Application of Molecular Dynamics Simulations to Determine Interactions between Canary Seed (Phalaris canariensis L.) Bioactive Peptides and Skin-Aging Enzymes
Abstract
:1. Introduction
2. Results and Discussion
2.1. In Vitro Inhibition of Aging-Enzymes by Peptide Fractions
2.2. Molecular Dynamics Simulations for Multi-Level Skin Aging Targets
2.2.1. Identification and Prediction of Bioactive Peptides from Canary Seed
2.2.2. Identification and Prediction of Canary Seed Peptides with Antioxidant Activity
2.2.3. Bioactive Canary Seed Peptides as Inhibitors of Skin-Aging Enzymes
2.2.4. Stratum Corneum Passive Permeability of Bioactive Peptides Using Molecular Dynamics Simulations
3. Materials and Methods
3.1. Preparation of Canary Seed Peptides
3.2. Evaluation of In Vitro Anti-Aging Bioactive Properties
3.2.1. Elastase Inhibition Assay
3.2.2. Tyrosinase Inhibition Assay
3.2.3. Identification of Elastase and Tyrosinase Inhibitory Peptides
3.3. Molecular Dynamics Simulations for Multi-Level Skin Aging Targets
3.3.1. Prediction of Physicochemical Descriptors from Bioactive Peptide Fractions
3.3.2. Prediction of Physicochemical Descriptors from Bioactive Peptide Fractions
3.3.3. Model Preparation of Skin-Aging Related Enzymes
3.3.4. Ensemble Docking of Skin-Aging Enzymes
3.3.5. Molecular Dynamics Simulations of Stratum Corneum–Peptide Complexes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aguilar-Toalá, J.E.; Vidal-Limon, A.; Liceaga, A.M. Nutricosmetics: A new frontier in bioactive peptides’ research toward skin aging. In Advances in Food and Nutrition Research; Academic Press: Cambridge, MA, USA, 2022. [Google Scholar]
- Ramos-e-Silva, M.; Celem, L.R.; Ramos-e-Silva, S.; Fucci-da-Costa, A.P. Anti-aging cosmetics: Facts and controversies. Clin. Dermatol. 2013, 31, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Hou, H.; Fan, Y.; Wang, S.; Chen, Q.; Si, L.; Li, B. Protective effect of gelatin peptides from pacific cod skin against photoaging by inhibiting the expression of MMPs via MAPK signaling pathway. J. Photochem. Photobiol. B Biol. 2016, 165, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Hakuta, A.; Yamaguchi, Y.; Okawa, T.; Yamamoto, S.; Sakai, Y.; Aihara, M. Anti-inflammatory effect of collagen tripeptide in atopic dermatitis. J. Dermatol. Sci. 2017, 88, 357–364. [Google Scholar] [CrossRef]
