Exosomal MicroRNA and Protein Profiles of Hepatitis B Virus-Related Hepatocellular Carcinoma Cells
Abstract
1. Introduction
2. Results
2.1. Isolation and Identification of Exosomes
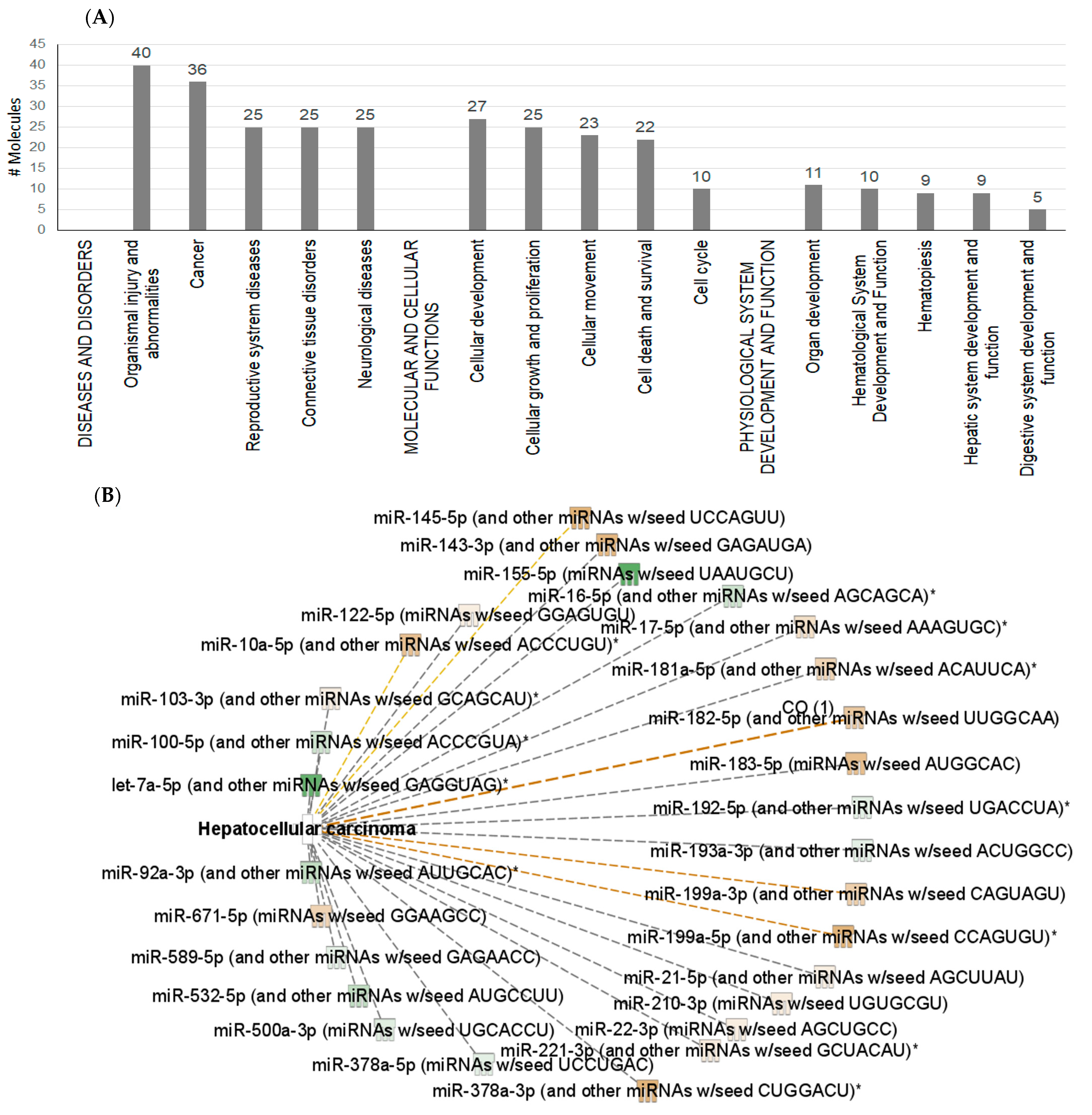
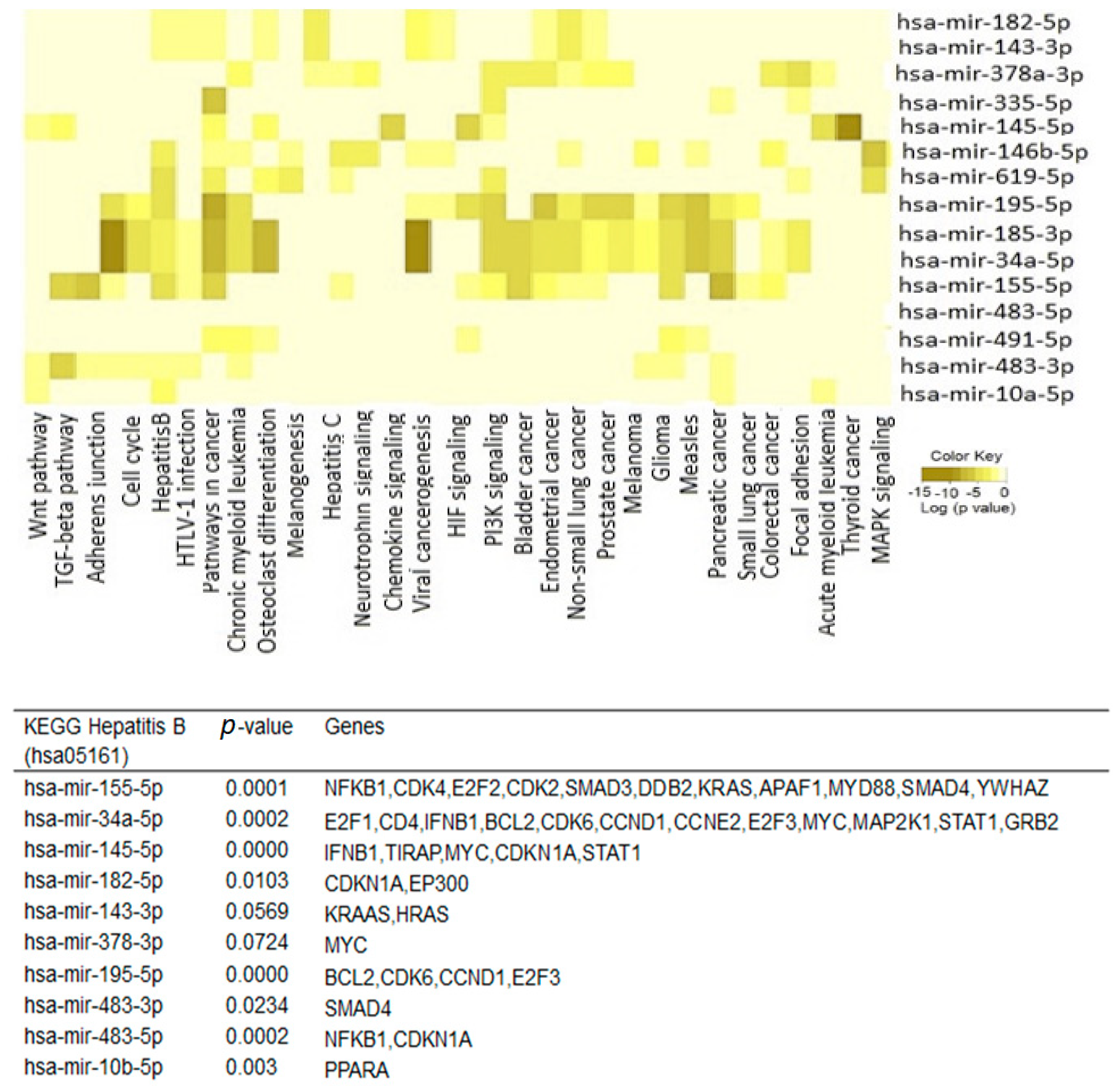
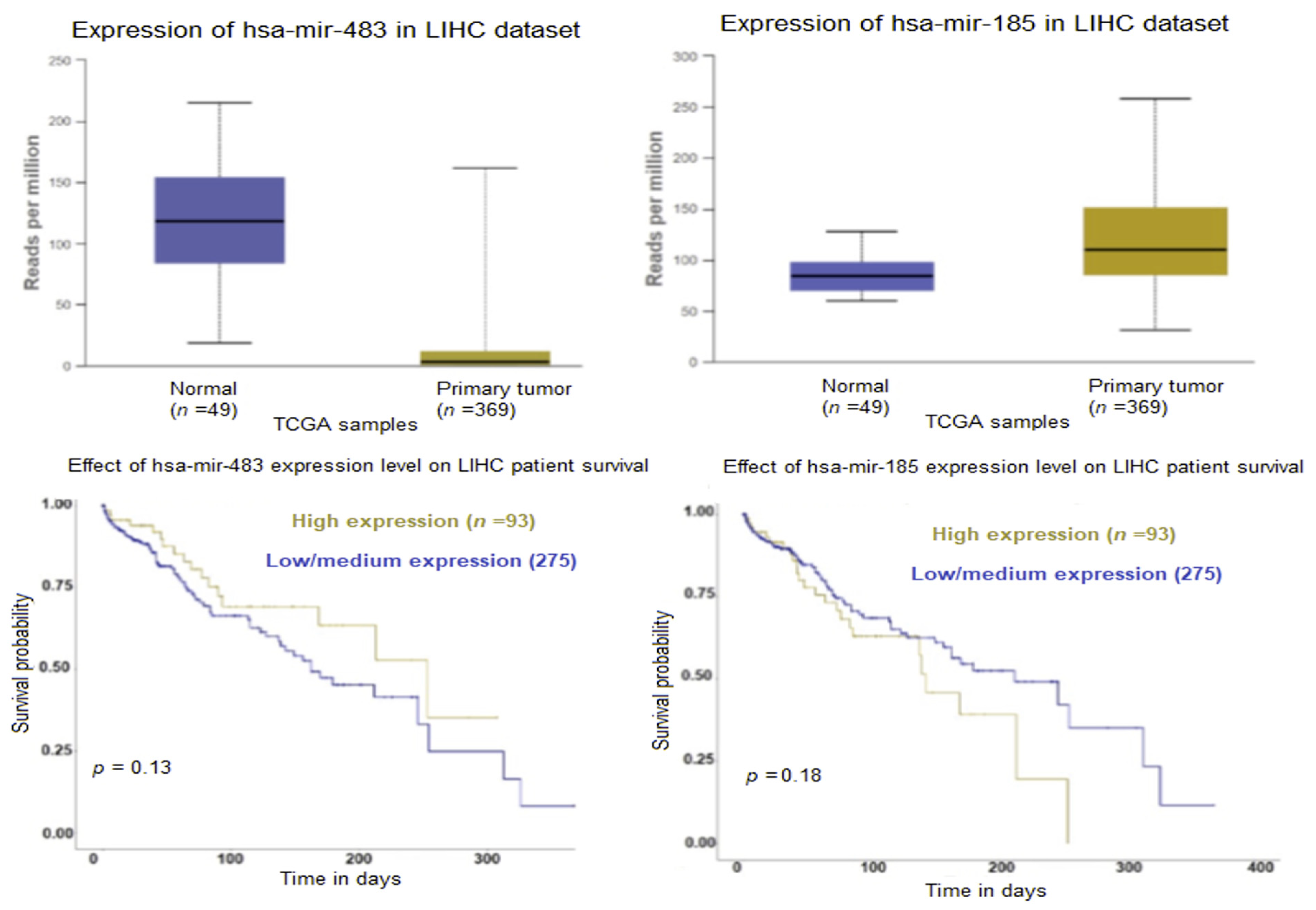
2.2. Exosomal microRNA
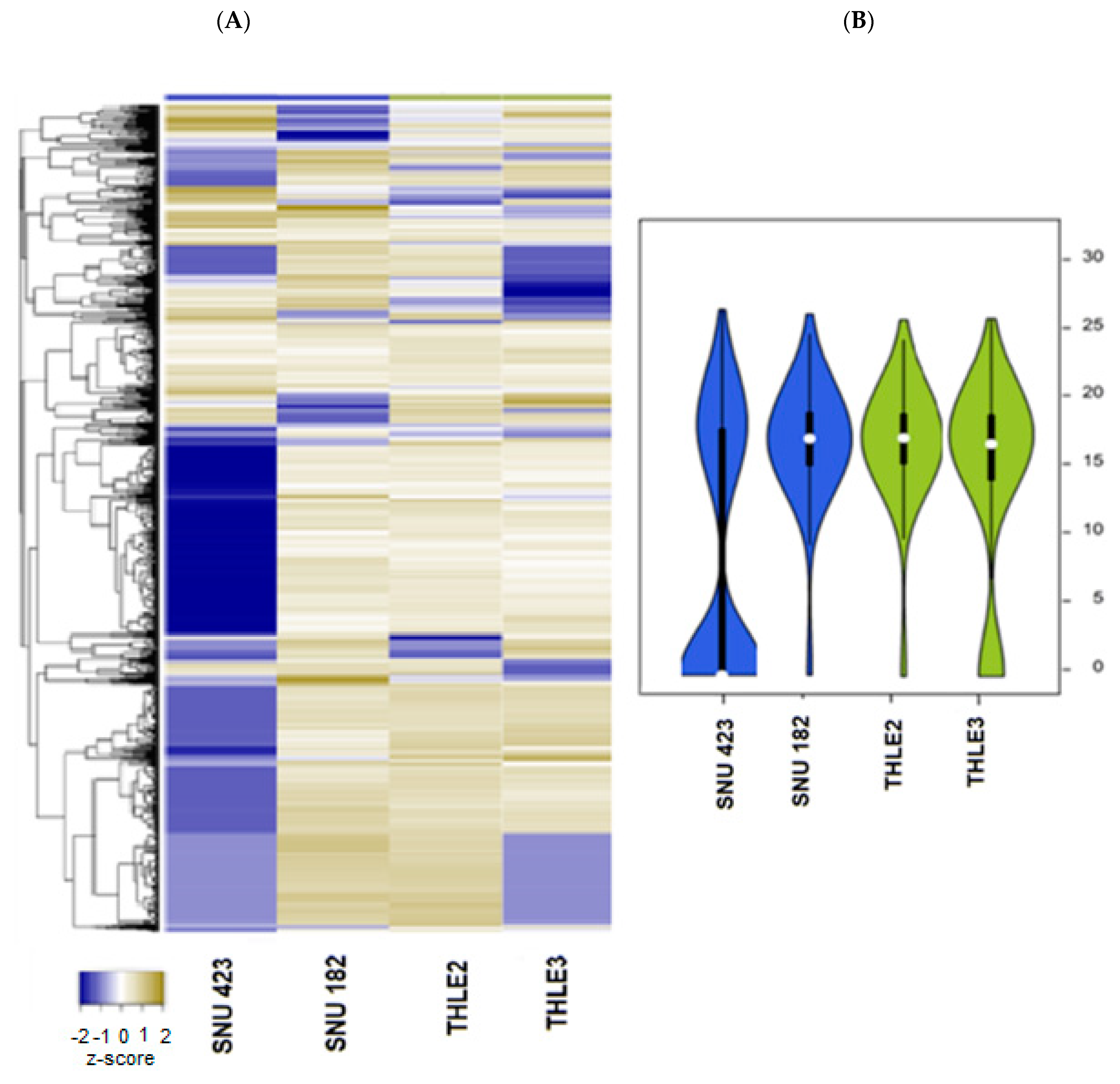
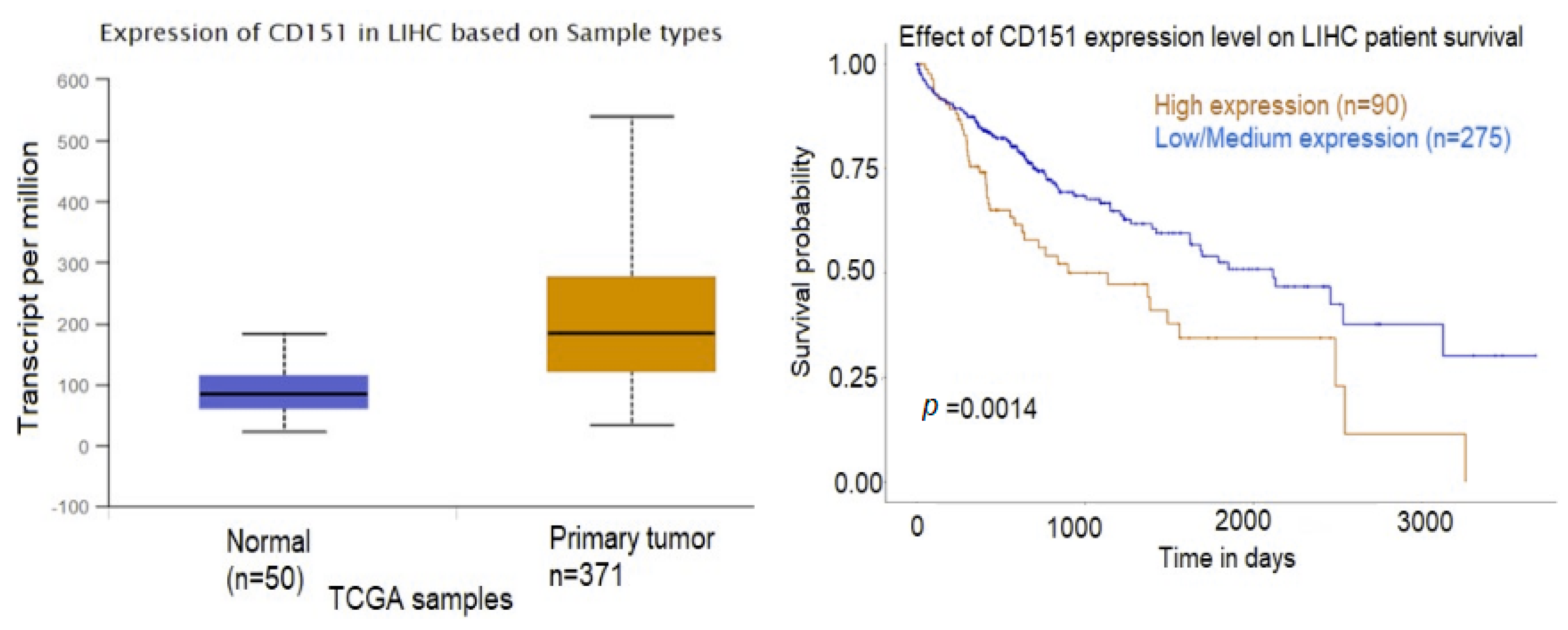
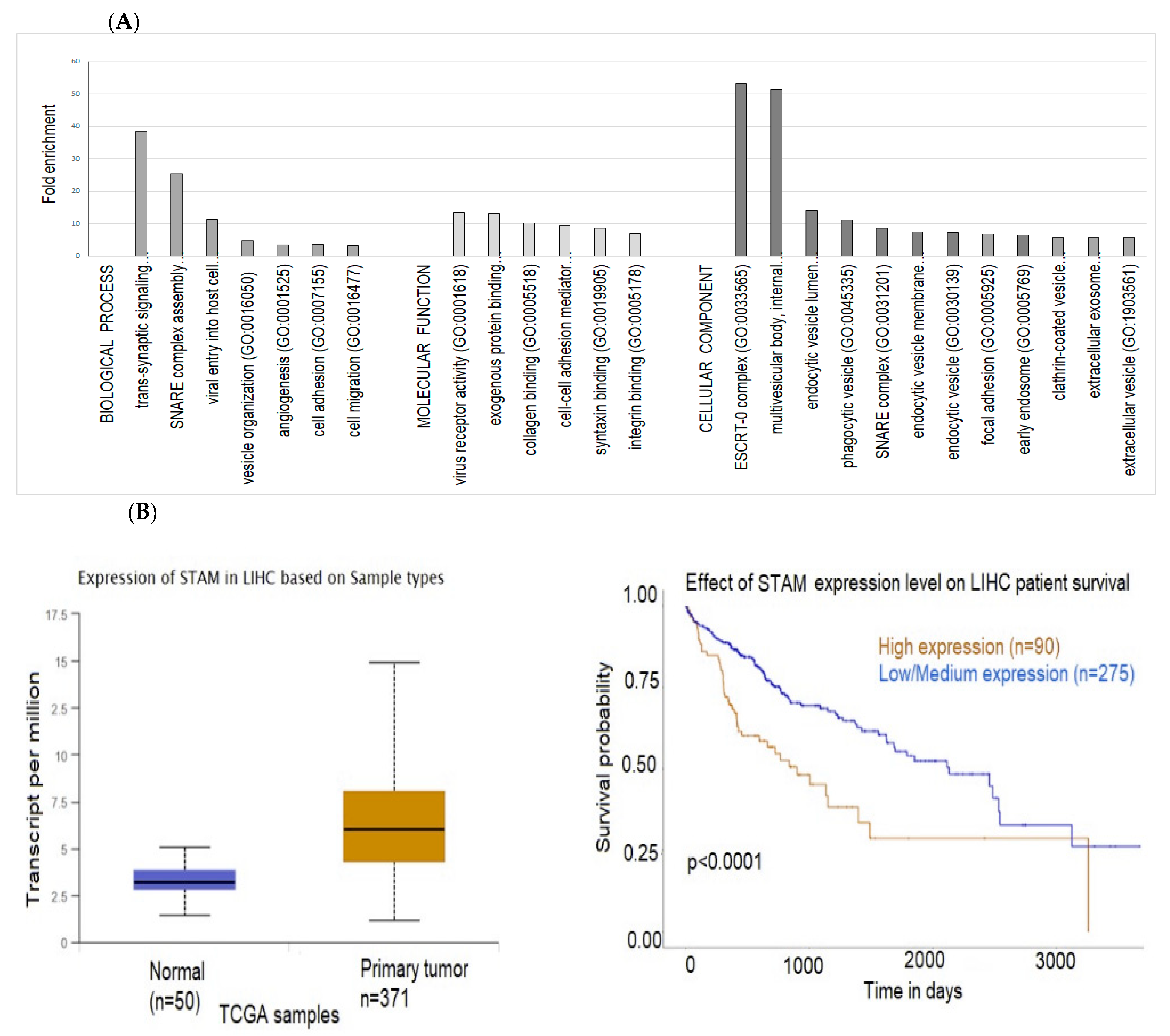
2.3. Exosomal Proteins
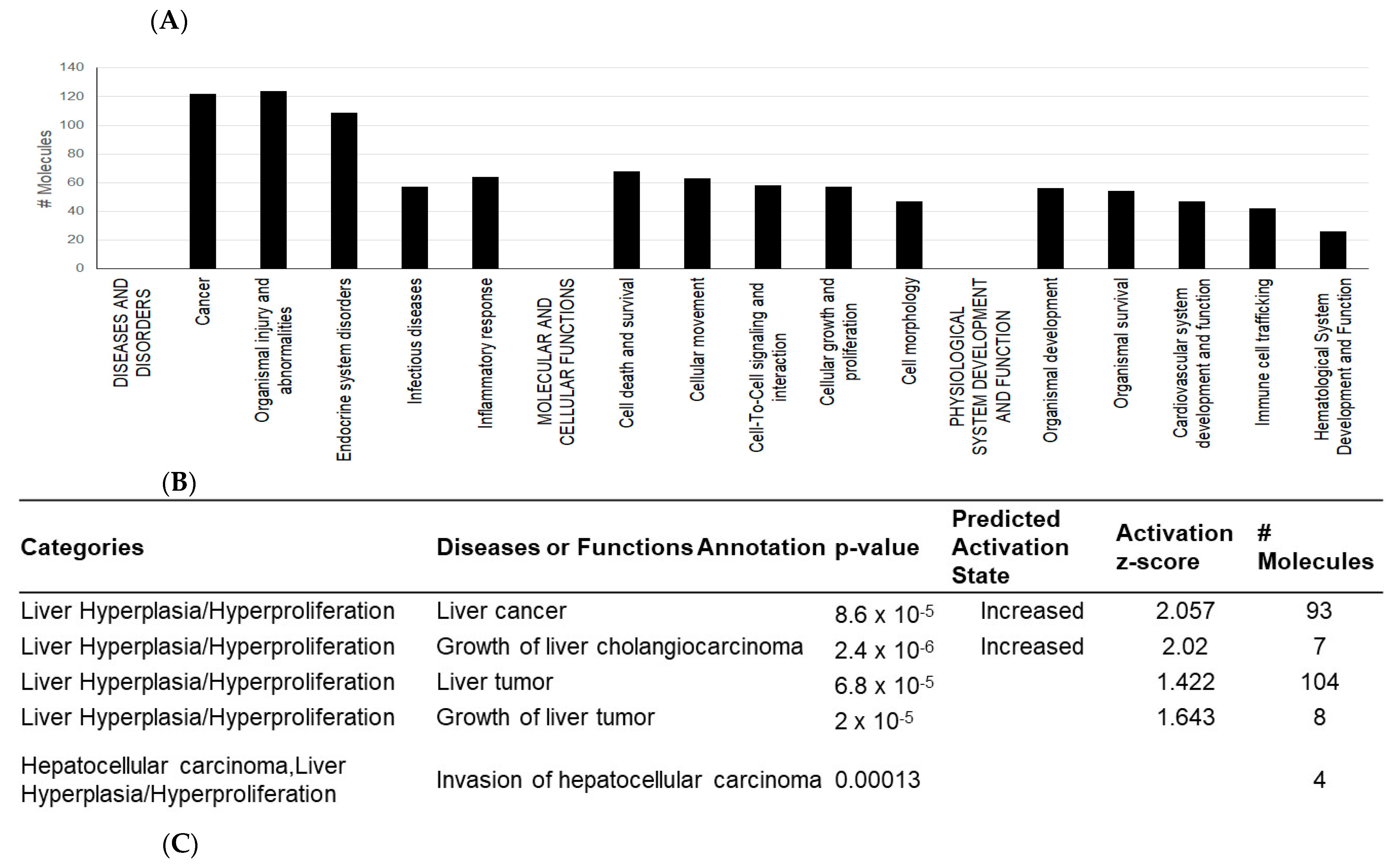
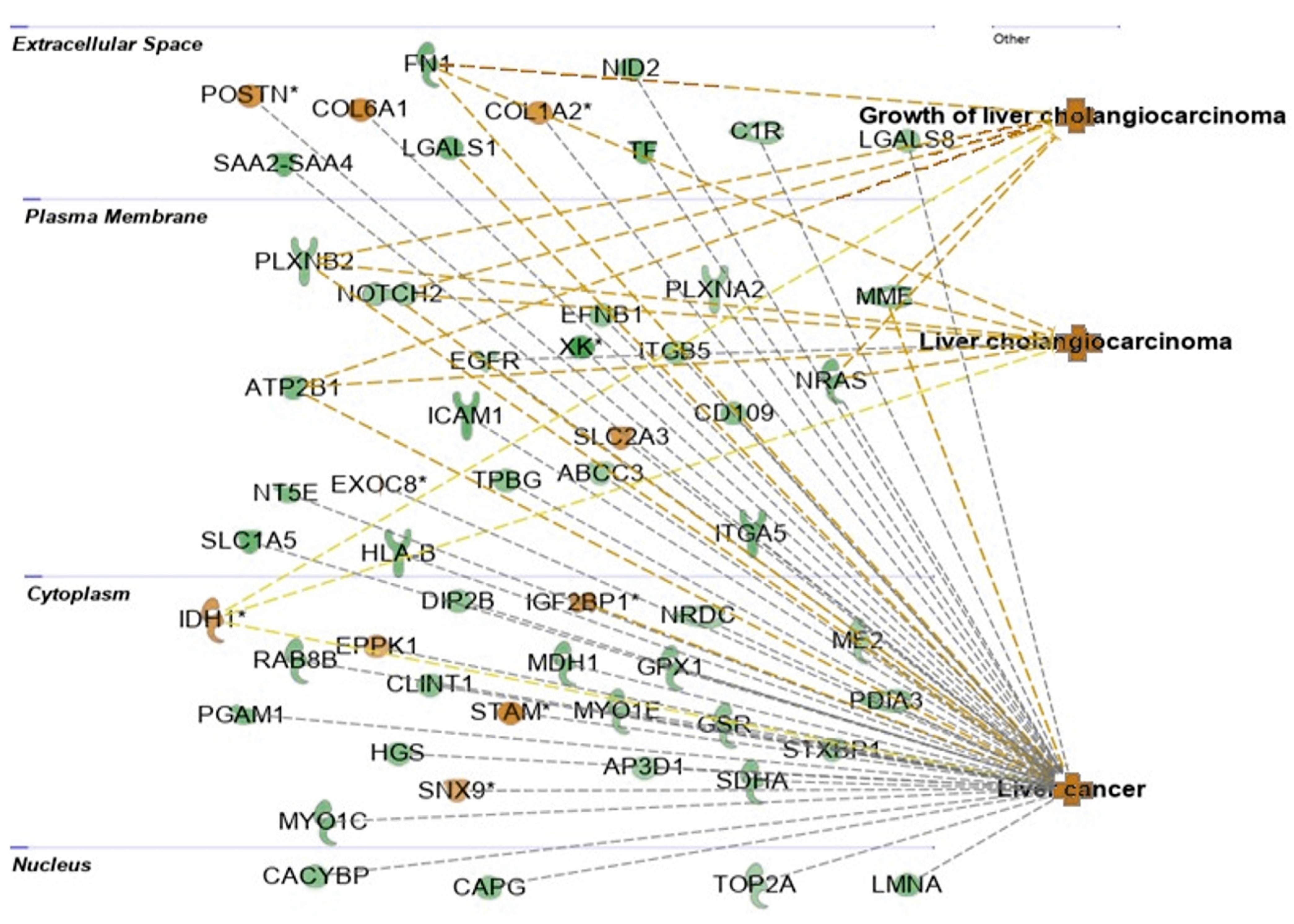
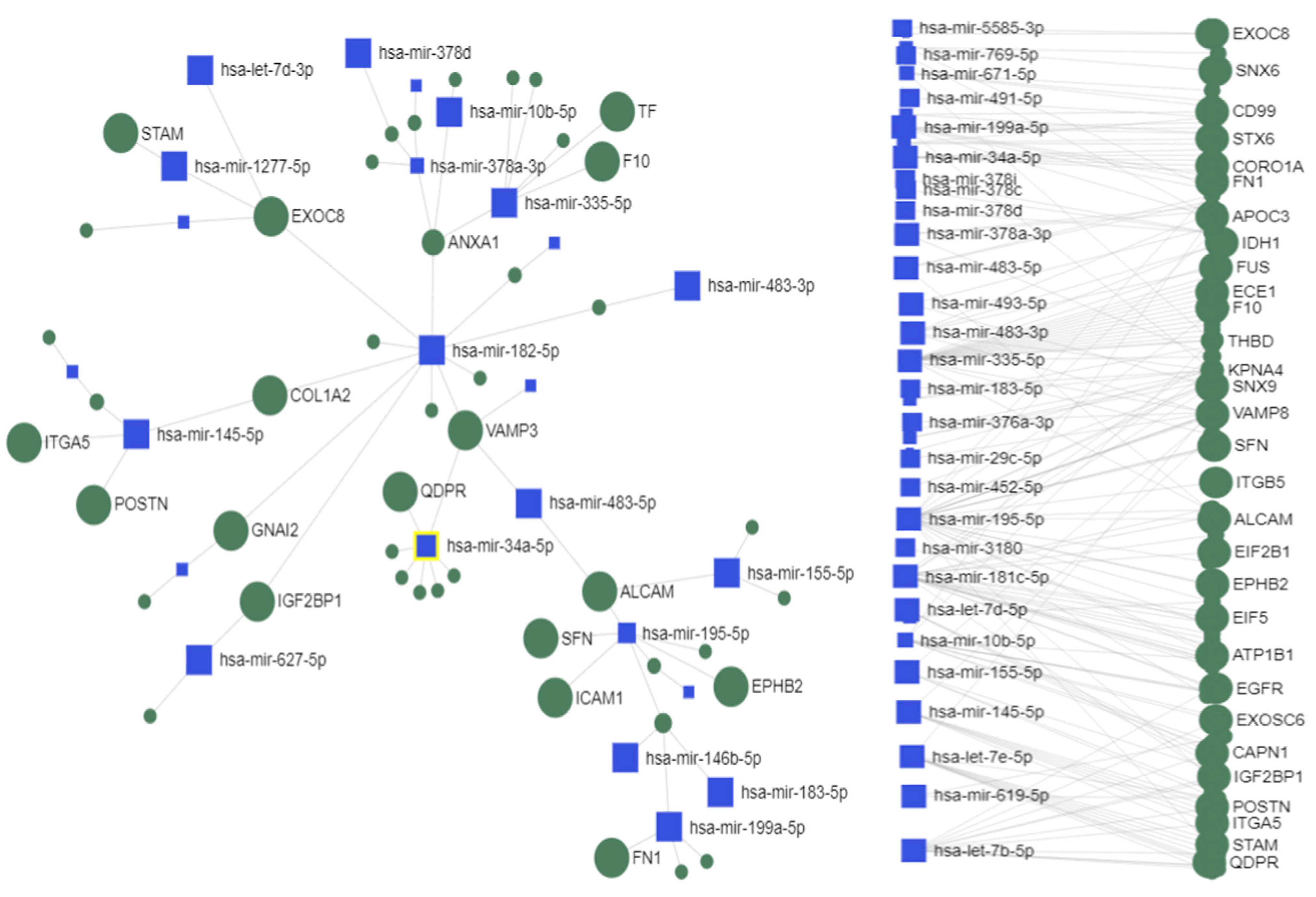
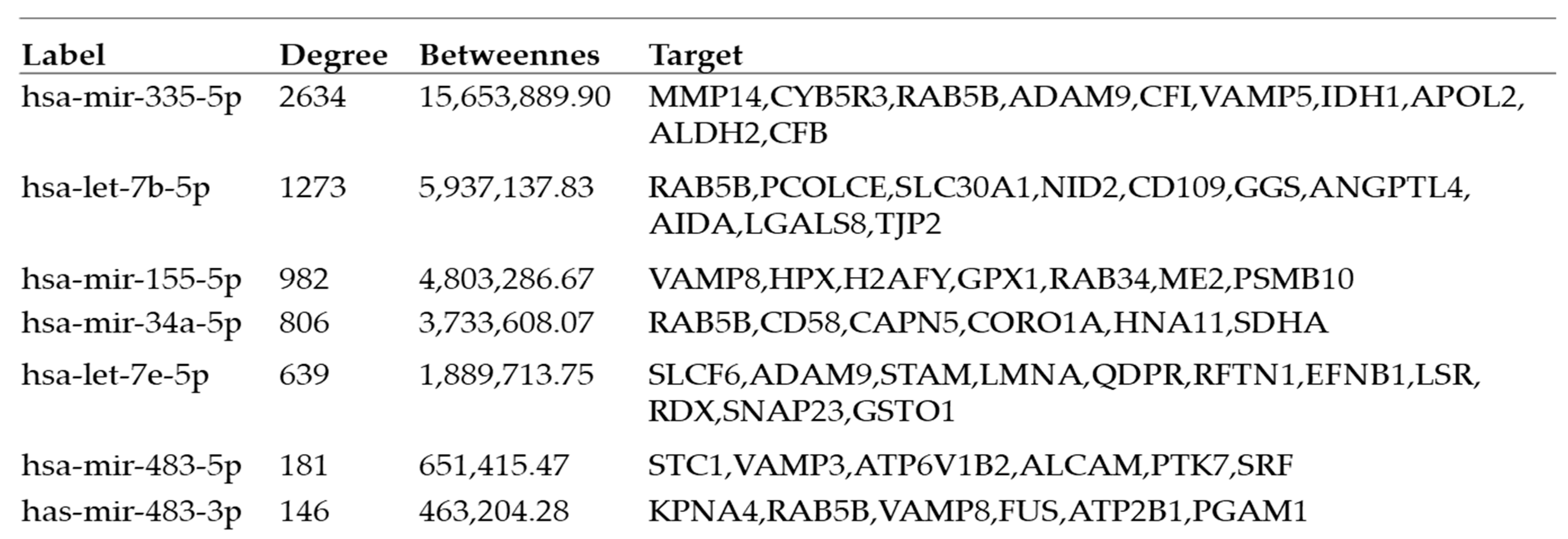
2.4. Interactions between Exosomal miRNAs and Proteins
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture
4.2. Exosome Purification
4.3. Nanoparticle Tracking Analysis (NTA)
4.4. Exosome microRNA Analysis
4.4.1. MicroRNA Isolation
4.4.2. Next Generation Sequencing (NGS)
4.4.3. Data Analysis
4.5. Exosome Proteomic Analysis
4.5.1. Liquid Chromatography with Tandem Mass Spectrometry (LC–MS)/MS)
4.5.2. Data Analysis
4.6. Functional and Pathway Analysis of Differentially Expressed Exosomal miRNAs and Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021, 73 (Suppl. S1), 4–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Golabi, P.; Fazel, S.; Otgonsuren, M.; Sayiner, M.; Locklear, C.T.; Younossi, Z.M. Mortality assessment of patients with hepatocellular carcinoma according to underlying disease and treatment modalities. Medicine 2017, 96, e5904. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; He, M.; Li, J.; Pessentheiner, A.; Wang, C.; Zhang, J.; Sun, Y.; Wang, W.T.; Zhang, Y.; Liu, J.; et al. microRNA-483 ameliorates hypercholesterolemia by inhibiting PCSK9 production. JCI Insight 2020, 5, e143812. [Google Scholar] [CrossRef]
- Zapatka, M.; Borozan, I.; Brewer, D.S.; Iskar, M.; Grundhoff, A.; Alawi, M.; Desai, N.; Sültmann, H.; Moch, H.; Cooper, C.S.; et al. The landscape of viral associations in human cancers. Nat. Genet. 2020, 52, 320–330. [Google Scholar] [CrossRef]
- Bouchard, M.J.; Navas-Martin, S. Hepatitis B and C virus hepatocarcinogenesis: Lessons learned and future challenges. Cancer Lett. 2011, 305, 123–143. [Google Scholar] [CrossRef]
- Rawat, S.; Bouchard, M.J. The hepatitis B virus (HBV) HBx protein activates AKT to simultaneously regulate HBV replication and hepatocyte survival. J. Virol. 2015, 89, 999–1012. [Google Scholar] [CrossRef]
- Seeger, C.; Mason, W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000, 64, 51–68. [Google Scholar] [CrossRef]
- Rawat, S.; Clippinger, A.J.; Bouchard, M.J. Modulation of apoptotic signaling by the hepatitis B virus X protein. Viruses 2012, 4, 2945–2972. [Google Scholar] [CrossRef]
- Liu, C.J.; Kao, J.H. Global perspective on the natural history of chronic hepatitis B: Role of hepatitis B virus genotypes A to J. Semin. Liver Dis. 2013, 33, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Paskeh, M.D.A.; Entezari, M.; Mirzaei, S.; Zabolian, A.; Saleki, H.; Naghdi, M.J.; Sabet, S.; Khoshbakht, M.A.; Hashemi, M.; Hushmandi, K.; et al. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J. Hematol. Oncol. 2022, 15, 83. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Wang, X.; Gong, Z.; Yu, M.; Wu, H.; Zhang, D. Exosome-mediated metabolic reprogramming: The emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target. Ther. 2020, 5, 242. [Google Scholar] [CrossRef] [PubMed]
- Osaki, M.; Okada, F. Exosomes and Their Role in Cancer Progression. Yonago Acta Med. 2019, 62, 182–190. [Google Scholar] [CrossRef]
- Jia, Y.; Chen, Y.; Wang, Q.; Jayasinghe, U.; Luo, X.; Wei, Q.; Wang, J.; Xiong, H.; Chen, C.; Xu, B.; et al. Exosome: Emerging biomarker in breast cancer. Oncotarget 2017, 8, 41717–41733. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.H.; Kim, J.H. A Comprehensive Review on Factors Influences Biogenesis, Functions, Therapeutic and Clinical Implications of Exosomes. Int. J. Nanomed. 2021, 16, 1281–1312. [Google Scholar] [CrossRef]
