TRIM67 Implicates in Regulating the Homeostasis and Synaptic Development of Mitral Cells in the Olfactory Bulb
Abstract
:1. Introduction
2. Results
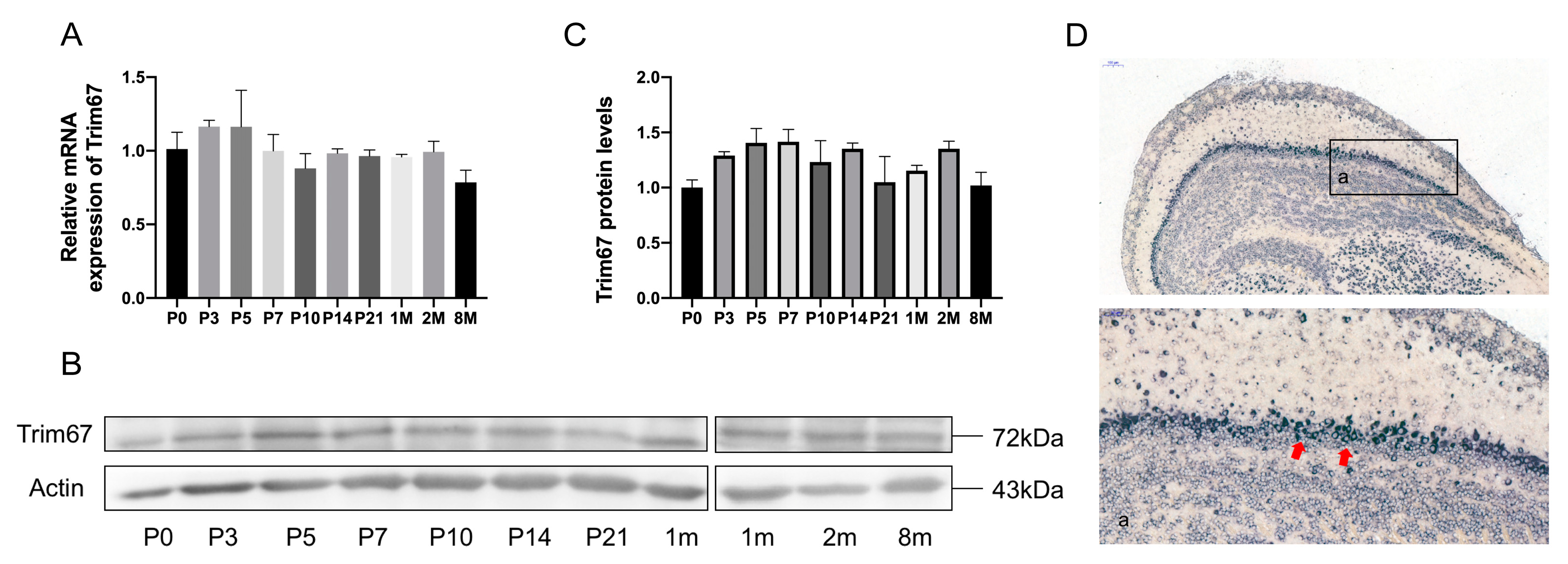
2.1. TRIM67 Highly Concentrates in the Mitral Cell Layer of the Olfactory Bulb
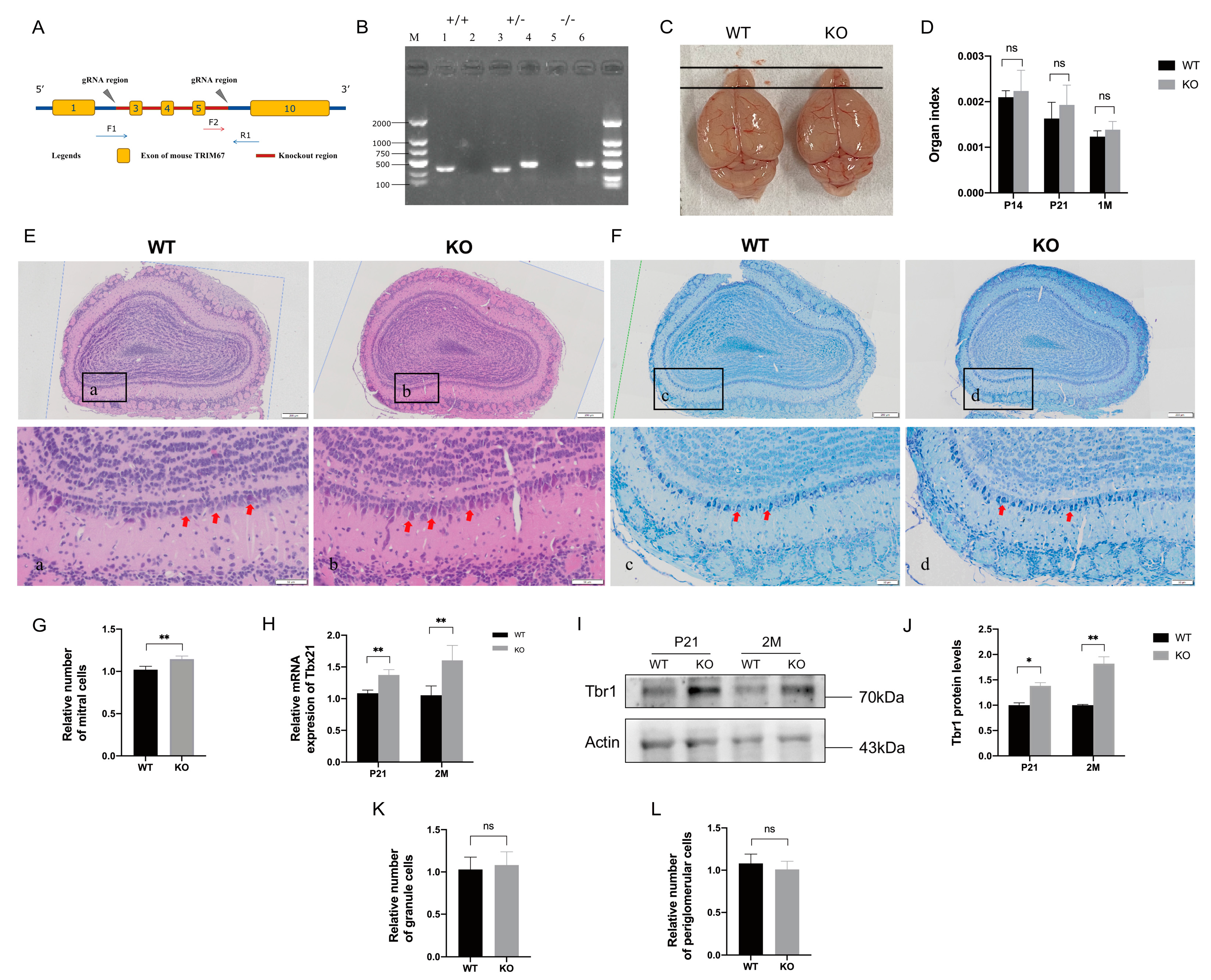
2.2. TRIM67 KO Affects the Homeostasis of Mitral Cells in the Olfactory Bulb
2.3. TRIM67 Affects the Proliferation of Mitral Cells in Embryonic Mouse
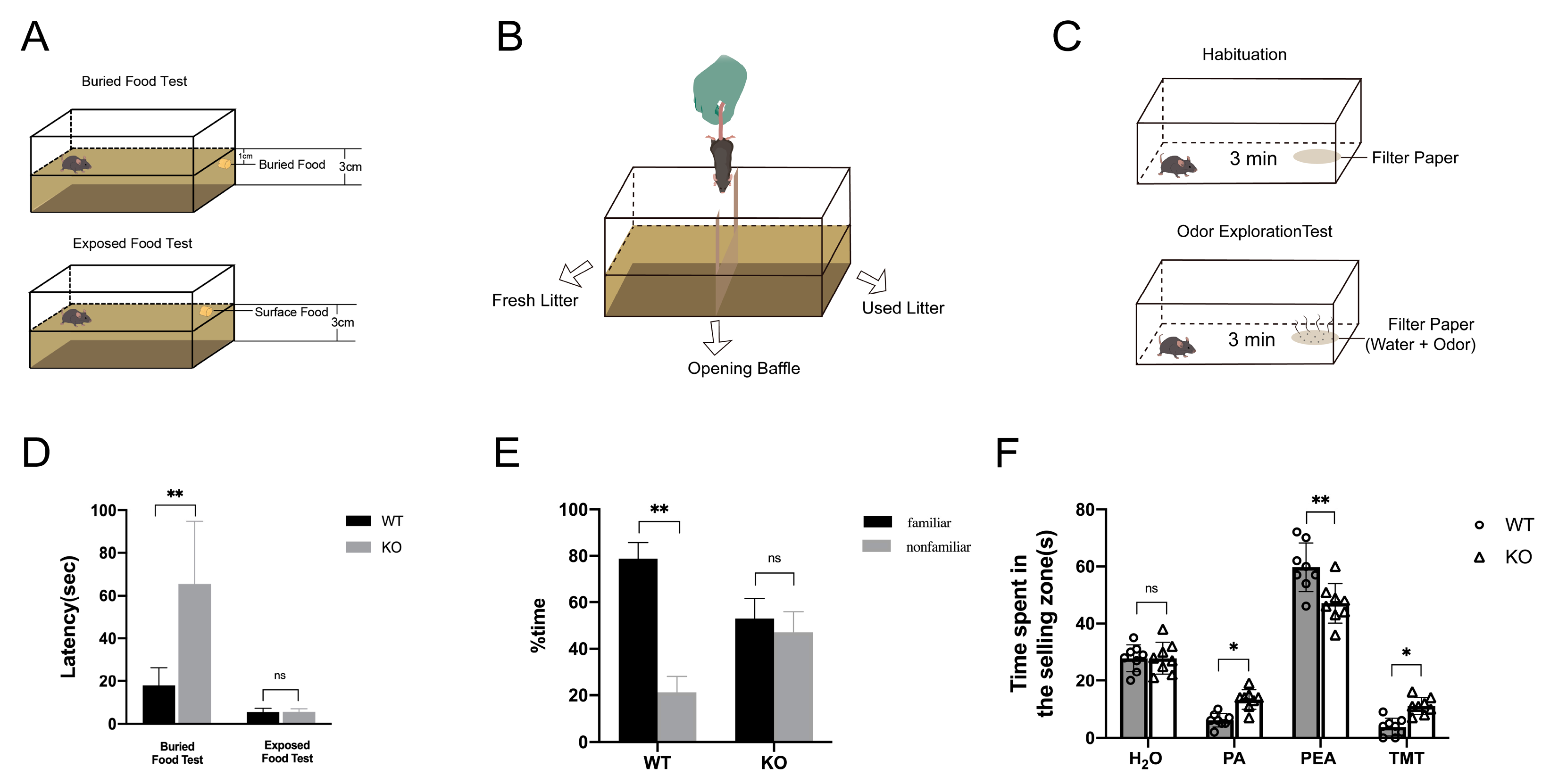
2.4. Loss of TRIM67 Causes Olfactory Dysfunction in Mice
2.5. Impaired Synapse Formation in Mitral Cells of TRIM67 KO Mice
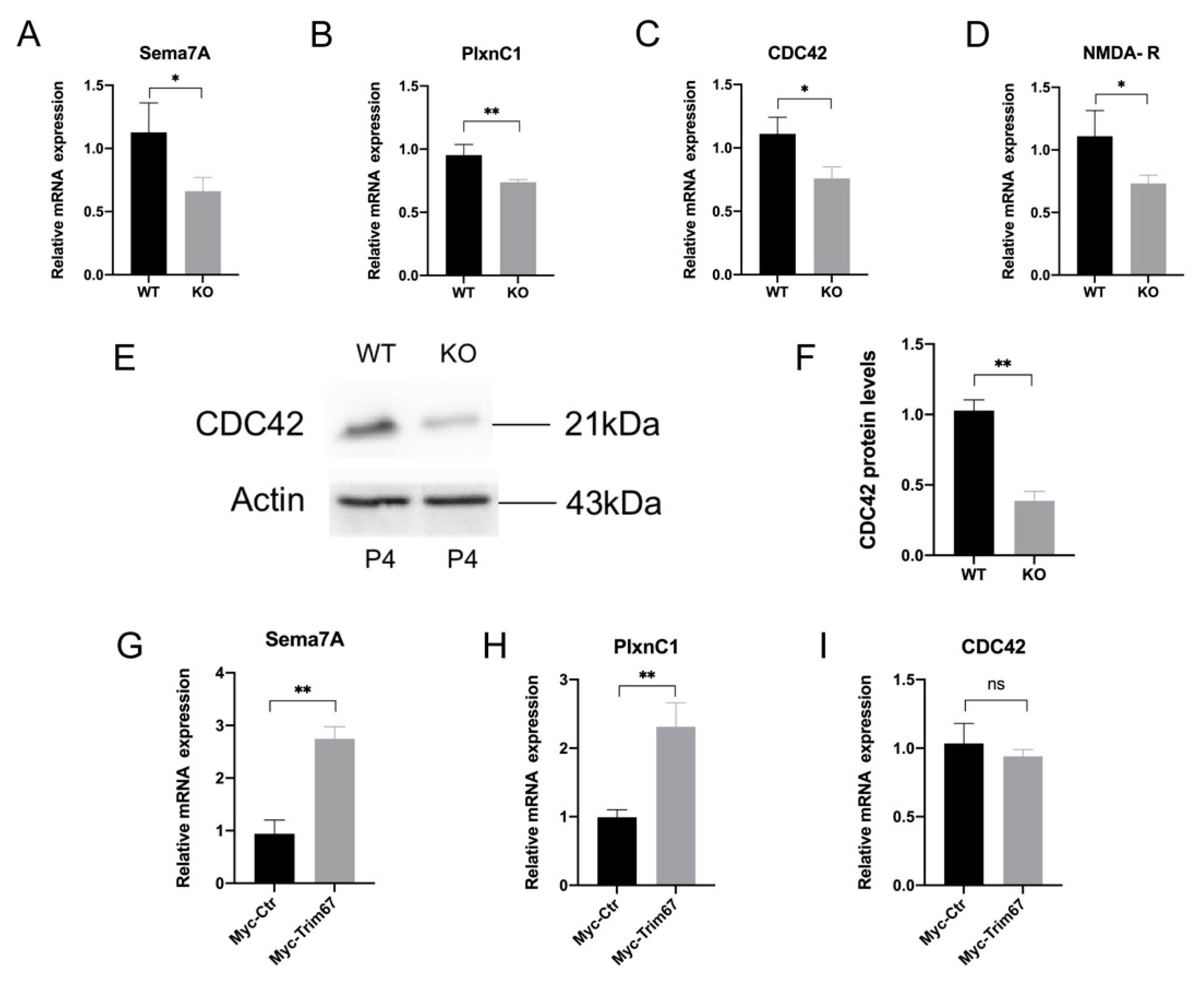
2.6. Sema7A/PlxnC1 Signaling Implicates in TRIM67-Regulated Synapse Development of Mitral Cells
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. RT-qPCR
4.3. Western Blots
4.4. Histological Staining
4.5. Immunohistochemical Staining
4.6. Immunofluorescence Staining
4.7. EDU Labeling Experiment
4.8. Golgi Staining
4.9. Electron Microscope Experiment
4.10. Food Burial Experiment
4.11. Odor Probing Test
4.12. Odor Discrimination Test
4.13. Cell Culture
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brunert, D.; Rothermel, M. Extrinsic neuromodulation in the rodent olfactory bulb. Cell Tissue Res. 2021, 383, 507–524. [Google Scholar] [CrossRef] [PubMed]
- Graven, S.N.; Browne, J.V. Sensory development in the fetus, neonate, and infant: Introduction and overview. Newborn Infant Nurs. Rev. 2008, 8, 169–172. [Google Scholar] [CrossRef]
- Hinds, J.W. Autoradiographic study of histogenesis in the mouse olfactory bulb. I. Time of origin of neurons and neuroglia. J. Comp. Neurol. 1968, 134, 287–304. [Google Scholar] [CrossRef] [PubMed]
- Sabiniewicz, A.; Hoffmann, L.; Haehner, A.; Hummel, T. Symptoms of depression change with olfactory function. Sci. Rep. 2022, 12, 5656. [Google Scholar] [CrossRef]