- Oba, C.; Ohara, H.; Morifuji, M.; Ito, K.; Ichikawa, S.; Kawahata, K.; Koga, J. Collagen hydrolysate intake improves the loss of epidermal barrier function and skin elasticity induced by UVB irradiation in hairless mice. Photodermatol. Photoimmunol. Photomed. 2013, 29, 204–211. [Google Scholar] [CrossRef]
- Hou, H.; Li, B.; Zhang, Z.; Xue, C.; Yu, G.; Wang, J.; Bao, Y.; Bu, L.; Sun, J.; Peng, Z.; et al. Moisture absorption and retention properties, and activity in alleviating skin photodamage of collagen polypeptide from marine fish skin. Food Chem. 2012, 135, 1432–1439. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.; Liceaga, A. Identification of chia seed (Salvia hispanica L.) peptides with enzyme inhibition activity towards skin-aging enzymes. Amino Acids 2020, 52, 1149–1159. [Google Scholar] [CrossRef]
- Urbizo-Reyes, U.; Aguilar-Toalá, J.; Liceaga, A. Hairless canary seeds (Phalaris canariensis L.) as a potential source of antioxidant, antihypertensive, antidiabetic, and antiobesity biopeptides. Food Prod. Process. Nutr. 2021, 3, 1–12. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.E.; Vidal-Limon, A.; Liceaga, A.M. Multifunctional analysis of chia seed (Salvia hispanica L.) bioactive peptides using peptidomics and molecular dynamics simulations approaches. Int. J. Mol. Sci. 2022, 23, 7288. [Google Scholar] [CrossRef]
- Gunalan, S.; Somarathinam, K.; Bhattacharya, J.; Srinivasan, S.; Jaimohan, S.M.; Manoharan, R.; Ramachandran, S.; Kanagaraj, S.; Kothandan, G. Understanding the dual mechanism of bioactive peptides targeting the enzymes involved in Renin Angiotensin System (RAS): An in-silico approach. J. Biomol. Struct. Dyn. 2020, 38, 5044–5061. [Google Scholar] [CrossRef]
- Mudgil, P.; Gan, C.-Y.; Affan Baig, M.; Hamdi, M.; Mohteshamuddin, K.; Aguilar-Toalá, J.E.; Vidal-Limon, A.M.; Liceaga, A.M.; Maqsood, S. In-depth peptidomic profile and molecular simulation studies on ACE-inhibitory peptides derived from probiotic fermented milk of different farm animals. Food Res. Int. 2023, 168, 112706. [Google Scholar] [CrossRef]
- Urbizo-Reyes, U.; Liceaga, A.M.; Reddivari, L.; Kim, K.-H.; Anderson, J.M. Enzyme kinetics, molecular docking, and in silico characterization of canary seed (Phalaris canariensis L.) peptides with ACE and pancreatic lipase inhibitory activity. J. Funct. Foods 2022, 88, 104892. [Google Scholar] [CrossRef]