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef]
- Zhang, Q.; Higginbotham, J.N.; Jeppesen, D.K.; Yang, Y.P.; Li, W.; McKinley, E.T.; Graves-Deal, R.; Ping, J.; Britain, C.M.; Dorsett, K.A.; et al. Transfer of Functional Cargo in Exomeres. Cell Rep. 2019, 27, 940–954.e946. [Google Scholar] [CrossRef]
- Al Halawani, A.; Mithieux, S.M.; Yeo, G.C.; Hosseini-Beheshti, E.; Weiss, A.S. Extracellular Vesicles: Interplay with the Extracellular Matrix and Modulated Cell Responses. Int. J. Mol. Sci. 2022, 23, 3389. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Ratajczak, M.Z.; Ratajczak, J. Extracellular microvesicles/exosomes: Discovery, disbelief, acceptance, and the future? Leukemia 2020, 34, 3126–3135. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, e1413. [Google Scholar] [CrossRef] [PubMed]
- Bandopadhyay, M.; Bharadwaj, M. Exosomal miRNAs in hepatitis B virus related liver disease: A new hope for biomarker. Gut Pathog. 2020, 12, 23. [Google Scholar] [CrossRef]
- Sanderson, R.D.; Bandari, S.K.; Vlodavsky, I. Proteases and glycosidases on the surface of exosomes: Newly discovered mechanisms for extracellular remodeling. Matrix Biol. 2019, 75–76, 160–169. [Google Scholar] [CrossRef]
- Tai, Y.L.; Chen, K.C.; Hsieh, J.T.; Shen, T.L. Exosomes in cancer development and clinical applications. Cancer Sci. 2018, 109, 2364–2374. [Google Scholar] [CrossRef]
- Sharma, A.; Khatun, Z.; Shiras, A. Tumor exosomes: Cellular postmen of cancer diagnosis and personalized therapy. Nanomedicine 2016, 11, 421–437. [Google Scholar] [CrossRef]
- Hu, M.; Li, J.; Liu, C.G.; Goh, R.; Yu, F.; Ma, Z.; Wang, L. Noncoding RNAs of Extracellular Vesicles in Tumor Angiogenesis: From Biological Functions to Clinical Significance. Cells 2022, 11, 947. [Google Scholar] [CrossRef]
- Olejarz, W.; Kubiak-Tomaszewska, G.; Chrzanowska, A.; Lorenc, T. Exosomes in Angiogenesis and Anti-angiogenic Therapy in Cancers. Int. J. Mol. Sci. 2020, 21, 5840. [Google Scholar] [CrossRef]
- Jamshidi-Parsian, A.; Griffin, R.J.; Kore, R.A.; Todorova, V.K.; Makhoul, I. Tumor-endothelial cell interaction in an experimental model of human hepatocellular carcinoma. Exp. Cell Res. 2018, 372, 16–24. [Google Scholar] [CrossRef]
- Zhou, H.; Yan, Z.H.; Yuan, Y.; Xing, C.; Jiang, N. The Role of Exosomes in Viral Hepatitis and Its Associated Liver Diseases. Front. Med. 2021, 8, 782485. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hou, L.; Li, A.; Duan, Y.; Gao, H.; Song, X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. BioMed Res. Int. 2014, 2014, 864894. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, Y.; Takagi, R.; Naito, Y.; Kiniwa, T.; Tanaka, Y.; Hamada-Tsutsumi, S.; Kawano, M.; Matsushita, S.; Ochiya, T.; Miyajima, A. Identification of the novel 3’ UTR sequences of human IL-21 mRNA as potential targets of miRNAs. Sci. Rep. 2017, 7, 7780. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Fan, J.; Lyon, C.; Wan, M.; Hu, Y. Role of Extracellular Vesicles in Viral and Bacterial Infections: Pathogenesis, Diagnostics, and Therapeutics. Theranostics 2018, 8, 2709–2721. [Google Scholar] [CrossRef]
- Pérez, P.S.; Romaniuk, M.A.; Duette, G.A.; Zhao, Z.; Huang, Y.; Martin-Jaular, L.; Witwer, K.W.; Théry, C.; Ostrowski, M. Extracellular vesicles and chronic inflammation during HIV infection. J. Extracell. Vesicles 2019, 8, 1687275. [Google Scholar] [CrossRef]
- Arzumanian, V.A.; Kiseleva, O.I.; Poverennaya, E.V. The Curious Case of the HepG2 Cell Line: 40 Years of Expertise. Int. J. Mol. Sci. 2021, 22, 13135. [Google Scholar] [CrossRef]
