Anaphase-Promoting Complex Subunit 1 Associates with Bone Mineral Density in Human Osteoporotic Bone
Abstract
1. Introduction
2. Results
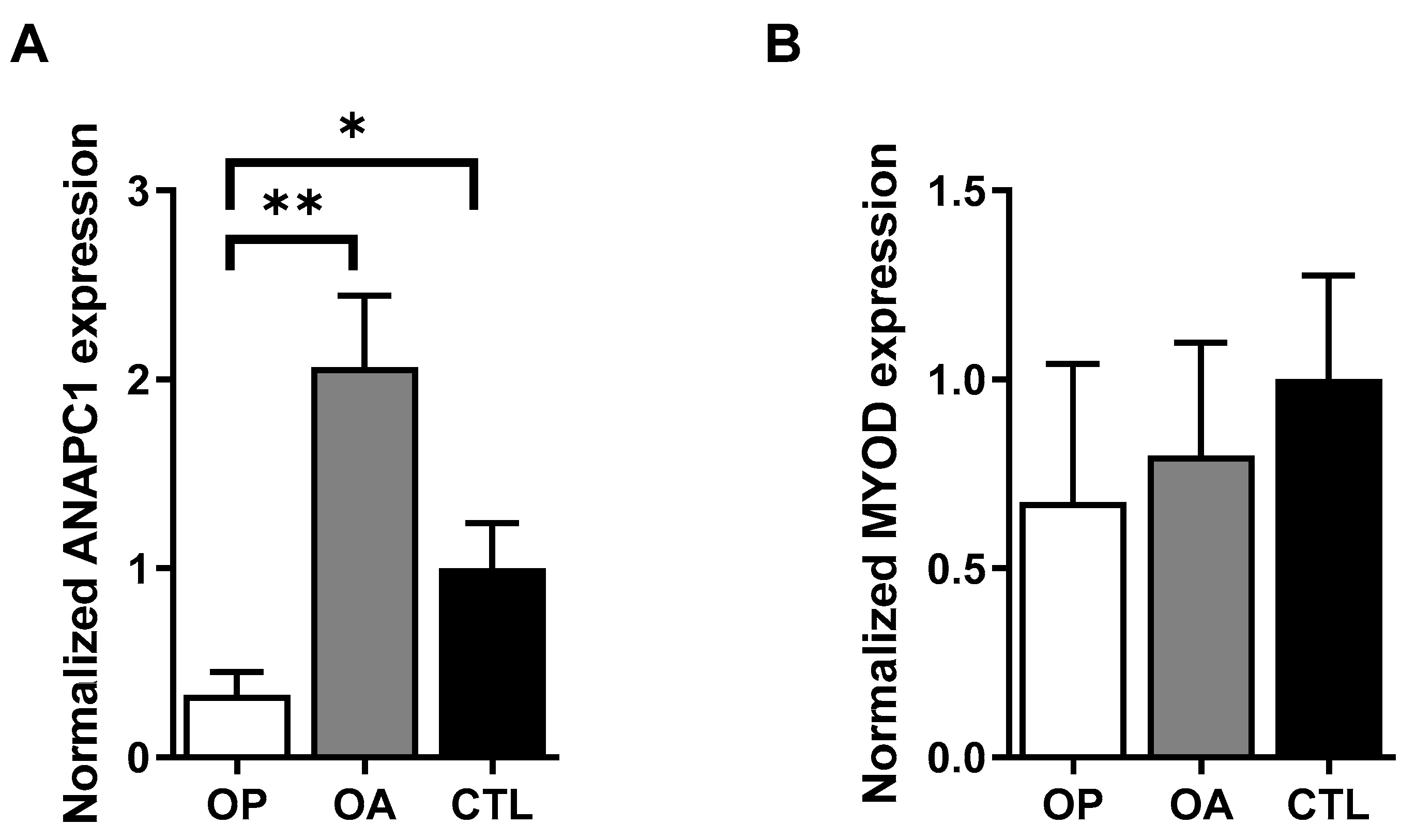
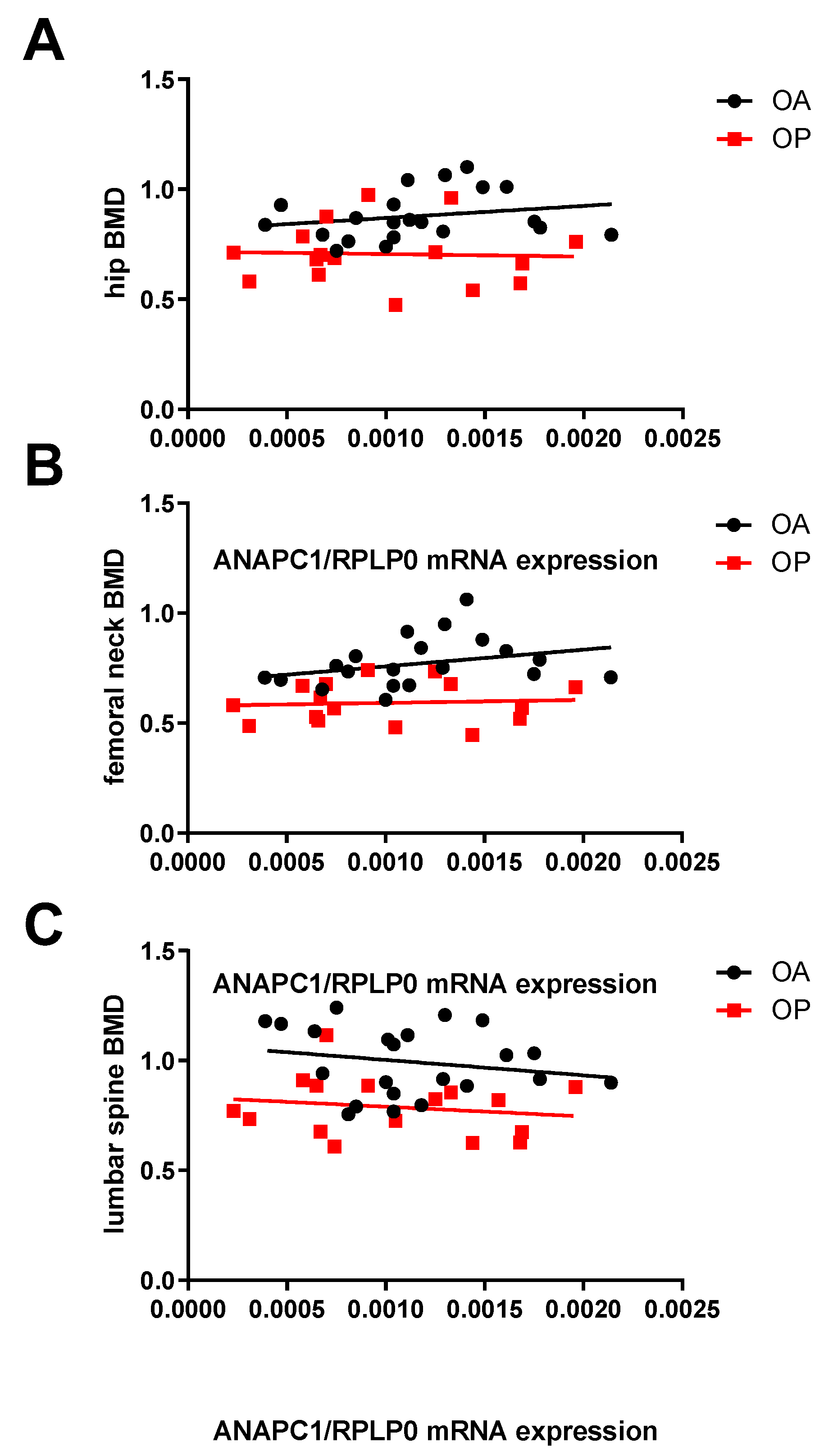
2.1. Expression of ANAPC1 Is Decreased in Osteoporotic Bone Samples
2.2. Expression of ANAPC1 Is Decreased in Osteoporotic Muscle Samples
2.3. The Expression of ANAPC1 Is a Part of the Osteogenic Differentiation Process
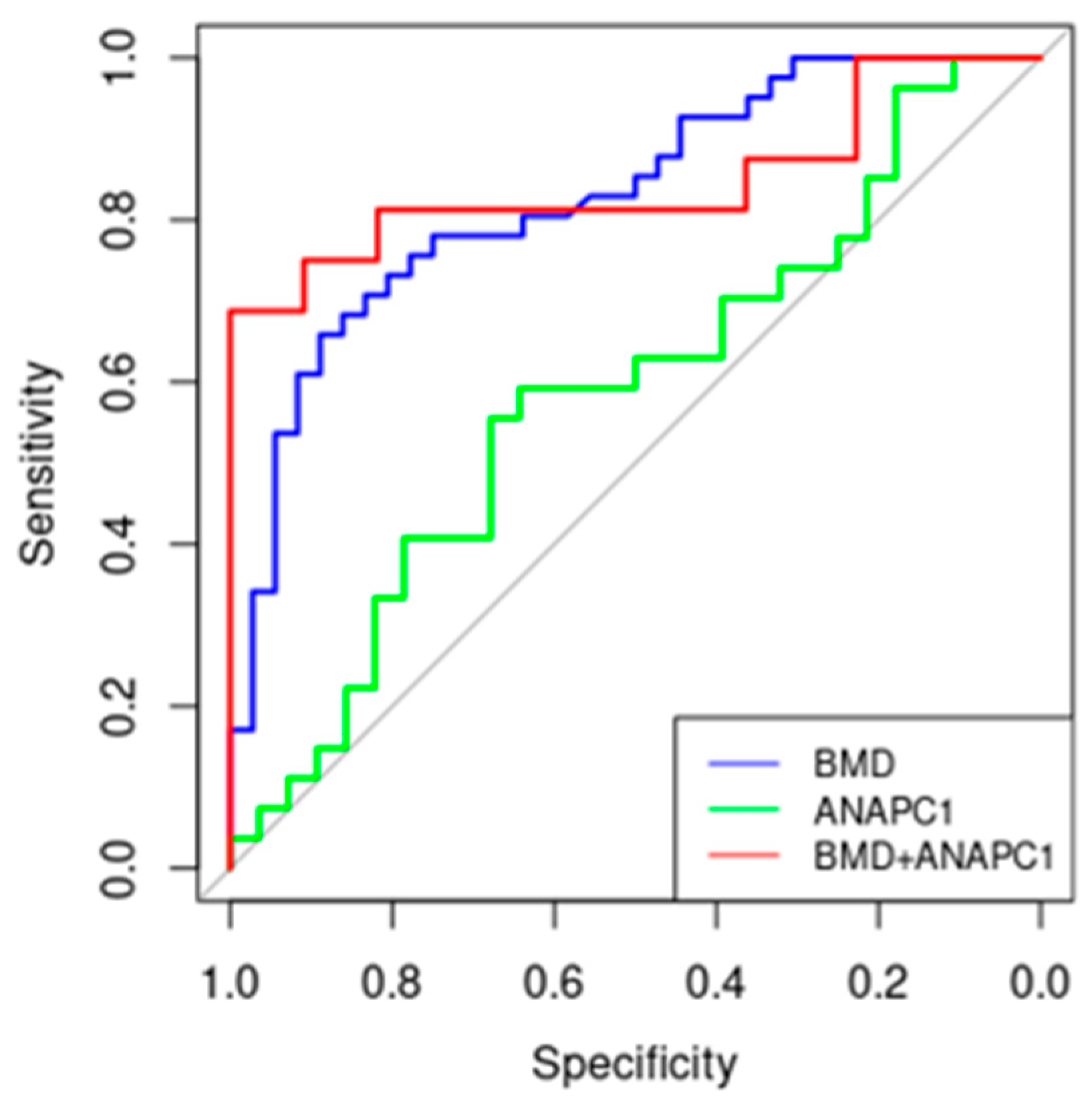
2.4. ANAPC1 as a Potential New Biomarker of Osteoporosis
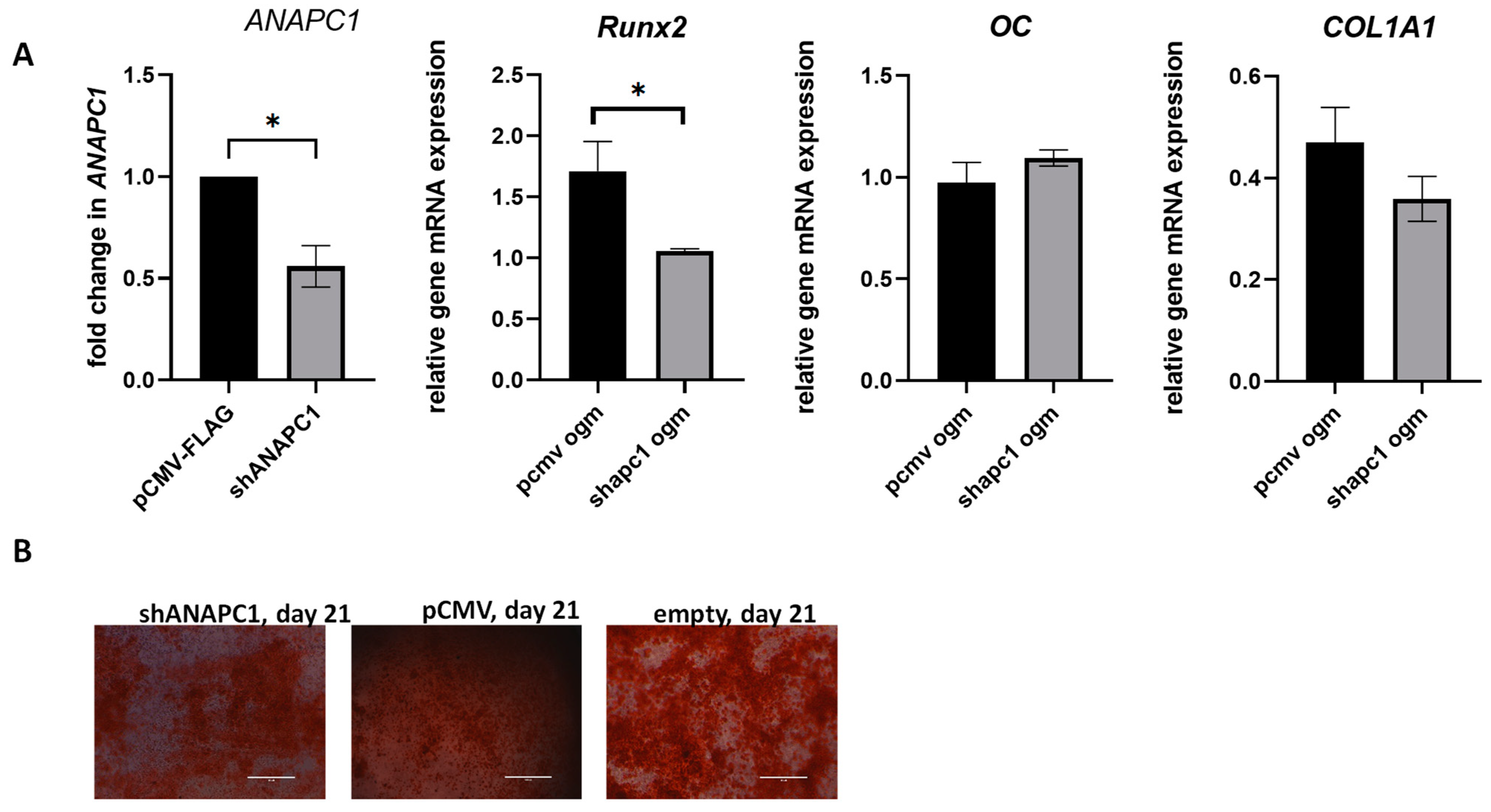
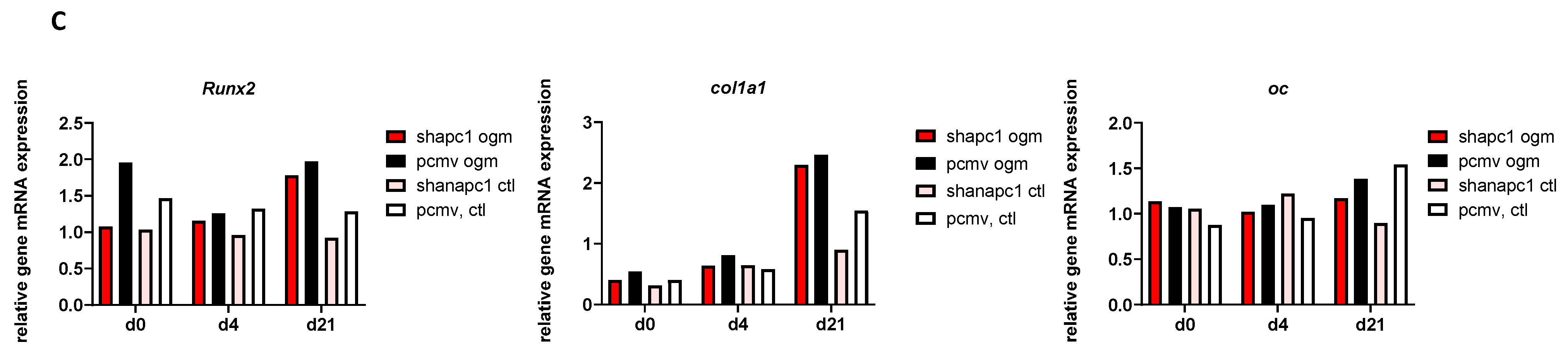
2.5. Silencing of ANAPC1 in Human Osteosarcoma (HOS) Cells Caused a Decrease in the Expression of RUNX2
3. Discussion
4. Materials and Methods
4.1. Bone and Muscle Tissue Samples Collection
4.2. Isolation of RNA from Bones
4.3. Isolation of RNA from Muscles
4.4. Cell Culturing and Transfections
4.5. Quantitative PCR
4.6. Osteogenic and Adipogenic Differentiation of Cells
4.7. Isolation of RNA from Cells Culture
4.8. Reverse Transcription
4.9. Alizarin Red S Staining
4.10. Oil Red O Staining
4.11. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Koromani, F.; Trajanoska, K.; Rivadeneira, F.; Oei, L. Recent Advances in the Genetics of Fractures in Osteoporosis. Front. Endocrinol. 2019, 10, 337. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Bai, W.; Zheng, H. Twelve years of GWAS discoveries for osteoporosis and related traits: Advances, challenges and applications. Bone Res. 2021, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Lovšin, N. Copy Number Variation and Osteoporosis. Curr. Osteoporos. Rep. 2023, 21, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Lovšin, N.; Zupan, J.; Marc, J. Genetic effects on bone health. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 233–239. [Google Scholar] [CrossRef]
