The Application of 3base™ Technology to Diagnose Eight of the Most Clinically Important Gastrointestinal Protozoan Infections
Abstract
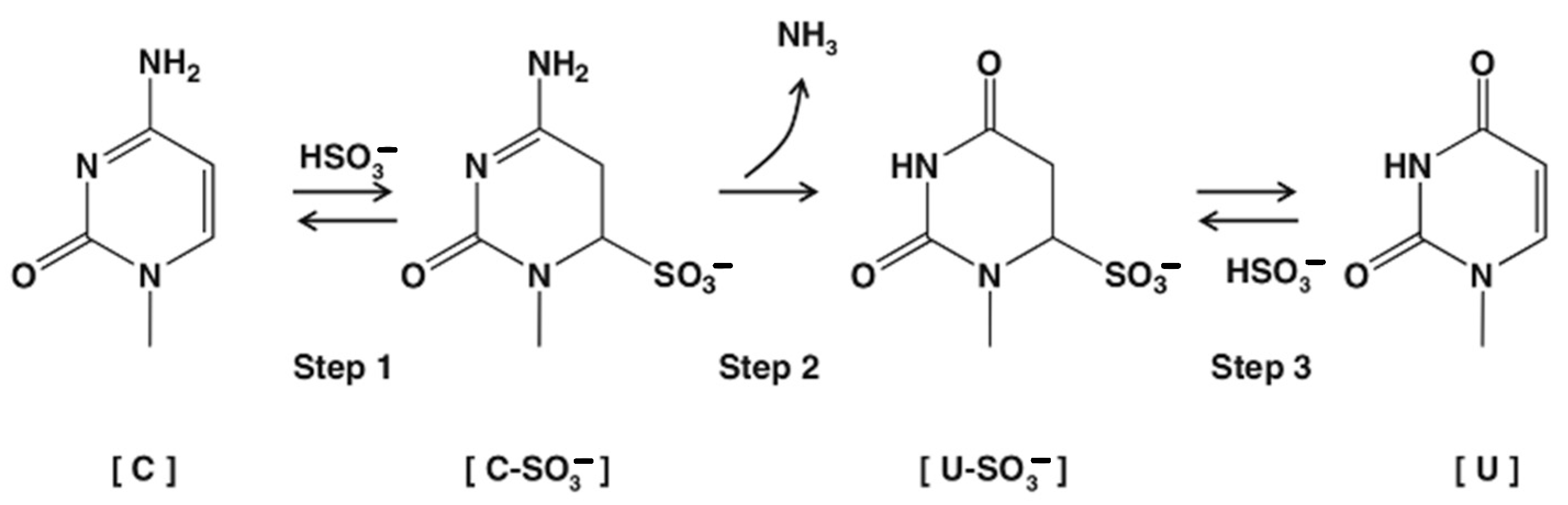
1. Introduction
2. Results
2.1. Assay Sensitivity
2.2. Assay specificity
2.3. Microbial Interference Studies
2.4. Quality Assurance Panels
2.5. Clinical Studies
3. Discussion
4. Materials and Methods
4.1. EP005 PCR Assays
4.2. Preparation of Negative Stool
4.3. LLOD and Inclusivity Studies
4.4. Cross-Reactivity Studies
4.5. Clinical Samples
4.6. Confirmatory Nucleic Acid Amplification Techniques
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cox, F.E. History of human parasitic diseases. Infect. Dis. Clin. N. Am. 2004, 18, 171–188. [Google Scholar] [CrossRef]
- Hajare, S.T.; Gobena, R.K.; Chauhan, N.M.; Erniso, F. Prevalence of Intestinal Parasite Infections and Their Associated Factors among Food Handlers Working in Selected Catering Establishments from Bule Hora, Ethiopia. BioMed Res. Int. 2021, 2021, 6669742. [Google Scholar] [CrossRef]
- Osman, M.; El Safadi, D.; Cian, A.; Benamrouz, S.; Nourrisson, C.; Poirier, P.; Pereira, B.; Razakandrainibe, R.; Pinon, A.; Lambert, C.; et al. Prevalence and Risk Factors for Intestinal Protozoan Infections with Cryptosporidium, Giardia, Blastocystis and Dientamoeba among Schoolchildren in Tripoli, Lebanon. PLoS Neglected Trop. Dis. 2016, 10, e0004496. [Google Scholar] [CrossRef]
- Harhay, M.O.; Horton, J.; Olliaro, P.L. Epidemiology and control of human gastrointestinal parasites in children. Expert Rev. Anti-Infect. Ther. 2010, 8, 219–234. [Google Scholar] [CrossRef]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Mbae, C.; Mulinge, E.; Guleid, F.; Wainaina, J.; Waruru, A.; Njiru, Z.K.; Kariuki, S. Molecular Characterization of Giardia duodenalis in Children in Kenya. BMC Infect. Dis. 2016, 16, 135. [Google Scholar] [CrossRef]