- Pfeifer, A.M.; Cole, K.E.; Smoot, D.T.; Weston, A.; Groopman, J.D.; Shields, P.G.; Vignaud, J.M.; Juillerat, M.; Lipsky, M.M.; Trump, B.F.; et al. Simian virus 40 large tumor antigen-immortalized normal human liver epithelial cells express hepatocyte characteristics and metabolize chemical carcinogens. Proc. Natl. Acad. Sci. USA 1993, 90, 5123–5127. [Google Scholar] [CrossRef]
- Wu, Q.; Beland, F.A.; Chang, C.W.; Fang, J.L. Role of DNA Repair Pathways in Response to Zidovudine-induced DNA Damage in Immortalized Human Liver THLE2 Cells. Int. J. Biomed. Sci. 2013, 9, 18–25. [Google Scholar]
- Ramboer, E.; De Craene, B.; De Kock, J.; Vanhaecke, T.; Berx, G.; Rogiers, V.; Vinken, M. Strategies for immortalization of primary hepatocytes. J. Hepatol. 2014, 61, 925–943. [Google Scholar] [CrossRef]
- Ramos, M.J.; Bandiera, L.; Menolascina, F.; Fallowfield, J.A. In vitro models for non-alcoholic fatty liver disease: Emerging platforms and their applications. iScience 2022, 25, 103549. [Google Scholar] [CrossRef]
- Wilkening, S.; Stahl, F.; Bader, A. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab. Dispos. 2003, 31, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Zeilinger, K.; Freyer, N.; Damm, G.; Seehofer, D.; Knöspel, F. Cell sources for in vitro human liver cell culture models. Exp. Biol. Med. 2016, 241, 1684–1698. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.C.; Hendriks, D.F.; Moro, S.M.; Ellis, E.; Walsh, J.; Renblom, A.; Fredriksson Puigvert, L.; Dankers, A.C.; Jacobs, F.; Snoeys, J.; et al. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci. Rep. 2016, 6, 25187. [Google Scholar] [CrossRef] [PubMed]
- Pirim, H.; Ekşioğlu, B.; Perkins, A.; Yüceer, C. Clustering of High Throughput Gene Expression Data. Comput. Oper. Res. 2012, 39, 3046–3061. [Google Scholar] [CrossRef] [PubMed]
- Ku, J.L.; Park, J.G. Biology of SNU cell lines. Cancer Res. Treat. 2005, 37, 1–19. [Google Scholar] [CrossRef]
- Park, J.G.; Lee, J.H.; Kang, M.S.; Park, K.J.; Jeon, Y.M.; Lee, H.J.; Kwon, H.S.; Park, H.S.; Yeo, K.S.; Lee, K.U.; et al. Characterization of cell lines established from human hepatocellular carcinoma. Int. J. Cancer 1995, 62, 276–282. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Peterson, M.F.; Otoc, N.; Sethi, J.K.; Gupta, A.; Antes, T.J. Integrated systems for exosome investigation. Methods 2015, 87, 31–45. [Google Scholar] [CrossRef]
- Malla, R.R.; Pandrangi, S.; Kumari, S.; Gavara, M.M.; Badana, A.K. Exosomal tetraspanins as regulators of cancer progression and metastasis and novel diagnostic markers. Asia Pac. J. Clin. Oncol. 2018, 14, 383–391. [Google Scholar] [CrossRef]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e1018. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Aheget, H.; Tristán-Manzano, M.; Mazini, L.; Cortijo-Gutierrez, M.; Galindo-Moreno, P.; Herrera, C.; Martin, F.; Marchal, J.A.; Benabdellah, K. Exosome: A New Player in Translational Nanomedicine. J. Clin. Med. 2020, 9, 2380. [Google Scholar] [CrossRef]
- Li, Z.; Qin, T.; Wang, K.; Hackenberg, M.; Yan, J.; Gao, Y.; Yu, L.R.; Shi, L.; Su, Z.; Chen, T. Integrated microRNA, mRNA, and protein expression profiling reveals microRNA regulatory networks in rat kidney treated with a carcinogenic dose of aristolochic acid. BMC Genom. 2015, 16, 365. [Google Scholar] [CrossRef]
- Grigoryev, Y.A.; Kurian, S.M.; Hart, T.; Nakorchevsky, A.A.; Chen, C.; Campbell, D.; Head, S.R.; Yates, J.R., 3rd.; Salomon, D.R. MicroRNA regulation of molecular networks mapped by global microRNA, mRNA, and protein expression in activated T lymphocytes. J. Immunol. 2011, 187, 2233–2243. [Google Scholar] [CrossRef] [PubMed]