- Yang, T.-L.; Shen, H.; Liu, A.; Dong, S.-S.; Zhang, L.; Deng, F.-Y.; Zhao, Q.; Deng, H.-W. A road map for understanding molecular and genetic determinants of osteoporosis. Nat. Rev. Endocrinol. 2020, 16, 91–103. [Google Scholar] [CrossRef]
- Estrada, K.; Styrkarsdottir, U.; Evangelou, E.; Hsu, Y.-H.; Duncan, E.L.; Ntzani, E.E.; Oei, L.; Albagha, O.M.E.; Amin, N.; Kemp, J.P.; et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 2012, 44, 491–501. [Google Scholar] [CrossRef]
- Rivadeneira, F.; Styrkársdottir, U.; Estrada, K.; Halldórsson, B.V.; Hsu, Y.-H.; Richards, J.B.; Zillikens, M.C.; Kavvoura, F.K.; Amin, N.; Aulchenko, Y.S.; et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat. Genet. 2009, 41, 1199–1206. [Google Scholar] [CrossRef]
- Sabik, O.L.; Farber, C.R. Using GWAS to identify novel therapeutic targets for osteoporosis. Transl. Res. 2017, 181, 15–26. [Google Scholar] [CrossRef]
- Rauner, M.; Foessl, I.; Formosa, M.M.; Kague, E.; Prijatelj, V.; Lopez, N.A.; Banerjee, B.; Bergen, D.; Busse, B.; Calado, Â.; et al. Perspective of the GEMSTONE Consortium on Current and Future Approaches to Functional Validation for Skeletal Genetic Disease Using Cellular, Molecular and Animal-Modeling Techniques. Front. Endocrinol. 2021, 12, 731217. [Google Scholar] [CrossRef]
- Medina-Gomez, C.; Kemp, J.P.; Trajanoska, K.; Luan, J.A.; Chesi, A.; Ahluwalia, T.S.; Mook-Kanamori, D.O.; Ham, A.; Hartwig, F.P.; Evans, D.S.; et al. Life-Course Genome-wide Association Study Meta-analysis of Total Body BMD and Assessment of Age-Specific Effects. Am. J. Hum. Genet. 2018, 102, 88–102. [Google Scholar] [CrossRef]
- Kim, B.J.; Ahn, S.H.; Kim, H.M.; Ikegawa, S.; Yang, T.L.; Guo, Y.; Deng, H.W.; Koh, J.M.; Lee, S.H. Replication of Caucasian Loci Associated with Osteoporosis-related Traits in East Asians. J. Bone Metab. 