- Maikai, B.V.; Umoh, J.U.; Lawal, I.A.; Kudi, A.C.; Ejembi, C.L.; Xiao, L. Molecular characterizations of Cryptosporidium, Giardia, and Enterocytozoon in humans in Kaduna State, Nigeria. Exp. Parasitol. 2012, 131, 452–456. [Google Scholar] [CrossRef]
- Guillén, N. Pathogenicity and virulence of Entamoeba histolytica, the agent of amoebiasis. Virulence 2023, 14, 2158656. [Google Scholar] [CrossRef]
- Lo, Y.C.; Ji, D.D.; Hung, C.C. Prevalent and incident HIV diagnoses among Entamoeba histolytica-infected adult males: A changing epidemiology associated with sexual transmission--Taiwan, 2006–2013. PLoS Neglected Trop. Dis. 2014, 8, e3222. [Google Scholar] [CrossRef] [PubMed]
- Stark, D.; Barratt, J.; Chan, D.; Ellis, J.T. Dientamoeba fragilis, the Neglected Trichomonad of the Human Bowel. Clin. Microbiol. Rev. 2016, 29, 553–580. [Google Scholar] [CrossRef]
- Vandenberg, O.; Peek, R.; Souayah, H.; Dediste, A.; Buset, M.; Scheen, R.; Retore, P.; Zissis, G.; van Gool, T. Clinical and microbiological features of dientamoebiasis in patients suspected of suffering from a parasitic gastrointestinal illness: A comparison of Dientamoeba fragilis and Giardia lamblia infections. Int. J. Infect. Dis. 2006, 10, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Basak, S.; Rajurkar, M.N.; Mallick, S.K. Detection of Blastocystis hominis: A controversial human pathogen. Parasitol. Res. 2013, 113, 261–265. [Google Scholar] [CrossRef]
- Wawrzyniak, I.; Poirier, P.; Viscogliosi, E.; Dionigia, M.; Texier, C.; Delbac, F.; Alaoui, H.E. Blastocystis, an unrecognized parasite: An overview of pathogenesis and diagnosis. Ther. Adv. Infect. Dis. 2013, 1, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Stenzel, D.J.; Boreham, P.F. Blastocystis hominis revisited. Clin. Microbiol. Rev. 1996, 9, 563–584. [Google Scholar] [CrossRef] [PubMed]
- WHO. Guidelines for Drinking-Water Quality (3rd Edition, Incorporating First and Second Addenda); World Health Organization: Geneva, Switzerland, 2008.