- Son, G.; Jahanshahi, A.; Yoo, S.J.; Boonstra, J.T.; Hopkins, D.A.; Steinbusch, H.W.M.; Moon, C. Olfactory neuropathology in Alzheimer’s disease: A sign of ongoing neurodegeneration. BMB Rep. 2021, 54, 295–304. [Google Scholar] [CrossRef]
- Murphy, C.; Schubert, C.R.; Cruickshanks, K.J.; Klein, B.E.; Klein, R.; Nondahl, D.M. Prevalence of olfactory impairment in older adults. JAMA 2002, 288, 2307–2312. [Google Scholar] [CrossRef]
- Yang, D.; Li, Q.; Fang, L.; Cheng, K.; Zhang, R.; Zheng, P.; Zhan, Q.; Qi, Z.; Zhong, S.; Xie, P. Reduced neurogenesis and pre-synaptic dysfunction in the olfactory bulb of a rat model of depression. Neuroscience 2011, 192, 609–618. [Google Scholar] [CrossRef]
- Huisman, E.; Uylings, H.B.; Hoogland, P.V. A 100% increase of dopaminergic cells in the olfactory bulb may explain hyposmia in Parkinson’s disease. Mov. Disord. 2004, 19, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem. Sci. 2017, 42, 297–311. [Google Scholar] [CrossRef]
- Di Rienzo, M.; Romagnoli, A.; Antonioli, M.; Piacentini, M.; Fimia, G.M. TRIM proteins in autophagy: Selective sensors in cell damage and innate immune responses. Cell Death Differ. 2020, 27, 887–902. [Google Scholar] [CrossRef]
- Kimura, T.; Jain, A.; Choi, S.W.; Mandell, M.A.; Johansen, T.; Deretic, V. TRIM-directed selective autophagy regulates immune activation. Autophagy 2017, 13, 989–990. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ren, H.; Xu, Y.; Wudu, M.; Wang, Q.; Liu, Z.; Su, H.; Jiang, X.; Zhang, Y.; Zhang, B.; et al. TRIM67 Promotes the Proliferation, Migration, and Invasion of Non-Small-Cell Lung Cancer by Positively Regulating the Notch Pathway. J. Cancer 2020, 11, 1240–1249. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, Y.; Huang, J.; Wong, C.C.; Zhai, J.; Li, C.; Wei, G.; Zhao, L.; Wang, G.; Wei, H.; et al. TRIM67 Activates p53 to Suppress Colorectal Cancer Initiation and Progression. Cancer Res. 2019, 79, 4086–4098. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Liu, X.; Zhang, J.; Qin, L.; Du, J.; Li, X.; Qian, S.; Chen, H.; Qian, P. TRIM67 Suppresses TNFalpha-Triggered NF-kB Activation by Competitively Binding Beta-TrCP to IkBa. Front. Immunol. 2022, 13, 793147. [Google Scholar] [CrossRef]
- Boyer, N.P.; Monkiewicz, C.; Menon, S.; Moy, S.S.; Gupton, S.L. Mammalian TRIM67 Functions in Brain Development and Behavior. eNeuro 2018, 5. [Google Scholar] [CrossRef]
- Menon, S.; Goldfarb, D.; Ho, C.T.; Cloer, E.W.; Boyer, N.P.; Hardie, C.; Bock, A.J.; Johnson, E.C.; Anil, J.; Major, M.B.; et al. The TRIM9/TRIM67 neuronal interactome reveals novel activators of morphogenesis. Mol. Biol. Cell 2021, 32, 314–330. [Google Scholar] [CrossRef]
- Yaguchi, H.; Okumura, F.; Takahashi, H.; Kano, T.; Kameda, H.; Uchigashima, M.; Tanaka, S.; Watanabe, M.; Sasaki, H.; Hatakeyama, S. TRIM67 protein negatively regulates Ras activity through degradation of 80K-H and induces neuritogenesis. J. Biol. Chem. 2012, 287, 12050–12059. [Google Scholar] [CrossRef]
- Jia, L.; Chen, Z.; Pan, T.; Xia, Y.; He, J.; Jahangir, A.; Wei, X.; Liu, W.; Shi, R.; Huang, C.; et al. TRIM67 Deficiency Exacerbates Hypothalamic Inflammation and Fat Accumulation in Obese Mice. Int. J. Mol. Sci. 2022, 23, 9438. [Google Scholar] [CrossRef]
- Huang, C.; Wei, X.; Luo, Q.; Xia, Y.; Pan, T.; He, J.; Jahangir, A.; Jia, L.; Liu, W.; Zou, Y.; et al. Loss of TRIM67 Attenuates the Progress of Obesity-Induced Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2022, 23, 7475. [Google Scholar] [CrossRef] [PubMed]