2016, 23, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Shen, H.; Fu, X.; Xu, C.; Tian, Q.; Deng, H. Integrative genomic analysis predicts novel functional enhancer-SNPs for bone mineral density. Hum. Genet. 2019, 138, 167–185. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Braz, M.G.; Ferraz-de-Souza, B. Genetics of osteoporosis: Searching for candidate genes for bone fragility. Arch. Endocrinol. Metab. 2016, 60, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Thornton, B.R.; Ng, T.M.; Matyskiela, M.E.; Carroll, C.W.; Morgan, D.O.; Toczyski, D.P. An architectural map of the anaphase-promoting complex. Genes Dev. 2006, 20, 449–460. [Google Scholar] [CrossRef]
- Jorgensen, P.M.; Brundell, E.; Starborg, M.; Hoog, C. A subunit of the anaphase-promoting complex is a centromere-associated protein in mammalian cells. Mol. Cell. Biol. 1998, 18, 468–476. [Google Scholar] [CrossRef]
- Kurasawa, Y.; Todokoro, K. Identification of human APC10/Doc1 as a subunit of anaphase promoting complex. Oncogene 1999, 18, 5131–5137. [Google Scholar] [CrossRef]
- Melloy, P.G. The anaphase-promoting complex: A key mitotic regulator associated with somatic mutations occurring in cancer. Genes Chromosomes Cancer 2020, 59, 189–202. [Google Scholar] [CrossRef]
- Sharp, S.P.; Malizia, R.A.; Walrath, T.; D’Souza, S.S.; Booth, C.J.; Kartchner, B.J.; Lee, E.C.; Stain, S.C.; O’Connor, W., Jr. DNA damage response genes mark the early transition from colitis to neoplasia in colitis-associated colon cancer. Gene 2018, 677, 299–307. [Google Scholar] [CrossRef]
- Alfieri, C.; Zhang, S.; Barford, D. Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (APC/C). Open Biol. 2017, 7, 170204. [Google Scholar] [CrossRef]
- Yamano, H. APC/C: Current understanding and future perspectives. F1000 Res. 2019, 8, 725. [Google Scholar] [CrossRef]