- Taghipour, A.; Rayatdoost, E.; Bairami, A.; Bahadory, S.; Abdoli, A. Are Blastocystis hominis and Cryptosporidium spp. playing a positive role in colorectal cancer risk? A systematic review and meta-analysis. Infect. Agents Cancer 2022, 17, 32. [Google Scholar] [CrossRef]
- Almeria, S.; Cinar, H.N.; Dubey, J.P. Cyclospora cayetanensis and Cyclosporiasis: An Update. Microorganisms 2019, 7, 317. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xiao, L. Ecological and public health significance of Enterocytozoon bieneusi. One Health 2020, 12, 100209. [Google Scholar] [CrossRef]
- Didier, E.S.; Weiss, L.M. Microsporidiosis: Current status. Curr. Opin. Infect. Dis. 2006, 19, 485–492. [Google Scholar] [CrossRef]
- Li, J.; Wang, Z.; Karim, M.R.; Zhang, L. Detection of human intestinal protozoan parasites in vegetables and fruits: A review. Parasites Vectors 2020, 13, 380. [Google Scholar] [CrossRef]
- Hanson, K.L.; Cartwright, C.P. Use of an enzyme immunoassay does not eliminate the need to analyze multiple stool specimens for sensitive detection of Giardia lamblia. J. Clin. Microbiol. 2001, 39, 474–477. [Google Scholar] [CrossRef]
- Jahan, N.; Khatoon, R.; Ahmad, S. A Comparison of Microscopy and Enzyme Linked Immunosorbent Assay for Diagnosis of Giardia lamblia in Human Faecal Specimens. J. Clin. Diagn. Res. 2014, 8, DC04-6. [Google Scholar] [CrossRef]
- Christy, N.C.V.; Hencke, J.D.; Cadiz, A.E.-D.; Nazib, F.; von Thien, H.; Yagita, K.; Ligaba, S.; Haque, R.; Nozaki, T.; Tannich, E.; et al. Multisite performance evaluation of an enzyme-linked immunosorbent assay for detection of Giardia, Cryptosporidium, and Entamoeba histolytica antigens in human stool. J. Clin. Microbiol. 2012, 50, 1762–1763. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ryan, U.; Paparini, A.; Oskam, C. New Technologies for Detection of Enteric Parasites. Trends Parasitol. 2017, 33, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Friesen, J.; Fuhrmann, J.; Kietzmann, H.; Tannich, E.; Müller, M.; Ignatius, R. Evaluation of the Roche LightMix Gastro parasites multiplex PCR assay detecting Giardia duodenalis, Entamoeba histolytica, cryptosporidia, Dientamoeba fragilis, and Blastocystis hominis. Clin. Microbiol. Infect. 2018, 24, 1333–1337. [Google Scholar] [CrossRef] [PubMed]
- Laude, A.; Valot, S.; Desoubeaux, G.; Argy, N.; Nourrisson, C.; Pomares, C.; Machouart, M.; Le Govic, Y.; Dalle, F.; Botterel, F.; et al. Is real-time PCR-based diagnosis similar in performance to routine parasitological examination for the identification of Giardia intestinalis, Cryptosporidium parvum/Cryptosporidium hominis and Entamoeba histolytica from stool samples? Evaluation of a new commercial multiplex PCR assay and literature review. Clin. Microbiol. Infect. 2016, 22, 190.e1–190.e8. [Google Scholar] [CrossRef]
- Basmaciyan, L.; François, A.; Vincent, A.; Valot, S.; Bonnin, A.; Costa, D.; Razakandrainibe, R.; Morio, F.; Favennec, L.; Dalle, F. Commercial Simplex and Multiplex PCR Assays for the Detection of Intestinal Parasites Giardia intestinalis, Entamoeba spp., and Cryptosporidium spp.: Comparative Evaluation of Seven Commercial PCR Kits with Routine In-House Simplex PCR Assays. Microorganisms 2021, 9, 2325. [Google Scholar] [CrossRef]