- Dou, X.; Chen, L.; Lei, M.; Zellmer, L.; Jia, Q.; Ling, P.; He, Y.; Yang, W.; Liao, D.J. Evaluating the Remote Control of Programmed Cell Death, with or without a Compensatory Cell Proliferation. Int. J. Biol. Sci. 2018, 14, 1800–1812. [Google Scholar] [CrossRef]
- Elmaci, İ.; Altinoz, M.A.; Sari, R.; Bolukbasi, F.H. Phosphorylated Histone H3 (PHH3) as a Novel Cell Proliferation Marker and Prognosticator for Meningeal Tumors: A Short Review. Appl. Immunohistochem. Mol. Morphol. 2018, 26, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Chen, Y.; Shou, T.; Hu, J.; Chen, J.; Qing, C. TRIM67 promotes NF-κB pathway and cell apoptosis in GA-13315-treated lung cancer cells. Mol. Med. Rep. 2019, 20, 2936–2944. [Google Scholar] [CrossRef] [PubMed]
- Schambra, U. Prenatal Mouse Brainatlas; Springer Science + Business Media: New York, NY, USA, 2008; p. 129. [Google Scholar]
- Chidambaram, S.B.; Rathipriya, A.G.; Bolla, S.R.; Bhat, A.; Ray, B.; Mahalakshmi, A.M.; Manivasagam, T.; Thenmozhi, A.J.; Essa, M.M.; Guillemin, G.J.; et al. Dendritic spines: Revisiting the physiological role. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 92, 161–193. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Li, M.X.; Xu, C.; Chen, H.B.; An, S.C.; Ma, X.M. Dendritic Spines in Depression: What We Learned from Animal Models. Neural Plast. 2016, 2016, 8056370. [Google Scholar] [CrossRef]
- Nishiyama, J. Plasticity of dendritic spines: Molecular function and dysfunction in neurodevelopmental disorders. Psychiatry Clin. Neurosci. 2019, 73, 541–550. [Google Scholar] [CrossRef]
- Marmolejo, N.; Paez, J.; Levitt, J.B.; Jones, L.B. Early postnatal lesion of the medial dorsal nucleus leads to loss of dendrites and spines in adult prefrontal cortex. Dev. Neurosci. 2012, 34, 463–476. [Google Scholar] [CrossRef]
