H.; Baisch, C.; Gilbert, M.; Harth, V.; et al. Common variants in the UBC9 gene encoding the SUMO-conjugating enzyme are associated with breast tumor grade. Int. J. Cancer 2009, 125, 596–602. [Google Scholar] [CrossRef]
- Zhang, H.; Ying, H.; Wang, X. Methyltransferase DNMT3B in leukemia. Leuk. Lymphoma 2020, 61, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.C.; Su, Y.T.; Chi, C.C.; Kuo, Y.C.; Lee, K.F.; Wu, Y.C.; Huang, Y.H. DNMT3b/OCT4 expression confers sorafenib resistance and poor prognosis of hepatocellular carcinoma through IL-6/STAT3 regulation. J. Exp. Clin. Cancer Res. 2019, 38, 474. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Chen, B.; Li, B.; Li, C.; Zhang, Y.; Jiang, N.; Lang, B. DNMT3B silencing suppresses migration and invasion by epigenetically promoting miR-34a in bladder cancer. Aging 2020, 12, 23668–23683. [Google Scholar] [CrossRef]
- Zhu, X.; Xue, C.; Kang, X.; Jia, X.; Wang, L.; Younis, M.H.; Liu, D.; Huo, N.; Han, Y.; Chen, Z.; et al. DNMT3B-mediated FAM111B methylation promotes papillary thyroid tumor glycolysis, growth and metastasis. Int. J. Biol. Sci. 2022, 18, 4372–4387. [Google Scholar] [CrossRef]
- Schulze, I.; Rohde, C.; Scheller-Wendorff, M.; Bäumer, N.; Krause, A.; Herbst, F.; Riemke, P.; Hebestreit, K.; Tschanter, P.; Lin, Q.; et al. Increased DNA methylation of Dnmt3b targets impairs leukemogenesis. Blood 2016, 127, 1575–1586. [Google Scholar] [CrossRef][Green Version]
- Zheng, Y.; Zhang, H.; Wang, Y.; Li, X.; Lu, P.; Dong, F.; Pang, Y.; Ma, S.; Cheng, H.; Hao, S.; et al. Loss of Dnmt3b accelerates MLL-AF9 leukemia progression. Leukemia 2016, 30, 2373–2384. [Google Scholar] [CrossRef]
- Singal, R.; Das, P.M.; Manoharan, M.; Reis, I.M.; Schlesselman, J.J. Polymorphisms in the DNA methyltransferase 3b gene and prostate cancer risk. Oncol. Rep. 2005, 14, 569–573. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sugiura, M.; Sato, H.; Okabe, A.; Fukuyo, M.; Mano, Y.; Shinohara, K.-I.; Rahmutulla, B.; Higuchi, K.; Maimaiti, M.; Kanesaka, M.; et al. Identification of AR-V7 downstream genes commonly targeted by AR/AR-V7 and specifically targeted by AR-V7 in castration resistant prostate cancer. Transl. Oncol. 2021, 14, 100915. [Google Scholar] [CrossRef] [PubMed]
- Borlido, J.; Sakuma, S.; Raices, M.; Carrette, F.; Tinoco, R.; Bradley, L.M.; D’angelo, M.A. Nuclear pore complex-mediated modulation of TCR signaling is required for naïve CD4+ T cell homeostasis. Nat. Immunol. 2018, 19, 594–605. [Google Scholar] [CrossRef]
- Chan, T.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 2018, 30, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 Mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2009, 2, a001008. [Google Scholar] [CrossRef]
- Stehmeier, P.; Muller, S. Regulation of p53 family members by the ubiquitin-like SUMO system. DNA Repair 2009, 8, 491–498. [Google Scholar] [CrossRef]