- Hayatsu, H.; Wataya, Y.; Kai, K.; Iida, S. Reaction of sodium bisulfite with uracil, cytosine, and their derivatives. Biochemistry 1970, 9, 2858–2865. [Google Scholar] [CrossRef]
- Shapiro, R.; Weisgras, J.M. Bisulfite-catalyzed transamination of cytosine and cytidine. Biochem. Biophys. Res. Commun. 1970, 40, 839–843. [Google Scholar] [CrossRef]
- Baleriola, C.; Millar, D.; Melki, J.; Coulston, N.; Altman, P.; Rismanto, N.; Rawlinson, W. Comparison of a novel HPV test with the Hybrid Capture II (hcII) and a reference PCR method shows high specificity and positive predictive value for 13 high-risk human papillomavirus infections. J. Clin. Virol. 2008, 42, 22–26. [Google Scholar] [CrossRef]
- Siah, S.P.; Merif, J.; Kaur, K.; Nair, J.; Huntington, P.G.; Karagiannis, T.; Stark, D.; Rawlinson, W.; Olma, T.; Thomas, L.; et al. Improved detection of gastrointestinal pathogens using generalised sample processing and amplification panels. Pathology 2014, 46, 53–59. [Google Scholar] [CrossRef]
- Garae, C.; Kalo, K.; Pakoa, G.J.; Baker, R.; Isaacs, P.; Millar, D.S. Validation of the easyscreen flavivirus dengue alphavirus detection kit based on 3base amplification technology and its application to the 2016/17 Vanuatu dengue outbreak. PLoS ONE 2020, 15, e0227550. [Google Scholar] [CrossRef]
- Rochelle, P.A.; De Leon, R.; Stewart, M.H.; Wolfe, R.L. Comparison of primers and optimization of PCR conditions for detection of Cryptosporidium parvum and Giardia lamblia in water. Appl. Environ. Microbiol. 1997, 63, 106–114. [Google Scholar] [CrossRef]
- Dirani, G.; Zannoli, S.; Paesini, E.; Farabegoli, P.; Dalmo, B.; Vocale, C.; Liguori, G.; Varani, S.; Sambri, V. Easyscreen™ Enteric Protozoa Assay For the Detection of Intestinal Parasites: A Retrospective Bi-Center Study. J. Parasitol. 2019, 105, 58–63. [Google Scholar] [CrossRef]
- Stark, D.; Roberts, T.; Ellis, J.T.; Marriott, D.; Harkness, J. Evaluation of the EasyScreen™ enteric parasite detection kit for the detection of Blastocystis spp., Cryptosporidium spp., Dientamoeba fragilis, Entamoeba complex, and Giardia intestinalis from clinical stool samples. Diagn. Microbiol. Infect. Dis. 2014, 78, 149–152. [Google Scholar] [CrossRef]
| Organism | Symptoms | Infections |
|---|---|---|
| G. intestinalis | Acute, watery diarrhea. Chronic infections can result in weight loss and malabsorption and can cause long-term complications, including irritable bowel syndrome and chronic fatigue. | >200 million people annually. |
| Cryptosporidium spp. | Symptoms include abdominal pain, fever, vomiting, malabsorption, and self-limiting diarrhea. | >2.9 million cases of cryptosporidiosis in children aged <2 years old in sub-Saharan Africa alone annually. |
| E. histolytica | Asymptomatic to severe symptoms, including dysentery (bloody diarrhea) and extra-intestinal abscesses. | Amebiasis results in 100,000 deaths annually on a global basis. |