- Trutzer, I.M.; García-Cabezas, M.; Zikopoulos, B. Postnatal development and maturation of layer 1 in the lateral prefrontal cortex and its disruption in autism. Acta Neuropathol. Commun. 2019, 7, 40. [Google Scholar] [CrossRef]
- Kolodkin, A.L.; Matthes, D.J.; Goodman, C.S. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell 1993, 75, 1389–1399. [Google Scholar] [CrossRef]
- Inoue, N.; Nishizumi, H.; Naritsuka, H.; Kiyonari, H.; Sakano, H. Sema7A/PlxnCl signaling triggers activity-dependent olfactory synapse formation. Nat. Commun. 2018, 9, 1842. [Google Scholar] [CrossRef]
- Hillje, A.L.; Pavlou, M.A.; Beckmann, E.; Worlitzer, M.M.; Bahnassawy, L.; Lewejohann, L.; Palm, T.; Schwamborn, J.C. TRIM32-dependent transcription in adult neural progenitor cells regulates neuronal differentiation. Cell Death Dis. 2013, 4, e976. [Google Scholar] [CrossRef]
- Hillje, A.L.; Beckmann, E.; Pavlou, M.A.; Jaeger, C.; Pacheco, M.P.; Sauter, T.; Schwamborn, J.C.; Lewejohann, L. The neural stem cell fate determinant TRIM32 regulates complex behavioral traits. Front. Cell. Neurosci. 2015, 9, 75. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, G.; Jiang, X.; Li, W.; Zhai, C.; Shang, F.; Chen, S.; Zhao, Z.; Yu, W. TRIM67 inhibits tumor proliferation and metastasis by mediating MAPK11 in Colorectal Cancer. J. Cancer 2020, 11, 6025–6037. [Google Scholar] [CrossRef]
- Yu, Y.; Xia, Q.; Zhan, G.; Gao, S.; Han, T.; Mao, M.; Li, X.; Wang, Y. TRIM67 alleviates cerebral ischemia-reperfusion injury by protecting neurons and inhibiting neuroinflammation via targeting IκBα for K63-linked polyubiquitination. Cell Biosci. 2023, 13, 99. [Google Scholar] [CrossRef] [PubMed]