- Tanaka-Matakatsu, M.; Thomas, B.J.; Du, W. Mutation of the Apc1 homologue shattered disrupts normal eye development by disrupting G1 cell cycle arrest and progression through mitosis. Dev. Biol. 2007, 309, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Teodoro, J.G.; Heilman, D.W.; Parker, A.E.; Green, M.R. The viral protein Apoptin associates with the anaphase-promoting complex to induce G2/M arrest and apoptosis in the absence of p53. Genes Dev. 2004, 18, 1952–1957. [Google Scholar] [CrossRef] [PubMed]
- Eom, T.-Y.; Stanco, A.; Guo, J.; Wilkins, G.; Deslauriers, D.; Yan, J.; Monckton, C.; Blair, J.; Oon, E.; Perez, A.; et al. Differential Regulation of Microtubule Severing by APC Underlies Distinct Patterns of Projection Neuron and Interneuron Migration. Dev. Cell 2014, 31, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Ajeawung, N.F.; Nguyen, T.T.M.; Lu, L.; Kucharski, T.J.; Rousseau, J.; Molidperee, S.; Atienza, J.; Gamache, I.; Jin, W.; Plon, S.E.; et al. Mutations in ANAPC1, Encoding a Scaffold Subunit of the Anaphase-Promoting Complex, Cause Rothmund-Thomson Syndrome Type 1. Am. J. Hum. Genet. 2019, 105, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Dragojevic, J.; Zupan, J.; Haring, G.; Herman, S.; Komadina, R.; Marc, J. Triglyceride metabolism in bone tissue is associated with osteoblast and osteoclast differentiation: A gene expression study. J. Bone Miner. Metab. 2013, 31, 512–519. [Google Scholar] [CrossRef]
- Trajanoska, K.; Rivadeneira, F.; Kiel, D.P.; Karasik, D. Genetics of Bone and Muscle Interactions in Humans. Curr. Osteoporos. Rep. 2019, 17, 86–95. [Google Scholar] [CrossRef]
- Meunier, P.; Aaron, J.; Edouard, C.; Vignon, G. Osteoporosis and the replacement of cell populations of the marrow by adipose tissue. A quantitative study of 84 iliac bone biopsies. Clin. Orthop. Relat. Res. 1971, 80, 147–154. [Google Scholar] [CrossRef]
- Kuo, T.-R.; Chen, C.-H. Bone biomarker for the clinical assessment of osteoporosis: Recent developments and future perspectives. Biomark. Res. 2017, 5, 18. [Google Scholar] [CrossRef]