| C. cayetanensis | Watery diarrhea, weight loss, and cramping. | Outbreaks of cyclosporiasis occur regularly in the US, mostly linked to infected food or drink. |
| Manufacturer | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Becton Dickinson (Franklin Lakes, NJ, USA) | Diagenode (Denville, NJ, USA) | R-Biopharm (Hessen, Germany) | Fast Track (Esch-sur-Alzette, Luxembourg) | Savyon (Ashdod, Israel) | Certest (Zaragoza, Spain) | Biofire (Salt Lake City, UT, USA) | SeeGene (Seoul, Republic of Korea) | AusDiagnostics (Mascot, NSW, Australia) | Genetic Signatures (Newtown, NSW, Australia) | |
| Target | BD MAX™ | Gastro panel | RIDAGENE | FTD Stool | NanoCHIP | VIASURE | Filmarray | ALLplex | Parasite 8 | EP005 |
| G. intestinalis | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Cryptosporidium spp. | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| E. histolytica * | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| D. fragilis | No | No | No | No | No | Yes | No | Yes | Yes | Yes |
| B. hominis | No | No | No | No | No | Yes | No | Yes | Yes | Yes |
| C. cayetanensis | No | No | No | No | No | No | Yes | Yes | Yes | Yes |
| E. bieneusi | No | No | No | No | No | No | No | No | No | Yes |
| E. intestinalis | No | No | No | No | No | No | No | No | No | Yes |
| 4Base Sequence | 3Base™ Sequence | |||
|---|---|---|---|---|
| Target | Sequence | Tm °C | Sequence | Tm °C |
| Primer-1 | GTACACACCGCCCGTCGCTCCTACC | 66.6 | GTATATATTGTTTGTTGTTTTTATT | 51 |
| Primer-2 | GAGGAAGGTGAAGTCGTAACAAG | 55.2 | GAGGAAGGTGAAGTTGTAATAAG | 49.7 |
| Entamoeba spp. Probe | TGAATAAAGAGGTGAAATTCTAGG | 51.3 | TGAATAAGAGGTGAAATTTTAGG | 49 |
| G. intestinalis Probe | GCGCGAAGGGCCGCGAGCCCCCGCGC | 78.8 | GTGTGAAGGGTTGTGAGTTTTTGTGT | 57.9 |
| D. fragilis Probe | TTTAAATGAAAAGGTGATTAAATCACG | 51.2 | TTTAAATGAAAAGGTGATTAAATTATG | 47.7 |
| Cryptosporidium spp. Probe | GACCATACTTTGTAGCAATACATGTAAGGA | 56.6 | GATTATATTTTGTAGTAATATATGTAAGGA | 48.1 |
| B. hominis Probe | TATTGAAAGAAGTTGTGTAAATCTTACCATT | 53.9 | TATTGAAAGAAGTTGTGTAAATTTTATTATT | 50 |
| Target Organism; Strain (Accession Number) | Target Type (ATCC Cat. Number) |
|---|---|
| Blastocystis hominis NANDII | Culture (50177) |
| Blastocystis hominis NTY | Culture (50610) |
| Cryptosporidium parvum; (* CP082114.1) | Clinical stool |
| Cryptosporidium hominis; (* XM_661199.1) | Clinical stool |
| Cyclospora cayetanensis; (* AF111183.1) | Clinical stool |
| Dientamoeba fragilis; (* JQ677148.1) | Culture |
| Dientamoeba fragilis; (* JQ677148.1) | Clinical stool |
| Entamoeba histolytica HU-21:AMC | Culture (30457) |
| Enterocytozoon bieneusi; (* AF023245.1) | Clinical stool |
| Enterocytozoon bieneusi; MWC_m4 (MG976813.1) | Clinical stool |
| Encephalitozoon intestinalis | Culture (50506) |
| Giardia lamblia, Human isolate—H3 (AF023245.1) | Quantified non-viable oocysts |
| Giardia intestinalis; BE-1 | Culture PRA-42 |
| LLOD | Positive Replicates | Ct Value Average | |||
|---|---|---|---|---|---|
| Target | Concentration | Isolate 1 | Isolate 2 | Isolate 1 | Isolate 2 |
| Dientamoeba fragilis | 125 org/mL | 20/20 | 20/20 | 32.45 | 30.66 |