- Osorio, D.; Rondón-Villarreal, P.; Torres, R. Peptides: A package for data mining of antimicrobial peptides. R J. 2015, 7, 4. [Google Scholar] [CrossRef]
- Kirchner, L.A.; Moody, R.P.; Doyle, E.; Bose, R.; Jeffery, J.; Chu, I. The prediction of skin permeability by using physicochemical data. Altern. Lab. Anim. 1997, 25, 359–370. [Google Scholar] [CrossRef]
- Potts, R.O.; Guy, R.H. A predictive algorithm for skin permeability: The effects of molecular size and hydrogen bond activity. Pharm. Res. 1995, 12, 1628–1633. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, V.M.; Bellmann-Sickert, K.; Beck-Sickinger, A.G. Peptides and peptide conjugates: Therapeutics on the upward path. Future Med. Chem. 2012, 4, 1567–1586. [Google Scholar] [CrossRef]
- Osorio, D.; Rondón-Villarreal, P.; Torres, R. Stability analysis of antimicrobial peptides in solvation conditions by molecular dynamics. In Advances in Computational Biology; Castillo, L.F., Cristancho, M., Isaza, G., Pinzón, A., Rodríguez, J.M.C., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 127–131. [Google Scholar]
- Brancolini, G.; Bellucci, L.; Maschio, M.C.; Di Felice, R.; Corni, S. The interaction of peptides and proteins with nanostructures surfaces: A challenge for nanoscience. Curr. Opin. Colloid Interface Sci. 2019, 41, 86–94. [Google Scholar] [CrossRef]
- Dutta, S.; Corni, S.; Brancolini, G. Molecular dynamics simulations of a catalytic multivalent peptide–Nanoparticle complex. Int. J. Mol. Sci. 2021, 22, 3624. [Google Scholar] [CrossRef]
- Javadzadeh, Y.; Azharshekoufeh Bahari, L. Chapter 8—Therapeutic Nanostructures for Dermal and Transdermal Drug Delivery. In Nano- and Microscale Drug Delivery Systems; Grumezescu, A.M., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 131–146. [Google Scholar]
- Asserin, J.; Lati, E.; Shioya, T.; Prawitt, J. The effect of oral collagen peptide supplementation on skin moisture and the dermal collagen network: Evidence from an ex vivo model and randomized, placebo-controlled clinical trials. J. Cosmet. Dermatol. 2015, 14, 291–301. [Google Scholar] [CrossRef]