- Greenspan, S.L.; von Stetten, E.; Emond, S.K.; Jones, L.; Parker, R.A. Instant Vertebral Assessment: A Noninvasive Dual X-ray Absorptiometry Technique to Avoid Misclassification and Clinical Mismanagement of Osteoporosis. J. Clin. Densitom. 2001, 4, 373–380. [Google Scholar] [CrossRef]
- Morris, J.A.; Kemp, J.P.; Youlten, S.E.; Laurent, L.; Logan, J.G.; Chai, R.C.; Vulpescu, N.A.; Forgetta, V.; Kleinman, A.; Mohanty, S.T.; et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 2019, 51, 258–266. [Google Scholar] [CrossRef]
- Alonso, N.; Estrada, K.; Albagha, O.M.E.; Herrera, L.; Reppe, S.; Olstad, O.K.; Gautvik, K.M.; Ryan, N.M.; Evans, K.L.; Nielson, C.M.; et al. Identification of a novel locus on chromosome 2q13, which predisposes to clinical vertebral fractures independently of bone density. Ann. Rheum. Dis. 2018, 77, 378–385. [Google Scholar] [CrossRef]
- Wang, Y.; Wactawski-Wende, J.; Sucheston-Campbell, L.E.; Preus, L.; Hovey, K.M.; Nie, J.; Jackson, R.D.; Handelman, S.K.; Nassir, R.; Crandall, C.J.; et al. The influence of genetic susceptibility and calcium plus vitamin D supplementation on fracture risk. Am. J. Clin. Nutr. 2017, 105, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, G.M.; Mesner, L.D.; Stains, J.P.; Tommasini, S.M.; Horowitz, M.C.; Rosen, C.J.; Farber, C.R. Integrating GWAS and Co-expression Network Data Identifies Bone Mineral Density Genes SPTBN1 and MARK3 and an Osteoblast Functional Module. Cell Syst. 2017, 4, 46–59.e44. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.M.; Lee, E.H. Transcriptional regulatory cascades in Runx2-dependent bone development. Tissue Eng. Part B Rev. 2013, 19, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Schwickart, M.; Havlis, J.; Habermann, B.; Bogdanova, A.; Camasses, A.; Oelschlaegel, T.; Shevchenko, A.; Zachariae, W. Swm1/Apc13 is an evolutionarily conserved subunit of the anaphase-promoting complex stabilizing the association of Cdc16 and Cdc27. Mol. Cell. Biol. 2004, 24, 3562–3576. [Google Scholar] [CrossRef]