| Cryptosporidium parvum | 1600 org/mL | 19/20 | 20/20 | 32.54 | 31.95 |
| Cryptosporidium hominis | 1600 org/mL | 20/20 | 20/20 | 31.81 | 30.34 |
| Cyclospora cayetanensis | 25 GE/mL | 20/20 | 20/20 | 33.11 | 31.76 |
| Blastocystis hominis | 100 org/mL | 20/20 | 19/20 | 31.96 | 33.56 |
| Giardia intestinalis | 5700 GE/mL | 20/20 | 19/20 | 32.23 | 32.63 |
| Entamoeba histolytica | 3000 org/mL | 20/20 | 20/20 | 33.76 | 32.81 |
| Enterocytozoon bieneusi | 4 org/mL | 20/20 | 20/20 | 34.53 | 32.79 |
| Encephalitozoon intestinalis | 1200–2500 org/mL | 20/20 * | 20/20 # | 33.04 | 29.84 |
| Organism; Strain (Accession Number) | Target Type * | Organism; Strain (Accession Number) | Target Type * |
|---|---|---|---|
| Blastocystis hominis; BT1 | Culture (50608) | Dientamoeba fragilis | Clinical |
| Blastocystis hominis; NMH | Culture (50754) | Dientamoeba fragilis | Clinical |
| Blastocystis hominis; DL | Culture (50626) | Dientamoeba fragilis | Clinical |
| Cryptosporidium parvum | Clinical | Encephalitozoon intestinalis; CDC:V297 (50651) | Culture (50651) |
| Cryptosporidium parvum | Clinical | Encephalitozoon intestinalis; nasal isolate (50507) | Culture (50507) |
| Cryptosporidium parvum | Clinical | Encephalitozoon intestinalis; CDC:V307 (50603) | Culture (50603) |
| Cryptosporidium parvum | Clinical | Encephalitozoon intestinalis; CDC:V308 (50790) | Culture (50790) |
| Cryptosporidium parvum | Clinical | Enterocytozoon bieneusi | DNA |
| Cryptosporidium hominis | Clinical | Entamoeba histolytica; HB-301:NIH (30190) | Culture (30190) |
| Cryptosporidium hominis | Clinical | Entamoeba histolytica; HK-9 Clone 6 (50544) | Culture (50544) |
| Cryptosporidium hominis | Clinical | Entamoeba histolytica; HB-301:NIH CL-1-3 (50547) | Culture (50547) |
| Cryptosporidium hominis | Clinical | Giardia intestinalis; WB (30957) | Culture (30957) |
| Cryptosporidium hominis | Clinical | Giardia intestinalis; WB clone C6 (50803) | Culture (50803) |
| Dientamoeba fragilis | Clinical | Giardia intestinalis; GS clone H7 (50581) | Culture (50581) |
| Cyclospora cayetanensis (KX618190) | Clinical | Giardia intestinalis; Portland-1 (30888) | Culture (30888) |
| Cyclospora cayetanensis (MN316534) | Clinical | Giardia intestinalis; (Lambl) Alexeieff Strain PR-15 | Culture |
| Cyclospora cayetanensis (MN316534) | Clinical | Giardia lamblia; CM (PRA242) | Culture (PRA242) |
| Cyclospora cayetanensis (MN316535) | Clinical | ||
| Dientamoeba fragilis | Clinical |
| Cross-Reactivity Species Tested | |||
|---|---|---|---|
| Atopobium vaginae * | Cedecea davisae * | Human adenovirus 5 | Proteus vulgaris |
| Abiotrophia defective * | Chlamydia trachomatis | Klebsiella oxytoca | Providencia stuartii |
| Acinetobacter baumannii | Chryseobacterium gleum * | Lactobacillus acidophilus | Rotavirus A |
| Adenovirus | Citrobacter freundii | Lactococcus lactis | Saccharomyces cerevisiae * |
| Aeromonas hydrophila | Clostridium perfringens | Leminorella grimontii * | Salmonella enterica |
| Akkermansia muciniphila | Corynebacterium glutamicum * | Listeria monocytogenes | Sapovirus |