- Lippman, J.; Dunaevsky, A. Dendritic spine morphogenesis and plasticity. J. Neurobiol. 2005, 64, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Matus, A.; Brinkhaus, H.; Wagner, U. Actin dynamics in dendritic spines: A form of regulated plasticity at excitatory synapses. Hippocampus 2000, 10, 555–560. [Google Scholar] [CrossRef]
- Urbina, F.L.; Menon, S.; Goldfarb, D.; Edwards, R.; Ben Major, M.; Brennwald, P.; Gupton, S.L. TRIM67 regulates exocytic mode and neuronal morphogenesis via SNAP47. Cell Rep. 2021, 34, 108743. [Google Scholar] [CrossRef]
- Boyer, N.P.; McCormick, L.E.; Menon, S.; Urbina, F.L.; Gupton, S.L. A pair of E3 ubiquitin ligases compete to regulate filopodial dynamics and axon guidance. J. Cell Biol. 2020, 219, e201902088. [Google Scholar] [CrossRef]
- Demirdizen, E.; Al-Ali, R.; Narayanan, A.; Sun, X.; Varga, J.P.; Steffl, B.; Brom, M.; Krunic, D.; Schmidt, C.; Schmidt, G.; et al. TRIM67 drives tumorigenesis in oligodendrogliomas through Rho GTPase-dependent membrane blebbing. Neuro Oncol. 2023, 25, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Montell, D.J. TRIMing Neural Connections with Ubiquitin. Dev. Cell 2019, 48, 5–6. [Google Scholar] [CrossRef]
- de Melo, G.D.; Lazarini, F.; Levallois, S.; Hautefort, C.; Michel, V.; Larrous, F.; Verillaud, B.; Aparicio, C.; Wagner, S.; Gheusi, G.; et al. COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci. Transl. Med. 2021, 13, eabf8396. [Google Scholar] [CrossRef]
- Stankiewicz, T.R.; Linseman, D.A. Rho family GTPases: Key players in neuronal development, neuronal survival, and neurodegeneration. Front. Cell. Neurosci. 2014, 8, 314. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liang, Z.; Fei, E.; Chen, Y.; Zhou, X.; Fang, W.; Fu, W.Y.; Fu, A.K.; Ip, N.Y. Axin Regulates Dendritic Spine Morphogenesis through Cdc42-Dependent Signaling. PLoS ONE 2015, 10, e0133115. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, S.; Shen, L.; Wang, J.; Wu, X.; Li, J.; Tu, C.; Ye, X.; Ling, S. Impairment of Dendrodendritic Inhibition in the Olfactory Bulb of APP/PS1 Mice. Front. Aging Neurosci. 2019, 11, 2. [Google Scholar] [CrossRef] [PubMed]