- Lee, M.-T.; Hung, W.-C.; Chen, F.-Y.; Huang, H.W. Mechanism and kinetics of pore formation in membranes by water-soluble amphipathic peptides. Proc. Natl. Acad. Sci. USA 2008, 105, 5087–5092. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Takechi-Haraya, Y.; Ohgita, T.; Kotani, M.; Kono, H.; Saito, C.; Tamagaki-Asahina, H.; Nishitsuji, K.; Uchimura, K.; Sato, T.; Kawano, R.; et al. Effect of hydrophobic moment on membrane interaction and cell penetration of apolipoprotein E-derived arginine-rich amphipathic α-helical peptides. Sci. Rep. 2022, 12, 4959. [Google Scholar] [CrossRef]
- White, S.H.; Wimley, W.C. Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta (BBA)-Rev. Biomembr. 1998, 1376, 339–352. [Google Scholar] [CrossRef]
- Cardenas, A.E.; Shrestha, R.; Webb, L.J.; Elber, R. Membrane permeation of a peptide: It is better to be positive. J. Phys. Chem. B 2015, 119, 6412–6420. [Google Scholar] [CrossRef] [PubMed]
- Povilaitis, S.C.; Fathizadeh, A.; Kogan, M.; Elber, R.; Webb, L.J. Design of peptides for membrane insertion: The critical role of charge separation. J. Phys. Chem. B 2022, 126, 6454–6463. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-W.; Kwon, S.-H.; Choi, J.-Y.; Na, J.-I.; Huh, C.-H.; Choi, H.-R.; Park, K.-C. Molecular mechanisms of dermal aging and antiaging approaches. Int. J. Mol. Sci. 2019, 20, 2126. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Toalá, J.E.; Hernández-Mendoza, A.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Liceaga, A.M. Potential role of natural bioactive peptides for development of cosmeceutical skin products. Peptides 2019, 122, 170170. [Google Scholar] [CrossRef] [PubMed]
- Olsen, T.H.; Yesiltas, B.; Marin, F.I.; Pertseva, M.; García-Moreno, P.J.; Gregersen, S.; Overgaard, M.T.; Jacobsen, C.; Lund, O.; Hansen, E.B.; et al. AnOxPePred: Using deep learning for the prediction of antioxidative properties of peptides. Sci. Rep. 2020, 10, 21471. [Google Scholar] [CrossRef]
- Kongsompong, S.; E-kobon, T.; Taengphan, W.; Sangkhawasi, M.; Khongkow, M.; Chumnanpuen, P. Computer-aided virtual screening and in vitro validation of biomimetic tyrosinase inhibitory peptides from abalone peptidome. Int. J. Mol. Sci. 2023, 24, 3154. [Google Scholar] [CrossRef]