- Aure, M.R.; Jernström, S.; Krohn, M.; Vollan, H.K.; Due, E.U.; Rødland, E.; Kåresen, R.; Ram, P.; Lu, Y.; Mills, G.B.; et al. Integrated analysis reveals microRNA networks coordinately expressed with key proteins in breast cancer. Genome Med. 2015, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Shalgi, R.; Lieber, D.; Oren, M.; Pilpel, Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput. Biol. 2007, 3, e131. [Google Scholar] [CrossRef]
- Fan, Y.; Siklenka, K.; Arora, S.K.; Ribeiro, P.; Kimmins, S.; Xia, J. miRNet-dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016, 44, W135–W141. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Cabral, B.C.A.; Hoffmann, L.; Bottaro, T.; Costa, P.F.; Ramos, A.L.A.; Coelho, H.S.M.; Villela-Nogueira, C.A.; Ürményi, T.P.; Faffe, D.S.; Silva, R. Circulating microRNAs associated with liver fibrosis in chronic hepatitis C patients. Biochem. Biophys. Rep. 2020, 24, 100814. [Google Scholar] [CrossRef]
- Shen, J.; Wang, A.; Wang, Q.; Gurvich, I.; Siegel, A.B.; Remotti, H.; Santella, R.M. Exploration of genome-wide circulating microRNA in hepatocellular carcinoma: MiR-483-5p as a potential biomarker. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2364–2373. [Google Scholar] [CrossRef]
- Ferragut, F.; Vachetta, V.S.; Troncoso, M.F.; Rabinovich, G.A.; Elola, M.T. ALCAM/CD166: A pleiotropic mediator of cell adhesion, stemness and cancer progression. Cytokine Growth Factor Rev. 2021, 61, 27–37. [Google Scholar] [CrossRef] [PubMed]
- van Kempen, L.C.; Nelissen, J.M.; Degen, W.G.; Torensma, R.; Weidle, U.H.; Bloemers, H.P.; Figdor, C.G.; Swart, G.W. Molecular basis for the homophilic activated leukocyte cell adhesion molecule (ALCAM)-ALCAM interaction. J. Biol. Chem. 2001, 276, 25783–25790. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Quertermous, T. Molecular isolation and characterization of a soluble isoform of activated leukocyte cell adhesion molecule that modulates endothelial cell function. J. Biol. Chem. 2004, 279, 55315–55323. [Google Scholar] [CrossRef]
- Ofori-Acquah, S.F.; King, J.A. Activated leukocyte cell adhesion molecule: A new paradox in cancer. Transl. Res. 2008, 151, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Jezierska, A.; Olszewski, W.P.; Pietruszkiewicz, J.; Olszewski, W.; Matysiak, W.; Motyl, T. Activated Leukocyte Cell Adhesion Molecule (ALCAM) is associated with suppression of breast cancer cells invasion. Med. Sci. Monit. 2006, 12, Br245–Br256. [Google Scholar]
- Burkhardt, M.; Mayordomo, E.; Winzer, K.J.; Fritzsche, F.; Gansukh, T.; Pahl, S.; Weichert, W.; Denkert, C.; Guski, H.; Dietel, M.; et al. Cytoplasmic overexpression of ALCAM is prognostic of disease progression in breast cancer. J. Clin. Pathol. 2006, 59, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.Y.; Chen, D.; Gu, X.Y.; Ding, J.; Zhao, Y.J.; Zhao, Q.; Yao, M.; Chen, Z.; He, X.H.; Cong, W.M. Predicting Value of ALCAM as a Target Gene of microRNA-483-5p in Patients with Early Recurrence in Hepatocellular Carcinoma. Front. Pharmacol. 2017, 8, 973. [Google Scholar] [CrossRef]
- Sohn, W.; Kim, J.; Kang, S.H.; Yang, S.R.; Cho, J.Y.; Cho, H.C.; Shim, S.G.; Paik, Y.H. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp. Mol. Med. 2015, 47, e184. [Google Scholar] [CrossRef]
- Li, L.M.; Liu, H.; Liu, X.H.; Hu, H.B.; Liu, S.M. Clinical significance of exosomal miRNAs and proteins in three human cancers with high mortality in China. Oncol. Lett. 2019, 17, 11–22. [Google Scholar] [CrossRef]
- Wang, G.; Dong, F.; Xu, Z.; Sharma, S.; Hu, X.; Chen, D.; Zhang, L.; Zhang, J.; Dong, Q. MicroRNA profile in HBV-induced infection and hepatocellular carcinoma. BMC Cancer 2017, 17, 805. [Google Scholar] [CrossRef]