- Yang, S.; Wei, W.; Zhao, Q. B7-H3, a checkpoint molecule, as a target for cancer immunotherapy. Int. J. Biol. Sci. 2020, 16, 1767–1773. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, E.S.; Piulats, J.M.; Gross-Goupil, M.; Goh, J.; Ojamaa, K.; Hoimes, C.J.; Vaishampayan, U.; Berger, R.; Sezer, A.; Alanko, T.; et al. Pembrolizumab for Treatment-Refractory Metastatic Castration-Resistant Prostate Cancer: Multicohort, Open-Label Phase II KEYNOTE-199 Study. J. Clin. Oncol. 2020, 38, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Madden, E.; Logue, S.E.; Healy, S.J.; Manie, S.; Samali, A. The role of the unfolded protein response in cancer progression: From oncogenesis to chemoresistance. Biol. Cell 2019, 111, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Kwan, P.W.; Shi, D. Sparse kernel learning with LASSO and Bayesian inference algorithm. Neural Netw. 2010, 23, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef]

| Overall | TCGA_Train | TCGA_Test | GSE46602 | GSE70770 | p | |
|---|---|---|---|---|---|---|
| n | 663 | 256 | 168 | 36 | 203 | |
| Age (%) | <0.001 | |||||
| 41 | 2 (0.3) | 1 (0.4) | 0 (0.0) | 0 (0.0) | 1 (0.5) | |
| 42 | 1 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | |
| 43 | 1 (0.2) | 1 (0.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 44 | 3 (0.5) | 1 (0.4) | 1 (0.6) | 0 (0.0) | 1 (0.5) | |
| 46 | 6 (0.9) | 5 (2.0) | 0 (0.0) | 1 (2.8) | 0 (0.0) | |
| 47 | 6 (0.9) | 4 (1.6) | 1 (0.6) | 0 (0.0) | 1 (0.5) | |
| 48 | 6 (0.9) | 3 (1.2) | 1 (0.6) | 0 (0.0) | 2 (1.0) | |
| 49 | 6 (0.9) | 5 (2.0) | 1 (0.6) | 0 (0.0) | 0 (0.0) | |
| 50 | 9 (1.4) | 4 (1.6) | 2 (1.2) | 0 (0.0) | 3 (1.5) | |
| 51 | 11 (1.7) | 6 (2.3) | 3 (1.8) | 0 (0.0) | 2 (1.0) | |
| 52 | 11 (1.7) | 7 (2.7) | 1 (0.6) | 1 (2.8) | 2 (1.0) | |
| 53 | 16 (2.4) | 8 (3.1) | 6 (3.6) | 1 (2.8) | 1 (0.5) | |
| 54 | 18 (2.7) | 4 (1.6) | 9 (5.4) | 0 (0.0) | 5 (2.5) | |
| 55 | 25 (3.8) | 10 (3.9) | 8 (4.8) | 1 (2.8) | 6 (3.0) | |
| 56 | 23 (3.5) | 9 (3.5) | 9 (5.4) | 0 (0.0) | 5 (2.5) | |
| 57 | 35 (5.3) | 16 (6.2) | 11 (6.5) | 3 (8.3) | 5 (2.5) | |
| 58 | 26 (3.9) | 12 (4.7) | 7 (4.2) | 3 (8.3) | 4 (2.0) | |
| 59 | 26 (3.9) | 9 (3.5) | 9 (5.4) | 4 (11.1) | 4 (2.0) | |
| 60 | 22 (3.3) | 8 (3.1) | 8 (4.8) | 1 (2.8) | 5 (2.5) | |
| 61 | 32 (4.8) | 15 (5.9) | 9 (5.4) | 1 (2.8) | 7 (3.4) | |
| 62 | 33 (5.0) | 13 (5.1) | 8 (4.8) | 1 (2.8) | 11 (5.4) | |
| 63 | 38 (5.7) | 14 (5.5) | 11 (6.5) | 4 (11.1) | 9 (4.4) | |
| 64 | 32 (4.8) | 13 (5.1) | 12 (7.1) | 1 (2.8) | 6 (3.0) | |
| 65 | 26 (3.9) | 13 (5.1) | 7 (4.2) | 1 (2.8) | 5 (2.5) | |
| 66 | 33 (5.0) | 18 (7.0) | 11 (6.5) | 0 (0.0) | 4 (2.0) | |
| 67 | 29 (4.4) | 13 (5.1) | 5 (3.0) | 3 (8.3) | 8 (3.9) | |
| 68 | 29 (4.4) | 14 (5.5) | 8 (4.8) | 6 (16.7) | 1 (0.5) | |
| 69 | 19 (2.9) | 7 (2.7) | 4 (2.4) | 3 (8.3) | 5 (2.5) | |