- Thaha, A.; Wang, B.-S.; Chang, Y.-W.; Hsia, S.-M.; Huang, T.-C.; Shiau, C.-Y.; Hwang, D.-F.; Chen, T.-Y. Food-derived bioactive peptides with antioxidative capacity, xanthine oxidase and tyrosinase inhibitory activity. Processes 2021, 9, 747. [Google Scholar] [CrossRef]
- Hawkins, C.L.; Davies, M.J. Generation and propagation of radical reactions on proteins. Biochim. Biophys. Acta-Bioenerg. 2001. [Google Scholar] [CrossRef]
- Hawkins, C.L.; Davies, M.J. EPR studies on the selectivity of hydroxyl radical attack on amino acids and peptides. J. Chem. Soc. Perkin Trans. 1998, 2, 2617–2622. [Google Scholar] [CrossRef]
- Miao, Y.; Feixas, F.; Eun, C.; McCammon, J.A. Accelerated molecular dynamics simulations of protein folding. J. Comput. Chem. 2015, 36, 1536–1549. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S. Predicting protein–ligand binding and unbinding kinetics with biased MD simulations and coarse-graining of dynamics: Current state and challenges. J. Chem. Inf. Model. 2023, 63, 2902–2910. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Do, H.N.; Koirala, K.; Miao, Y. Predicting biomolecular binding kinetics: A review. J. Chem. Theory Comput. 2023, 19, 2135–2148. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, A.; Wang, J.; Miao, Y. Retrospective ensemble docking of allosteric modulators in an adenosine G-protein-coupled receptor. Biochim. Biophys. Acta (BBA) Gen. Subj. 2020, 1864, 129615. [Google Scholar] [CrossRef]
- Jaghoori, M.M.; Bleijlevens, B.; Olabarriaga, S.D. 1001 Ways to run AutoDock Vina for virtual screening. J. Comput.-Aided Mol. Des. 2016, 30, 237–249. [Google Scholar] [CrossRef]
- Vidal-Limon, A.; Aguilar-Toalá, J.E.; Liceaga, A.M. Integration of molecular docking analysis and molecular dynamics simulations for studying food proteins and bioactive peptides. J. Agric. Food Chem. 2022, 70, 934–943. [Google Scholar] [CrossRef]
- Badhe, Y.; Schmitt, T.; Gupta, R.; Rai, B.; Neubert, R.H.H. Investigating the nanostructure of a CER[NP]/CER[AP]-based stratum corneum lipid matrix model: A combined neutron diffraction & molecular dynamics simulations approach. Biochim. Biophys. Acta (BBA) Biomembr. 2022, 1864, 184007. [Google Scholar] [CrossRef]
- Sugita, M.; Sugiyama, S.; Fujie, T.; Yoshikawa, Y.; Yanagisawa, K.; Ohue, M.; Akiyama, Y. Large-scale membrane permeability prediction of cyclic peptides crossing a lipid bilayer based on enhanced sampling molecular dynamics simulations. J. Chem. Inf. Model. 2021, 61, 3681–3695. [Google Scholar] [CrossRef]