| Alcaligenes faecalis | Cronobacter sakazakii | Mycobacterium abscessus | Serratia marcescens |
| Anaerococcus tetradius * | Desulfovibrio pigerl * | Mycobacterium avium | Shigella flexneri |
| Arcobacter butzleri * | Edwardsiella tarda * | Mycobacterium spp. | Shigella sonnei |
| Astrovirus | Eggerthella lenta | Mycoplasma fermentans | Staphylococcus aureus |
| Atopogium vaginae * | Enterococcus faecalis | Mycoplasma hominis | Staphylococcus hominis |
| Bacillus cereus | Enterococcus faecium | Mycoplasma salivarium | Stenotrophomonas maltophila |
| Bacteroides fragilis | Enterovirus | Neisseria flava | Streptococcus mitis |
| Bifidobacterium adolescentis | Escherichia coli | Norovirus GI | Streptococcus pyogenes |
| Bocavirus | Eubacterium rectale * | Norovirus GII | Streptococcus salivarius |
| Campylobacter coli | Faecalibacterium prausnitzii * | Peptoniphilus asaccharolyticus * | Streptococcus sanguinis |
| Campylobacter hominis | Fusobacterium varium * | Peptostreptococcus anaerobius | Streptococcus thermophilus |
| Campylobacter jejuni | Gardnerella vaginalis | Porphyromonas asaccharolytica * | Veillonella parvula |
| Campylobacter lari | Gemella morbillorum * | Porphyromonas levii * | Yersinia pseudotuberculosis |
| Candida albicans | Hafnia alvei | Prevotella melaninogenica | |
| Capnocytophaga gingivalis | Helicobacter pylori | Proteus mirabilis | |
| Target Organism | Target Type (ATCC Cat. Number) |
|---|---|
| Pseudomonas aeruginosa | ATCC 47085DQ |
| Candida albicans | ATCC MYA-2876D-5 |
| Clostridioides perfringens | ATCC 13124DQ |
| non-pathogenic Escherichia coli | ATCC 25922DQ |
| Enterococcus faecalis | ATCC 700802DQ |
| Bacteroides fragilis | ATCC 25285D-5 |
| Klebsiella pneumoniae | ATCC 13883DQ |
| Saccharomyces cerevisiae * | ATCC MYA-796 |
| Sample | QCMD Result | EP005 Result |
|---|---|---|
| EP19S-01 | D. fragilis/B. hominis | D. fragilis/B. hominis |
| EP19S-02 | E. histolytica | E. histolytica |
| EP19S-03 | Negative | Negative |
| EP19S-04 | E. histolytica | E. histolytica |
| EP19S-05 | G. intestinalis | G. intestinalis |
| EP19S-06 | Cryptosporidium spp. | Cryptosporidium spp. |
| EP19S-07 | G. intestinalis | G. intestinalis |
| EP19S-08 | Cryptosporidium spp. | Cryptosporidium spp. |
| EP19S-09 | G. intestinalis | G. intestinalis |
| Target | Specimen Type | Samples Tested | EP005 Positive | Confirmatory Method | Positive with Confirmatory Method |
|---|---|---|---|---|---|
| Cryptosporidium spp. | Prospective | 380 | 10 | BD MAX™ | 9 |
| G. intestinalis | Prospective | 380 | 20 | BD MAX™ | 21 |
| E. histolytica | Prospective | 380 | 8 | BD MAX™ | 8 |
| B. hominis | Prospective | 380 | 24 | NAAT | 11/16 * |
| D. fragilis | Prospective | 380 | 37 | NAAT | 15/16 * |
| C. cayetanensis | Retrospective | 11 | 11 | NAAT | 11 |
| E. bieneusi | Retrospective | 21 | 21 | NAAT | 19 ** |
| E. intestinalis | Contrived/Retrospective | 5 | 5 | NAAT | 5 |
| Target | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|
| Cryptosporidium spp. | 100% (66.37–100) | 99.7% (98.51–99.99) | 90% (55.97–98.46) | 100% |