- Ivarsdottir, E.V.; Benonisdottir, S.; Thorleifsson, G.; Sulem, P.; Oddsson, A.; Styrkarsdottir, U.; Kristmundsdottir, S.; Arnadottir, G.A.; Thorgeirsson, G.; Jonsdottir, I.; et al. Sequence variation at ANAPC1 accounts for 24% of the variability in corneal endothelial cell density. Nat. Commun. 2019, 10, 1284. [Google Scholar] [CrossRef]
- Li, M.; Shin, Y.-H.; Hou, L.; Huang, X.; Wei, Z.; Klann, E.; Zhang, P. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat. Cell Biol. 2008, 10, 1083–1089. [Google Scholar] [CrossRef]
- Cappell, S.D.; Chung, M.; Jaimovich, A.; Spencer, S.L.; Meyer, T. Irreversible APCCdh1 Inactivation Underlies the Point of No Return for Cell-Cycle Entry. Cell 2016, 166, 167–180. [Google Scholar] [CrossRef]
- Eguren, M.; Manchado, E.; Malumbres, M. Non-mitotic functions of the Anaphase-Promoting Complex. Semin. Cell Dev. Biol. 2011, 22, 572–578. [Google Scholar] [CrossRef]
- Vodermaier, H.C.; Gieffers, C.; Maurer-Stroh, S.; Eisenhaber, F.; Peters, J.-M. TPR Subunits of the Anaphase-Promoting Complex Mediate Binding to the Activator Protein CDH1. Curr. Biol. 2003, 13, 1459–1468. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, M.; Liu, X.; Li, Z.; Hu, M.; Tian, Y.; Lv, L.; Zhang, X.; Liu, Y.; Zhang, P.; et al. CDC20 promotes bone formation via APC/C dependent ubiquitination and degradation of p65. EMBO Rep. 2021, 22, e52576. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Zou, W.; Gao, D.; Inuzuka, H.; Fukushima, H.; Berg, A.H.; Drapp, R.; Shaik, S.; Hu, D.; Lester, C.; et al. Cdh1 regulates osteoblast function through an APC/C-independent modulation of Smurf1. Mol. Cell 2011, 44, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Čamernik, K.; Mihelič, A.; Mihalič, R.; Haring, G.; Herman, S.; Marolt