- Tieleman, D.P. Computer simulations of transport through membranes: Passive diffusion, pores, channels and transporters. Clin. Exp. Pharmacol. Physiol. 2006, 33, 893–903. [Google Scholar] [CrossRef]
- Schiltz, M.; Shepard, W.; Fourme, R.; Prangé, T.; de la Fortelle, E.; Bricogne, G. High-pressure krypton gas and statistical heavy-atom refinement: A successful combination of tools for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, 78–92. [Google Scholar] [CrossRef]
- Pretzler, M.; Bijelic, A.; Rompel, A. Heterologous expression and characterization of functional mushroom tyrosinase (AbPPO4). Sci. Rep. 2017, 7, 1810. [Google Scholar] [CrossRef] [PubMed]
- Götz, A.W.; Williamson, M.J.; Xu, D.; Poole, D.; Le Grand, S.; Walker, R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012, 8, 1542–1555. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cheatham Iii, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- DCase, D.A.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Giambasu, D.; et al. AMBER 2020; University of California: Oakland, CA, USA, 2020. [Google Scholar]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Feher, V.A.; McCammon, J.A. Gaussian accelerated molecular dynamics: Unconstrained enhanced sampling and freeeEnergy calculation. J. Chem. Theory Comput. 2015, 11, 3584–3595. [Google Scholar] [CrossRef]
- Miao, Y.; McCammon, J.A. Unconstrained enhanced sampling for free energy calculations of biomolecules: A review. Mol. Simul. 2016, 42, 1046–1055. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; McCammon, J.A. Gaussian accelerated molecular dynamics: Theory, implementation, and applications. Annu. Rep. Comput. Chem. 2017, 13, 231–278. [Google Scholar] [CrossRef]
- Wang, J.; Alekseenko, A.; Kozakov, D.; Miao, Y. Improved modeling of peptide-protein binding through global docking and accelerated molecular dynamics simulations. Front. Mol. Biosci. 2019, 6, 112. [Google Scholar] [CrossRef]
- Quiroga, R.; Villarreal, M.A. Vinardo: A scoring function based on Autodock Vina improves scoring, docking, and virtual screening. PLoS ONE 2016, 11, e0155183. [Google Scholar] [CrossRef]