| G. intestinalis | 95.45% (77.16–99.88) | 100% (98.98–100) | 100% | 99.74% (98.14–99.96) |
| E. histolytica | 100% (63.06–100) | 100% (99.01–100) | 100% | 100% |
| Channel/Representative Fluorophore | Panel A | Panel B | Panel C |
|---|---|---|---|
| Fam | D. fragilis | B. hominis | E. bieneusi |
| Hex/Vic | EC | IPC | EC |
| Texas Red/Rox | C. cayetanensis | E. histolytica | E. intestinalis |
| Cy5 | Cryptosporidium. spp. | G. intestinalis | Not used |
| Target | Forward Primer | Reverse Primer | Gene | Anneal Temp |
|---|---|---|---|---|
| D. fragilis Region 1 | AATACCTTTTAATAGGTAATCCAATCGA | GCCCTCTGCTAGGTTACAATATAC | 18S rRNA | 64 °C |
| D. fragilis Region 2 | TTTAATGACTGATCAGGCTATAGG | CATCACGGACCTGTTATTGCTACCA | 18S rRNA | 64 °C |
| C. cayetanensis Region 1 | CGCATTTGGCTTTAGCCGGCGATA | TTACTCTGGAAGGATTTTAAATTCCT | 18S rRNA | 62 °C |
| C. cayetanensis Region 2 | CACGCTCTACCAATATTCGTTATCA | GGATCGTGTTGGCTAGGTGTACTAA | Mitochondria | 65 °C |
| B. hominis Region 1 | AGTAGTCATACGCTCGTCTCAAA | TCTTCGTTACCCGTTACTGC | SSU RNA | 65 °C |
| B. hominis Region 2 | CAATTGGAGGGCAAGTCTGGTGC | CACCTCTAACTATTGAATATGAATACC | SSU RNA | 64 °C |
| E. bieneusi Region 1 | TAGCGGAACGGATAGGGAGTGTAGT | CTTGCGAGCGTACTATCCCCAGAG | SSU RNA | 64 °C |
| E. bieneusi Region 2 | CATTCGTTGATCGAATACGTGAGAAT | GTTACTAGGAATTCCTTATTCACTACG | SSU RNA | 64 °C |
| E. intestinalis Region 1 | TCACGGCATCCATTTCAAACGG | GATGAAGGACGAAGGCTAGAGGA | 16S rRNA | 64 °C |
| E. intestinalis Region 2 | GTCCAAGAGCACAGCCTTCGCTTC | GGGATCGGGGTTTGATTCCGGA | 16S rRNA | 64 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aghazadeh, M.; Jones, M.; Perera, S.; Nair, J.; Tan, L.; Clark, B.; Curtis, A.; Jones, J.; Ellem, J.; Olma, T.; et al. The Application of 3base™ Technology to Diagnose Eight of the Most Clinically Important Gastrointestinal Protozoan Infections. Int. J. Mol. Sci. 2023, 24, 13387. https://doi.org/10.3390/ijms241713387
Aghazadeh M, Jones M, Perera S, Nair J, Tan L, Clark B, Curtis A, Jones J, Ellem J, Olma T, et al. The Application of 3base™ Technology to Diagnose Eight of the Most Clinically Important Gastrointestinal Protozoan Infections. International Journal of Molecular Sciences. 2023; 24(17):13387. https://doi.org/10.3390/ijms241713387
Chicago/Turabian StyleAghazadeh, Mahdis, Meghan Jones, Suneth Perera, Jiny Nair, Litty Tan, Brett Clark, Angela Curtis, Jackson Jones, Justin Ellem, Tom Olma, and et al. 2023. "The Application of 3base™ Technology to Diagnose Eight of the Most Clinically Important Gastrointestinal Protozoan Infections" International Journal of Molecular Sciences 24, no. 17: 13387. https://doi.org/10.3390/ijms241713387
APA StyleAghazadeh, M., Jones, M., Perera, S., Nair, J., Tan, L., Clark, B., Curtis, A., Jones, J., Ellem, J., Olma, T., Stark, D., Melki, J., Coulston, N., Baker, R., & Millar, D. (2023). The Application of 3base™ Technology to Diagnose Eight of the Most Clinically Important Gastrointestinal Protozoan Infections. International Journal of Molecular Sciences, 24(17), 13387. https://doi.org/10.3390/ijms241713387