- Morishita, A.; Fujita, K.; Iwama, H.; Chiyo, T.; Fujihara, S.; Oura, K.; Tadokoro, T.; Mimura, S.; Nomura, T.; Tani, J.; et al. Role of microRNA-210-3p in hepatitis B virus-related hepatocellular carcinoma. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G401–G409. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; An, P.; Winkler, C.A.; Yu, Y. Dysregulated microRNAs in Hepatitis B Virus-Related Hepatocellular Carcinoma: Potential as Biomarkers and Therapeutic Targets. Front. Oncol. 2020, 10, 1271. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Zhong, C.; Duan, S. miR-1269a and miR-1269b: Emerging Carcinogenic Genes of the miR-1269 Family. Front. Cell Dev. Biol. 2022, 10, 809132. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.X.; Lv, Y.R.; Shao, L.P.; Nong, X.Y.; Zhang, G.L.; Zhang, Y.; Fan, H.X.; Liu, M.; Li, X.; Tang, H. HBx-induced MiR-1269b in NF-κB dependent manner upregulates cell division cycle 40 homolog (CDC40) to promote proliferation and migration in hepatoma cells. J. Transl. Med. 2016, 14, 189. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, J.; Liu, Y.; Zeng, X.; Wei, M.; Wu, S.; Xiong, Q.; Song, F.; Yuan, X.; Xiao, Y.; et al. Hepatitis B Virus Induces Autophagy to Promote its Replication by the Axis of miR-192-3p-XIAP Through NF kappa B Signaling. Hepatology 2019, 69, 974–992. [Google Scholar] [CrossRef]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef]
- Edgar, J.R.; Eden, E.R.; Futter, C.E. Hrs- and CD63-dependent competing mechanisms make different sized endosomal intraluminal vesicles. Traffic 2014, 15, 197–211. [Google Scholar] [CrossRef]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef]
- van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef]
- Petersen, S.H.; Odintsova, E.; Haigh, T.A.; Rickinson, A.B.; Taylor, G.S.; Berditchevski, F. The role of tetraspanin CD63 in antigen presentation via MHC class II. Eur. J. Immunol. 2011, 41, 2556–2561. [Google Scholar] [CrossRef] [PubMed]
- Lammerding, J.; Kazarov, A.R.; Huang, H.; Lee, R.T.; Hemler, M.E. Tetraspanin CD151 regulates alpha6beta1 integrin adhesion strengthening. Proc. Natl. Acad. Sci. USA 2003, 100, 7616–7621. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Devi, G.t.; Badana, A.; Dasari, V.R.; Malla, R.R. CD151-A Striking Marker for Cancer Therapy. Biomark. Cancer 2015, 7, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Baleato, R.M.; Guthrie, P.L.; Gubler, M.C.; Ashman, L.K.; Roselli, S. Deletion of CD151 results in a strain-dependent glomerular disease due to severe alterations of the glomerular basement membrane. Am. J. Pathol. 2008, 173, 927–937. [Google Scholar] [CrossRef]
- Ke, A.W.; Shi, G.M.; Zhou, J.; Huang, X.Y.; Shi, Y.H.; Ding, Z.B.; Wang, X.Y.; Devbhandari, R.P.; Fan, J. CD151 amplifies signaling by integrin α6β1 to PI3K and induces the epithelial-mesenchymal transition in HCC cells. Gastroenterology 2011, 140, 1629–1641.e1615. [Google Scholar] [CrossRef]
- Shi, G.M.; Ke, A.W.; Zhou, J.; Wang, X.Y.; Xu, Y.; Ding, Z.B.; Devbhandari, R.P.; Huang, X.Y.; Qiu, S.J.; Shi, Y.H.; et al. CD151 modulates expression of matrix metalloproteinase 9 and promotes neoangiogenesis and progression of hepatocellular carcinoma. Hepatology 2010, 52, 183–196. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, Y.; Li, Y.; Shi, T.; Luan, Y.; Yin, C. Exosome and virus infection. Front. Immunol. 2023, 14, 1154217. [Google Scholar] [CrossRef]
- Shi, Y.; Du, L.; Lv, D.; Li, Y.; Zhang, Z.; Huang, X.; Tang, H. Emerging role and therapeutic application of exosome in hepatitis virus infection and associated diseases. J. Gastroenterol. 2021, 56, 336–349. [Google Scholar] [CrossRef]
- Jiang, Y.; Han, Q.; Zhao, H.; Zhang, J. The Mechanisms of HBV-Induced Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2021, 8, 435–450. [Google Scholar] [CrossRef]
- Wu, Q.; Zhou, L.; Lv, D.; Zhu, X.; Tang, H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J. Hematol. Oncol. 2019, 12, 53. [Google Scholar] [CrossRef]