- Dewan, A.; Pacifico, R.; Zhan, R.; Rinberg, D.; Bozza, T. Non-redundant coding of aversive odours in the main olfactory pathway. Nature 2013, 497, 486–489. [Google Scholar] [CrossRef]
- Prediger, R.D.; Batista, L.C.; Takahashi, R.N. Caffeine reverses age-related deficits in olfactory discrimination and social recognition memory in rats. Involvement of adenosine A1 and A2A receptors. Neurobiol. Aging 2005, 26, 957–964. [Google Scholar] [CrossRef]


| Gene | Sequence (5′-3′) |
|---|---|
| β-actin | F: AGAGGGAAATCGTGCGTGAC |
| R: CAATAGTGATGACCTGGCCGT | |
| TRIM67 | F: GGCGAAGGAGTTTCTGGTTC |
| R: TAGCTTCAGGGTGCAGTGATT | |
| TNF-α | F: ACGGCATGGATCTCAAAGAC |
| R: GTGGGTGAGGAGCACGTAG | |
| IL-21 | F: TCAAGCCATCAAACCCTGGA |
| R: CATACGAATCACAGGAAGGGC | |
| Tbx21 | F: TGTTCCCAGCCGTTTCTACC |
| R: GCTCATCTTGGGCGGGTATT | |
| Bax | F: TGGAGATGAACTGGACAGCA |
| R: TGAAGTTGCCATCAGCAAAC | |
| Bcl-2 | F: AGGATTGTGGCCTTCTTTGA |
| R: CAGATGCCGGTTCAGGTACT | |
| Caspase3 | F: TGGAGAAATTCAAAGGACGGG |
| R: AGCATGGACACAATACACGGG | |
| PSD95 | F: CAATGGTGTTGACCTCCGCA |
| R: GGGGTTGCTTCGCAGAGAT | |
| VGLUT2 | F: GGATCGTTCTTCTGGGGCTAT |
| R: GTAGAGGTGAGCAGTATCGCA | |
| cpg15 | F: GGGCAAGTGTGATGCAGGTCT |
| R: TCCTGGCAATCCGTAAGAGC | |
| GluR1 | F: CCCGTTGACACATCCAATCAG |
| R: GGTATCCTTCCTCCGTGGTT | |
| Sema7A | F: TGCGGGTAGCGAAGGTTTTC |
| R: CATGGTCCTGCCCTTTCCAG | |
| PlxnC1 | F: CAAGGGGATTGCACACATGCT |
| R: GGTCCAGTTCAGTTGGCTTC | |
| Cdc42 | F: GGAGAGCCATACACTCTTGGAC |
| R: ATGAGGATGGAGAGACCACTGA | |
| NMDA-R | F: AACGACCACTTCACTCCCAC |
| R: CGCGCATCATCTCAAACCAG | |