- Koes, D.R.; Baumgartner, M.P.; Camacho, C.J. Lessons learned in empirical scoring with smina from the CSAR 2011 Benchmarking exercise. J. Chem. Inf. Model. 2013, 53, 1893–1904. [Google Scholar] [CrossRef]
- Masters, L.; Eagon, S.; Heying, M. Evaluation of consensus scoring methods for AutoDock Vina, smina and idock. J. Mol. Graph. Model. 2020, 96, 107532. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016. [Google Scholar] [CrossRef] [PubMed]
- Dickson, C.J.; Walker, R.C.; Gould, I.R. Lipid21: Complex lipid membrane simulations with AMBER. J. Chem. Theory Comput. 2022, 18, 1726–1736. [Google Scholar] [CrossRef] [PubMed]
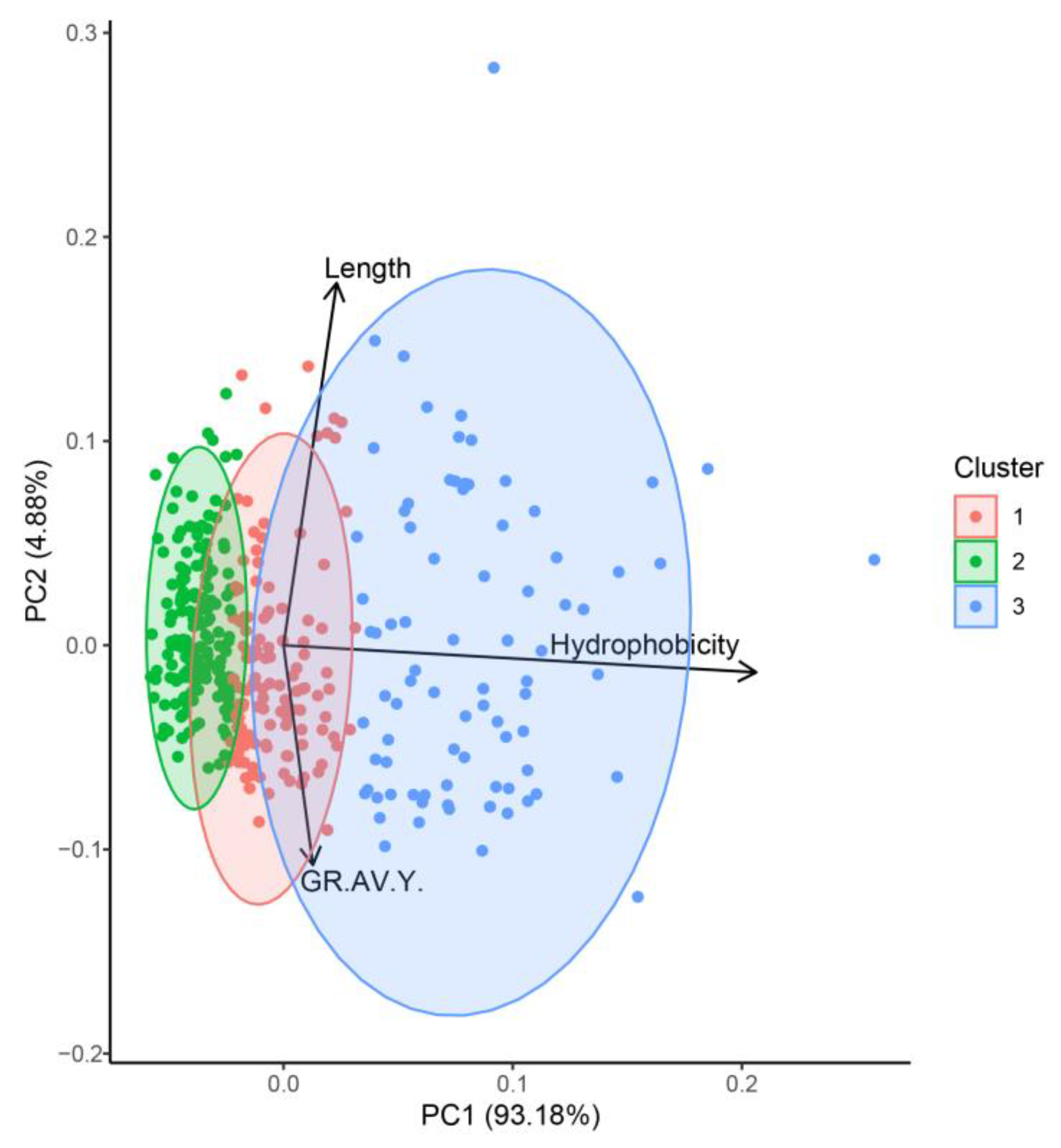

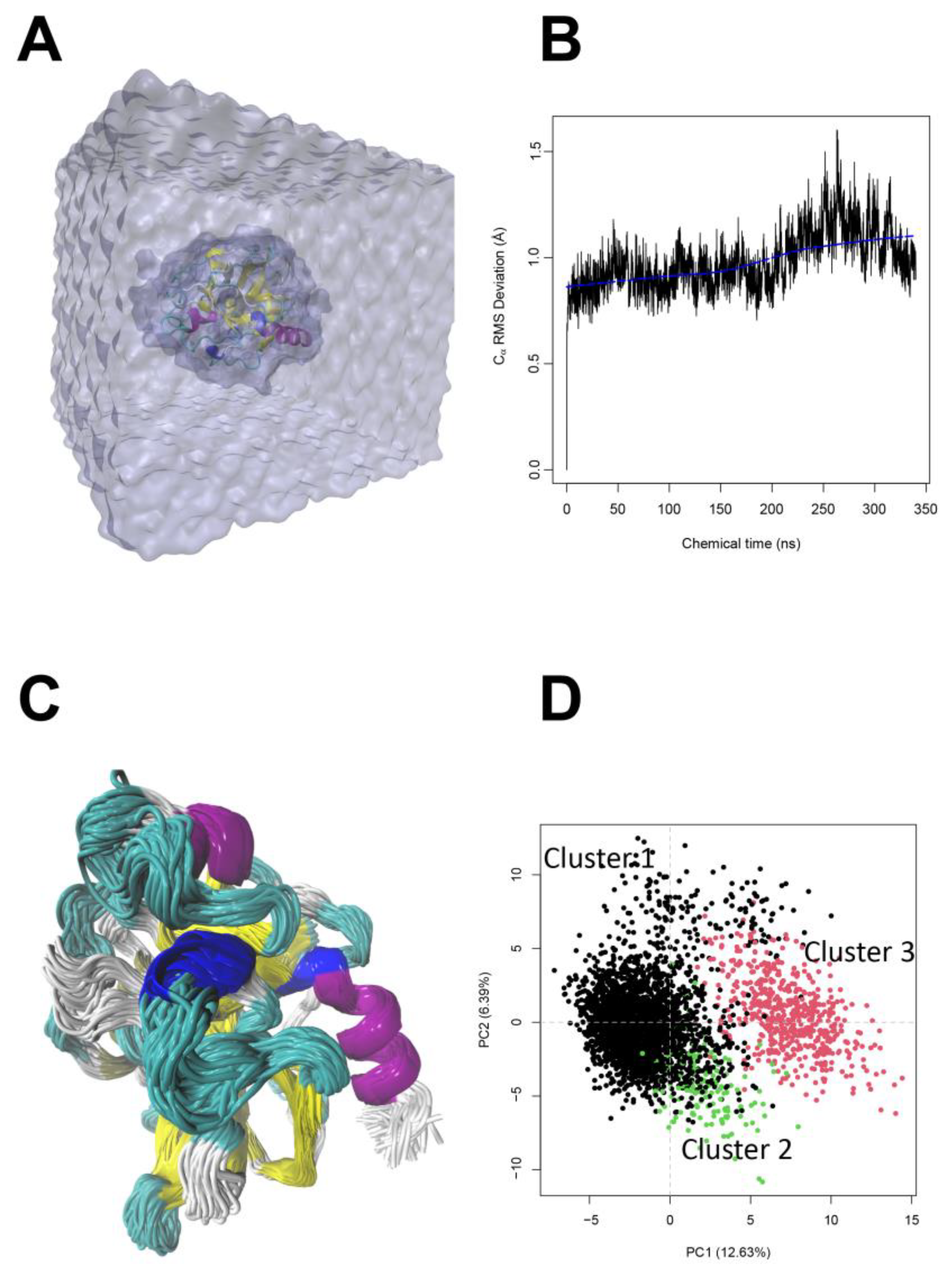

| Peptide Fraction | Elastase Inhibition (%) 1 | Tyrosinase Inhibition (%) 1 |
|---|---|---|
| <3 kDa (2.5 mg/mL) | 33.76 ± 0.012 | 29.95 ± 0.011 |
| <3 kDa (5 mg/mL) | 45.96 ± 0.018 | 54.19 ± 0.040 |
| <3 kDa (10 mg/mL) | 65.26 ± 0.008 | 62.63 ± 0.034 |
| IC50 | 6.24 mg/mL | 6.10 mg/mL |
| F-I | ND | ND |
| F-II | ND | ND |
| F-III | ND | ND |
| F-IV | 84.05 ± 0.002 | 14.96 ± 0.004 |
| F-V | 55.14 ± 0.004 | ND |
| F-VI | 92.82 ± 0.001 | 66.93 ± 0.001 |
| Tyrosinase | Elastase | ||
|---|---|---|---|
| Peptide | Docking Score 1 | Peptide | Docking Score 1 |
| GGWH | −9.36 ± 0.56 | VPPH | −8.00 ± 0.85 |
| VPPH | −9.18 ± 1.22 | EGLEPNHRVE | −7.92 ± 0.91 |
| FLPH | −9.08 ± 0.75 | GGWH | −7.70 ± 1.01 |
| VPHGAP | −8.94 ± 1.10 | FGPAGHT | −7.32 ± 0.99 |
| WAGW | −8.62 ± 1.24 | RPVNKYTPPQ | −7.24 ± 0.72 |
| MPYN | −8.54 ± 1.38 | FLPH | −7.12 ± 1.38 |
| ELHPQ | −8.54 ± 1.48 | LLPH | −7.00 ± 1.83 |
| FVPH | −8.46 ± 1.02 | LHPE | −6.92 ± 1.24 |
| EGLEPNHRVE | −8.44 ± 1.23 | VVPH | −6.90 ± 1.16 |
| LLPH | −8.40 ± 1.26 | VPPAH | −6.86 ± 0.81 |
| VYPN | −8.30 ± 0.92 | MPYN | −6.84 ± 1.55 |
| FHPQ | −8.26 ± 1.05 | NEEWPR | −6.80 ± 0.62 |
| LTPH | −8.20 ± 0.83 | VPHGAP | −6.78 ± 1.42 |
| FGPAGHT | −8.20 ± 0.85 | FVPH | −6.74 ± 1.31 |
| NEEWPR | −8.14 ± 0.99 | WGPALH | −6.68 ± 1.30 |
| VVPPGVPY | −8.04 ± 1.09 | WAGW | −6.66 ± 1.14 |
| WGPALH | −7.98 ± 0.99 | LTPH | −6.64 ± 0.91 |
| VPPAH | −7.86 ± 0.87 | KGGCEHEV | −6.64 ± 0.53 |
| VPPHQQ | −7.78 ± 0.66 | ELHPQ | −6.56 ± 1.09 |
| KGGCEHEV | −7.30 ± 0.66 | FHPQ | −6.50 ± 1.28 |
| PLGH | −6.94 ± 1.01 | VVPPGVPY | −6.50 ± 0.85 |
| NYPVG | −6.52 ± 0.44 | VYPN | −6.44 ± 1.07 |