Presen, D.; Janež, A.; Trebše, R.; Marc, J.; Zupan, J. Comprehensive analysis of skeletal muscle- and bone-derived mesenchymal stem/stromal cells in patients with osteoarthritis and femoral neck fracture. Stem Cell Res. Ther. 2020, 11, 146. [Google Scholar] [CrossRef] [PubMed]
- Čamernik, K.; Mihelič, A.; Mihalič, R.; Marolt Presen, D.; Janež, A.; Trebše, R.; Marc, J.; Zupan, J. Increased Exhaustion of the Subchondral Bone-Derived Mesenchymal Stem/ Stromal Cells in Primary Versus Dysplastic Osteoarthritis. Stem Cell Rev. Rep. 2020, 16, 742–754. [Google Scholar] [CrossRef]
- Kodrič, K.; Zupan, J.; Kranjc, T.; Komadina, R.; Mlakar, V.; Marc, J.; Lovšin, N. Sex-determining region Y (SRY) attributes to gender differences in RANKL expression and incidence of osteoporosis. Exp. Mol. Med. 2019, 51, 1–16. [Google Scholar] [CrossRef]
- Čamernik, K.; Zupan, J. Complete Assessment of Multilineage Differentiation Potential of Human Skeletal Muscle-Derived Mesenchymal Stem/Stromal Cells. Methods Mol. Biol. 2019, 2045, 131–144. [Google Scholar] [CrossRef]



| OA (n = 37) | OP (n = 47) | K (n = 10) | p (OA-OP) | |
|---|---|---|---|---|
| Age (years) | 71.1 (49 to 87) | 76.3 (53 to 88) | 68.1 (56 to 87) | 0.2513 |
| Women | 24 | 38 | 0 | |
| Men | 13 | 9 | 10 | |
| BMI (kg/m2) | 29.3 (22.8 to 43.7) | 25.1 (19.3 to 33.9) | 25.3 (19.0 to 31.2) | <0.001 |
| Hip BMD (g/cm2) | 0.909 (0.541 to 1.314) | 0.696 (0.402 to 0.974) | na | <0.001 |
| Hip t-score | −0.629 (−3.600 to 1.900) | −2.261 (−4.400 to 0.000) | na | <0.001 |
| Femoral neck BMD (g/cm2) | 0.791 (0.474 to 1.179) | 0.596 (0.386 to 0.798) | na | <0.001 |
| Femoral neck t-score | −1.192 (−4.160 to 1.800) | −2.681 (−4.200 to −0.500) | na | <0.001 |
| Lumbar spine BMD (g/cm2) | 1.015 (0.583 to 1.401) | 0.837 (0.601 to 1.461) | na | <0.001 |
| Lumbar spine t-score | −0.465 (−4.220 to 2.800) | −2.006 (−4.100 to 3.800) | na | <0.001 |