- Kumar, B.; Garcia, M.; Weng, L.; Jung, X.; Murakami, J.L.; Hu, X.; McDonald, T.; Lin, A.; Kumar, A.R.; DiGiusto, D.L.; et al. Acute myeloid leukemia transforms the bone marrow niche into a leukemia-permissive microenvironment through exosome secretion. Leukemia 2018, 32, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Stefanius, K.; Servage, K.; de Souza Santos, M.; Gray, H.F.; Toombs, J.E.; Chimalapati, S.; Kim, M.S.; Malladi, V.S.; Brekken, R.; Orth, K. Human pancreatic cancer cell exosomes, but not human normal cell exosomes, act as an initiator in cell transformation. eLife 2019, 8, e40226. [Google Scholar] [CrossRef] [PubMed]
- Abdouh, M.; Hamam, D.; Gao, Z.H.; Arena, V.; Arena, M.; Arena, G.O. Exosomes isolated from cancer patients’ sera transfer malignant traits and confer the same phenotype of primary tumors to oncosuppressor-mutated cells. J. Exp. Clin. Cancer Res. 2017, 36, 113. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef]
- Ju, Y.; Bai, H.; Ren, L.; Zhang, L. The Role of Exosome and the ESCRT Pathway on Enveloped Virus Infection. Int. J. Mol. Sci. 2021, 22, 9060. [Google Scholar] [CrossRef]
- Ahmed, I.; Akram, Z.; Iqbal, H.M.N.; Munn, A.L. The regulation of Endosomal Sorting Complex Required for Transport and accessory proteins in multivesicular body sorting and enveloped viral budding—An overview. Int. J. Biol. Macromol. 2019, 127, 1–11. [Google Scholar] [CrossRef]
- Martin-Serrano, J.; Neil, S.J. Host factors involved in retroviral budding and release. Nat. Rev. Microbiol. 2011, 9, 519–531. [Google Scholar] [CrossRef]
- Pincetic, A.; Leis, J. The Mechanism of Budding of Retroviruses From Cell Membranes. Adv. Virol. 2009, 2009, 6239691–6239699. [Google Scholar] [CrossRef]
- Kian Chua, P.; Lin, M.H.; Shih, C. Potent inhibition of human Hepatitis B virus replication by a host factor Vps4. Virology 2006, 354, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lambert, C.; Döring, T.; Prange, R. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma 2-adaptin. J. Virol. 2007, 81, 9050–9060. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Sorensen, E.M.; Naito, A.; Schott, M.; Kim, S.; Ahlquist, P. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc. Natl. Acad. Sci. USA 2007, 104, 10205–10210. [Google Scholar] [CrossRef] [PubMed]
- Stieler, J.T.; Prange, R. Involvement of ESCRT-II in hepatitis B virus morphogenesis. PLoS ONE 2014, 9, e91279. [Google Scholar] [CrossRef]
- Giannakis, M.; Mu, X.J.; Shukla, S.A.; Qian, Z.R.; Cohen, O.; Nishihara, R.; Bahl, S.; Cao, Y.; Amin-Mansour, A.; Yamauchi, M.; et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016, 15, 857–865. [Google Scholar] [CrossRef]
- Chen, G.; Nakamura, I.; Dhanasekaran, R.; Iguchi, E.; Tolosa, E.J.; Romecin, P.A.; Vera, R.E.; Almada, L.L.; Miamen, A.G.; Chaiteerakij, R.; et al. Transcriptional Induction of Periostin by a Sulfatase 2-TGFβ1-SMAD Signaling Axis Mediates Tumor Angiogenesis in Hepatocellular Carcinoma. Cancer Res. 2017, 77, 632–645. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, C.; Li, X.; Luo, W.; Roy, B.; Xiong, T.; Zhang, X.; Yang, H.; Wang, J.; Ye, Z.; et al. The landscape of somatic mutation in sporadic Chinese colorectal cancer. Oncotarget 2018, 9, 27412–27422. [Google Scholar] [CrossRef]
- Baril, P.; Gangeswaran, R.; Mahon, P.C.; Caulee, K.; Kocher, H.M.; Harada, T.; Zhu, M.; Kalthoff, H.; Crnogorac-Jurcevic, T.; Lemoine, N.R. Periostin promotes invasiveness and resistance of pancreatic cancer cells to hypoxia-induced cell death: Role of the beta4 integrin and the PI3k pathway. Oncogene 2007, 26, 2082–2094. [Google Scholar] [CrossRef]
- García-Palmero, I.; Torres, S.; Bartolomé, R.A.; Peláez-García, A.; Larriba, M.J.; Lopez-Lucendo, M.; Peña, C.; Escudero-Paniagua, B.; Muñoz, A.; Casal, J.I. Twist1-induced activation of human fibroblasts promotes matrix stiffness by upregulating palladin and collagen α1(VI). Oncogene 2016, 35, 5224–5236. [Google Scholar] [CrossRef]