| Iba1 | F: CTTTTGGACTGCTGAAGGC |
| R: GTTTCTCCAGCATTCGCTTC |
| Antibodies | Source | Catalog No./Dilution |
|---|---|---|
| Tbr1 Rabbit pAb | Proteintech | 20932-1-AP/WB 1:1000 |
| Iba1 Rabbit mAb | Abcam | ab178846/WB 1:1000 |
| Cleaved-caspase3 Rabbit pAb | CST | #9664/WB 1:1000 |
| VGLUT2 Rabbit mAb | Sigma | ZRB1170/WB 1:1000 |
| Synaptophysin Rabbit mAb | CST | #36406/WB 1:1000 |
| CDC42 Rabbit pAb | Proteintech | 10155-1-AP/WB 1:1000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, C.; Luo, Q.; Jia, L.; Xia, Y.; Lan, X.; Wei, X.; Shi, S.; Liu, Y.; Wang, Y.; Xiong, Z.; et al. TRIM67 Implicates in Regulating the Homeostasis and Synaptic Development of Mitral Cells in the Olfactory Bulb. Int. J. Mol. Sci. 2023, 24, 13439. https://doi.org/10.3390/ijms241713439
Cai C, Luo Q, Jia L, Xia Y, Lan X, Wei X, Shi S, Liu Y, Wang Y, Xiong Z, et al. TRIM67 Implicates in Regulating the Homeostasis and Synaptic Development of Mitral Cells in the Olfactory Bulb. International Journal of Molecular Sciences. 2023; 24(17):13439. https://doi.org/10.3390/ijms241713439
Chicago/Turabian StyleCai, Chunyu, Qihui Luo, Lanlan Jia, Yu Xia, Xinting Lan, Xiaoli Wei, Shuai Shi, Yucong Liu, Yao Wang, Zongliang Xiong, and et al. 2023. "TRIM67 Implicates in Regulating the Homeostasis and Synaptic Development of Mitral Cells in the Olfactory Bulb" International Journal of Molecular Sciences 24, no. 17: 13439. https://doi.org/10.3390/ijms241713439
APA StyleCai, C., Luo, Q., Jia, L., Xia, Y., Lan, X., Wei, X., Shi, S., Liu, Y., Wang, Y., Xiong, Z., Shi, R., Huang, C., & Chen, Z. (2023). TRIM67 Implicates in Regulating the Homeostasis and Synaptic Development of Mitral Cells in the Olfactory Bulb. International Journal of Molecular Sciences, 24(17), 13439. https://doi.org/10.3390/ijms241713439