| Symbol | Gene Name | Primer Sequence |
|---|---|---|
| ALPL | Alkaline phosphatase | F: 5′-CCAAGTACTGGCGAGACCAA-3′ R: 5′-GTGGAGACACCCATCCCATC-3′ |
| COL1A1 | Collagen type I alpha 1 chain | F: 5′-GCCAAGACGAAGACATCCCA-3′ R: 5′-GTTTCCACACGTCTCGGTCA-3′ |
| RUNX2 | RUNX family transcription factor 2 | F: 5′-AGCAAGGTTCAACGATCTGAGAT-3′ R: 5′-TTTGTGAAGACGGTTATGGTCAA-3′ |
| OC | Osteocalcin (bone gamma-carboxyglutamate protein) | F: 5′-AAGAGACCCAGGCGCTACCT-3′ R: 5′-AACTCGTCACAGTCCGGATTG-3′ |
| MYOD | Myogenic differentiation 1 | F: 5′-TGCCACAACGGACGACTTC-3′ R: 5′-CGGGTCCAGGTCTTCGAA-3′ |
| RPLP0 | Ribosomal protein lateral stalk subunit P0 | F: 5′-TCTACAACCCTGAAGTGCTTGAT-3′ R: 5′-CAATCTGCAGACAGACACTGG-3′ |
| ANAPC1 | Anaphase-promoting complex subunit 1 | F: 5′-AATGTCCACAGTGCTCCAGG-3′ R: 5′-TTTTTGGGCGCAATGACAGG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malavašič, P.; Polajžer, S.; Lovšin, N. Anaphase-Promoting Complex Subunit 1 Associates with Bone Mineral Density in Human Osteoporotic Bone. Int. J. Mol. Sci. 2023, 24, 12895. https://doi.org/10.3390/ijms241612895
Malavašič P, Polajžer S, Lovšin N. Anaphase-Promoting Complex Subunit 1 Associates with Bone Mineral Density in Human Osteoporotic Bone. International Journal of Molecular Sciences. 2023; 24(16):12895. https://doi.org/10.3390/ijms241612895
Chicago/Turabian StyleMalavašič, Petra, Sara Polajžer, and Nika Lovšin. 2023. "Anaphase-Promoting Complex Subunit 1 Associates with Bone Mineral Density in Human Osteoporotic Bone" International Journal of Molecular Sciences 24, no. 16: 12895. https://doi.org/10.3390/ijms241612895
APA StyleMalavašič, P., Polajžer, S., & Lovšin, N. (2023). Anaphase-Promoting Complex Subunit 1 Associates with Bone Mineral Density in Human Osteoporotic Bone. International Journal of Molecular Sciences, 24(16), 12895. https://doi.org/10.3390/ijms241612895