- van Huizen, N.A.; Coebergh van den Braak, R.R.J.; Doukas, M.; Dekker, L.J.M.; JNM, I.J.; Luider, T.M. Up-regulation of collagen proteins in colorectal liver metastasis compared with normal liver tissue. J. Biol. Chem. 2019, 294, 281–289. [Google Scholar] [CrossRef]
- Afik, R.; Zigmond, E.; Vugman, M.; Klepfish, M.; Shimshoni, E.; Pasmanik-Chor, M.; Shenoy, A.; Bassat, E.; Halpern, Z.; Geiger, T.; et al. Tumor macrophages are pivotal constructors of tumor collagenous matrix. J. Exp. Med. 2016, 213, 2315–2331. [Google Scholar] [CrossRef] [PubMed]
- Mouradov, D.; Sloggett, C.; Jorissen, R.N.; Love, C.G.; Li, S.; Burgess, A.W.; Arango, D.; Strausberg, R.L.; Buchanan, D.; Wormald, S.; et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014, 74, 3238–3247. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.; Mun, J.; Kim, J.H.; Kim, C.W.; Roh, S.A.; Cho, D.H.; Kim, Y.S.; Kim, S.Y.; Kim, J.C. Genome-wide mutation profiles of colorectal tumors and associated liver metastases at the exome and transcriptome levels. Oncotarget 2015, 6, 22179–22190. [Google Scholar] [CrossRef]
- Naganuma, S.; Whelan, K.A.; Natsuizaka, M.; Kagawa, S.; Kinugasa, H.; Chang, S.; Subramanian, H.; Rhoades, B.; Ohashi, S.; Itoh, H.; et al. Notch receptor inhibition reveals the importance of cyclin D1 and Wnt signaling in invasive esophageal squamous cell carcinoma. Am. J. Cancer Res. 2012, 2, 459–475. [Google Scholar] [PubMed]
- Gillan, L.; Matei, D.; Fishman, D.A.; Gerbin, C.S.; Karlan, B.Y.; Chang, D.D. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002, 62, 5358–5364. [Google Scholar]
- Planche, A.; Bacac, M.; Provero, P.; Fusco, C.; Delorenzi, M.; Stehle, J.C.; Stamenkovic, I. Identification of prognostic molecular features in the reactive stroma of human breast and prostate cancer. PLoS ONE 2011, 6, e18640. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Zhao, L.; Budhu, A.; Forgues, M.; Jia, H.L.; Qin, L.X.; Ye, Q.H.; Yu, J.; Shi, X.; Tang, Z.Y.; et al. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J. Hepatol. 2010, 52, 690–697. [Google Scholar] [CrossRef]
- Cheng, N.; Brantley, D.M.; Chen, J. The ephrins and Eph receptors in angiogenesis. Cytokine Growth Factor Rev. 2002, 13, 75–85. [Google Scholar] [CrossRef]
- Liu, W.; Yu, C.; Li, J.; Fang, J. The Roles of EphB2 in Cancer. Front. Cell Dev. Biol. 2022, 10, 788587. [Google Scholar] [CrossRef]
- Yu, G.; Gao, Y.; Ni, C.; Chen, Y.; Pan, J.; Wang, X.; Ding, Z.; Wang, J. Reduced expression of EphB2 is significantly associated with nodal metastasis in Chinese patients with gastric cancer. J. Cancer Res. Clin. Oncol. 2011, 137, 73–80. [Google Scholar] [CrossRef]
- Ramaswamy, S.; Ross, K.N.; Lander, E.S.; Golub, T.R. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2003, 33, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Tong, C.; Kazobinka, G.; Zhang, W.; Huang, X.; Huang, Y.; Zhang, Y. Expression of COL6A1 predicts prognosis in cervical cancer patients. Am. J. Transl. Res. 2016, 8, 2838–2844. [Google Scholar] [PubMed]
- Trastulla, L.; Noorbakhsh, J.; Vazquez, F.; McFarland, J.; Iorio, F. Computational estimation of quality and clinical relevance of cancer cell lines. Mol. Syst. Biol. 2022, 18, e11017. [Google Scholar] [CrossRef]
- Roerink, S.F.; Sasaki, N.; Lee-Six, H.; Young, M.D.; Alexandrov, L.B.; Behjati, S.; Mitchell, T.J.; Grossmann, S.; Lightfoot, H.; Egan, D.A.; et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 2018, 556, 457–462. [Google Scholar] [CrossRef]
- Raghavan, S.; Winter, P.S.; Navia, A.W.; Williams, H.L.; DenAdel, A.; Lowder, K.E.; Galvez-Reyes, J.; Kalekar, R.L.; Mulugeta, N.; Kapner, K.S.; et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell 2021, 184, 6119–6137.e6126. [Google Scholar] [CrossRef] [PubMed]