| GHDPK | −6.38 ± 0.13 | HTHL | −6.36 ± 1.91 |
| CPPLH | −6.34 ± 0.62 | PLGH | −6.06 ± 0.51 |
| LHPE | −6.26 ± 0.47 | VPPHQQ | −5.86 ± 0.63 |
| SLHPQ | −6.14 ± 0.90 | QAHPK | −5.76 ± 0.62 |
| RPVNKYTPPQ | −6.10 ± 0.33 | CPPLH | −5.74 ± 0.52 |
| QAHPK | −5.98 ± 0.44 | NYPVG | −5.68 ± 0.42 |
| HTHL | −5.68 ± 0.44 | SLHPQ | −5.44 ± 0.59 |
| VVPH | −5.66 ± 0.50 | GHDPK | −5.24 ± 0.38 |
| Lipid 1 | Upper Leaflet | Lower Leaflet |
|---|---|---|
| CER (Ceramides [NP] shingonoids) | 29 | 30 |
| CHOL (cholesterol) | 35 | 35 |
| POPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) | 25 | 27 |
| POPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine) | 28 | 29 |
| POPS (1,2-dioleoyl-sn-glycero-3-phospho-L-serine) | 30 | 29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguilar-Toalá, J.E.; Vidal-Limon, A.; Liceaga, A.M.; Zambrano-Zaragoza, M.L.; Quintanar-Guerrero, D. Application of Molecular Dynamics Simulations to Determine Interactions between Canary Seed (Phalaris canariensis L.) Bioactive Peptides and Skin-Aging Enzymes. Int. J. Mol. Sci. 2023, 24, 13420. https://doi.org/10.3390/ijms241713420
Aguilar-Toalá JE, Vidal-Limon A, Liceaga AM, Zambrano-Zaragoza ML, Quintanar-Guerrero D. Application of Molecular Dynamics Simulations to Determine Interactions between Canary Seed (Phalaris canariensis L.) Bioactive Peptides and Skin-Aging Enzymes. International Journal of Molecular Sciences. 2023; 24(17):13420. https://doi.org/10.3390/ijms241713420
Chicago/Turabian StyleAguilar-Toalá, José E., Abraham Vidal-Limon, Andrea M. Liceaga, Maria L. Zambrano-Zaragoza, and David Quintanar-Guerrero. 2023. "Application of Molecular Dynamics Simulations to Determine Interactions between Canary Seed (Phalaris canariensis L.) Bioactive Peptides and Skin-Aging Enzymes" International Journal of Molecular Sciences 24, no. 17: 13420. https://doi.org/10.3390/ijms241713420
APA StyleAguilar-Toalá, J. E., Vidal-Limon, A., Liceaga, A. M., Zambrano-Zaragoza, M. L., & Quintanar-Guerrero, D. (2023). Application of Molecular Dynamics Simulations to Determine Interactions between Canary Seed (Phalaris canariensis L.) Bioactive Peptides and Skin-Aging Enzymes. International Journal of Molecular Sciences, 24(17), 13420. https://doi.org/10.3390/ijms241713420