| 70 | 12 (1.8) | 5 (2.0) | 5 (3.0) | 0 (0.0) | 2 (1.0) | |
| 71 | 10 (1.5) | 6 (2.3) | 2 (1.2) | 1 (2.8) | 1 (0.5) | |
| 72 | 11 (1.7) | 4 (1.6) | 5 (3.0) | 0 (0.0) | 2 (1.0) | |
| 73 | 6 (0.9) | 1 (0.4) | 3 (1.8) | 0 (0.0) | 2 (1.0) | |
| 74 | 2 (0.3) | 1 (0.4) | 1 (0.6) | 0 (0.0) | 0 (0.0) | |
| 75 | 3 (0.5) | 3 (1.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 76 | 1 (0.2) | 1 (0.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 77 | 1 (0.2) | 1 (0.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 78 | 1 (0.2) | 1 (0.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| unknow | 92 (13.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 92 (45.3) | |
| T (%) | 0.386 | |||||
| T1–2 | 253 (38.2) | 97 (37.9) | 56 (33.3) | 19 (52.8) | 81 (39.9) | |
| T3–4 | 402 (60.6) | 157 (61.3) | 109 (64.9) | 17 (47.2) | 119 (58.6) | |
| unknow | 8 (1.2) | 2 (0.8) | 3 (1.8) | 0 (0.0) | 3 (1.5) | |
| BCR_status = BCR/non-BCR (%) | 142/521 (21.4/78.6) | 30/226 (11.7/88.3) | 26/142 (15.5/84.5) | 22/14 (61.1/38.9) | 64/139 (31.5/68.5) | <0.001 |
| DNMT3B (mean (SD)) | 2.71 (0.28) | 2.69 (0.28) | 2.71 (0.27) | 2.72 (0.36) | 2.72 (0.26) | 0.601 |
| NUP210 (mean (SD)) | 4.88 (0.54) | 4.86 (0.55) | 4.74 (0.56) | 5.07 (0.38) | 5.00 (0.48) | <0.001 |
| riskScore (median [IQR]) | 1.01 [0.69, 1.52] | 1.00 [0.62, 1.48] | 0.91 [0.58, 1.47] | 1.09 [0.83, 1.48] | 1.11 [0.81, 1.56] | 0.002 |
| Risk = high/low (%) | 339/324 (51.1/48.9) | 128/128 (50.0/50.0) | 74/94 (44.0/56.0) | 20/16 (55.6/44.4) | 117/86 (57.6/42.4) | 0.065 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.-X.; An, Y.; Xiang, J.-C.; Xu, J.-Z.; Hu, J.; Wang, S.-G.; Xia, Q.-D. The Prognosis-Predictive and Immunoregulatory Role of SUMOylation Related Genes: Potential Novel Targets in Prostate Cancer Treatment. Int. J. Mol. Sci. 2023, 24, 13603. https://doi.org/10.3390/ijms241713603
Sun J-X, An Y, Xiang J-C, Xu J-Z, Hu J, Wang S-G, Xia Q-D. The Prognosis-Predictive and Immunoregulatory Role of SUMOylation Related Genes: Potential Novel Targets in Prostate Cancer Treatment. International Journal of Molecular Sciences. 2023; 24(17):13603. https://doi.org/10.3390/ijms241713603
Chicago/Turabian StyleSun, Jian-Xuan, Ye An, Jia-Cheng Xiang, Jin-Zhou Xu, Jia Hu, Shao-Gang Wang, and Qi-Dong Xia. 2023. "The Prognosis-Predictive and Immunoregulatory Role of SUMOylation Related Genes: Potential Novel Targets in Prostate Cancer Treatment" International Journal of Molecular Sciences 24, no. 17: 13603. https://doi.org/10.3390/ijms241713603
APA StyleSun, J.-X., An, Y., Xiang, J.-C., Xu, J.-Z., Hu, J., Wang, S.-G., & Xia, Q.-D. (2023). The Prognosis-Predictive and Immunoregulatory Role of SUMOylation Related Genes: Potential Novel Targets in Prostate Cancer Treatment. International Journal of Molecular Sciences, 24(17), 13603. https://doi.org/10.3390/ijms241713603