- Salvadores, M.; Fuster-Tormo, F.; Supek, F. Matching cell lines with cancer type and subtype of origin via mutational, epigenomic, and transcriptomic patterns. Sci. Adv. 2020, 6, eaba1862. [Google Scholar] [CrossRef]
- Llerena, S.; García-Díaz, N.; Curiel-Olmo, S.; Agraz-Doblas, A.; García-Blanco, A.; Pisonero, H.; Varela, M.; Santibáñez, M.; Almaraz, C.; Cereceda, L.; et al. Applied diagnostics in liver cancer. Efficient combinations of sorafenib with targeted inhibitors blocking AKT/mTOR. Oncotarget 2018, 9, 30869–30882. [Google Scholar] [CrossRef][Green Version]
- Fuchs, B.C.; Fujii, T.; Dorfman, J.D.; Goodwin, J.M.; Zhu, A.X.; Lanuti, M.; Tanabe, K.K. Epithelial-to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res. 2008, 68, 2391–2399. [Google Scholar] [CrossRef]
- Guichard, C.; Amaddeo, G.; Imbeaud, S.; Ladeiro, Y.; Pelletier, L.; Maad, I.B.; Calderaro, J.; Bioulac-Sage, P.; Letexier, M.; Degos, F.; et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 2012, 44, 694–698. [Google Scholar] [CrossRef]
- Gennarino, V.A.; Sardiello, M.; Avellino, R.; Meola, N.; Maselli, V.; Anand, S.; Cutillo, L.; Ballabio, A.; Banfi, S. MicroRNA target prediction by expression analysis of host genes. Genome Res. 2009, 19, 481–490. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef] [PubMed]
- Thurman, T.J.; Washam, C.L.; Alkam, D.; Bird, J.T.; Gies, A.; Dhusia, K.; Roberson, M.S.; Byrum, S.D. proteoDA: A package for quantitative proteomics. J. Open Source Softw. 2023, 8, 5184. [Google Scholar] [CrossRef]
- Graw, S.; Tang, J.; Zafar, M.K.; Byrd, A.K.; Bolden, C.; Peterson, E.C.; Byrum, S.D. proteiNorm—A User-Friendly Tool for Normalization and Analysis of TMT and Label-Free Protein Quantification. ACS Omega 2020, 5, 25625–25633. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]


| Pathways | Molecules |
|---|---|
| ECM-receptor interaction and focal adhesion | COL1A2, COL6A1, FN1, ITGA5, ITGB3, ITGB1 |
| Cell adhesion and Axon guidance | ALCAM, EPHB2, ITGA5, ITGB3, NRAS |
| Vesicle-mediated transport | EXOC8, COL1A2, SFN, STAM |
| Cell surface interactions at the vascular wall | ATP1B3, COL1A2, FN1, ITGA5, ITGB3, NRAS |
| P53 signaling pathway | CD82, SFN |
| SNARE interactions in vesicular transport | VAMP3 |
| VEGF signaling | NRAS |
| MAPK signaling | NRAS, SRF |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Todorova, V.K.; Byrum, S.D.; Mackintosh, S.G.; Jamshidi-Parsian, A.; Gies, A.J.; Washam, C.L.; Jenkins, S.V.; Spiva, T.; Bowman, E.; Reyna, N.S.; et al. Exosomal MicroRNA and Protein Profiles of Hepatitis B Virus-Related Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2023, 24, 13098. https://doi.org/10.3390/ijms241713098
Todorova VK, Byrum SD, Mackintosh SG, Jamshidi-Parsian A, Gies AJ, Washam CL, Jenkins SV, Spiva T, Bowman E, Reyna NS, et al. Exosomal MicroRNA and Protein Profiles of Hepatitis B Virus-Related Hepatocellular Carcinoma Cells. International Journal of Molecular Sciences. 2023; 24(17):13098. https://doi.org/10.3390/ijms241713098
Chicago/Turabian StyleTodorova, Valentina K., Stephanie D. Byrum, Samuel G. Mackintosh, Azemat Jamshidi-Parsian, Allen J. Gies, Charity L. Washam, Samir V. Jenkins, Timothy Spiva, Emily Bowman, Nathan S. Reyna, and et al. 2023. "Exosomal MicroRNA and Protein Profiles of Hepatitis B Virus-Related Hepatocellular Carcinoma Cells" International Journal of Molecular Sciences 24, no. 17: 13098. https://doi.org/10.3390/ijms241713098
APA StyleTodorova, V. K., Byrum, S. D., Mackintosh, S. G., Jamshidi-Parsian, A., Gies, A. J., Washam, C. L., Jenkins, S. V., Spiva, T., Bowman, E., Reyna, N. S., Griffin, R. J., & Makhoul, I. (2023). Exosomal MicroRNA and Protein Profiles of Hepatitis B Virus-Related Hepatocellular Carcinoma Cells. International Journal of Molecular Sciences, 24(17), 13098. https://doi.org/10.3390/ijms241713098







