Biofilm and Cancer: Interactions and Future Directions for Cancer Therapy
Abstract
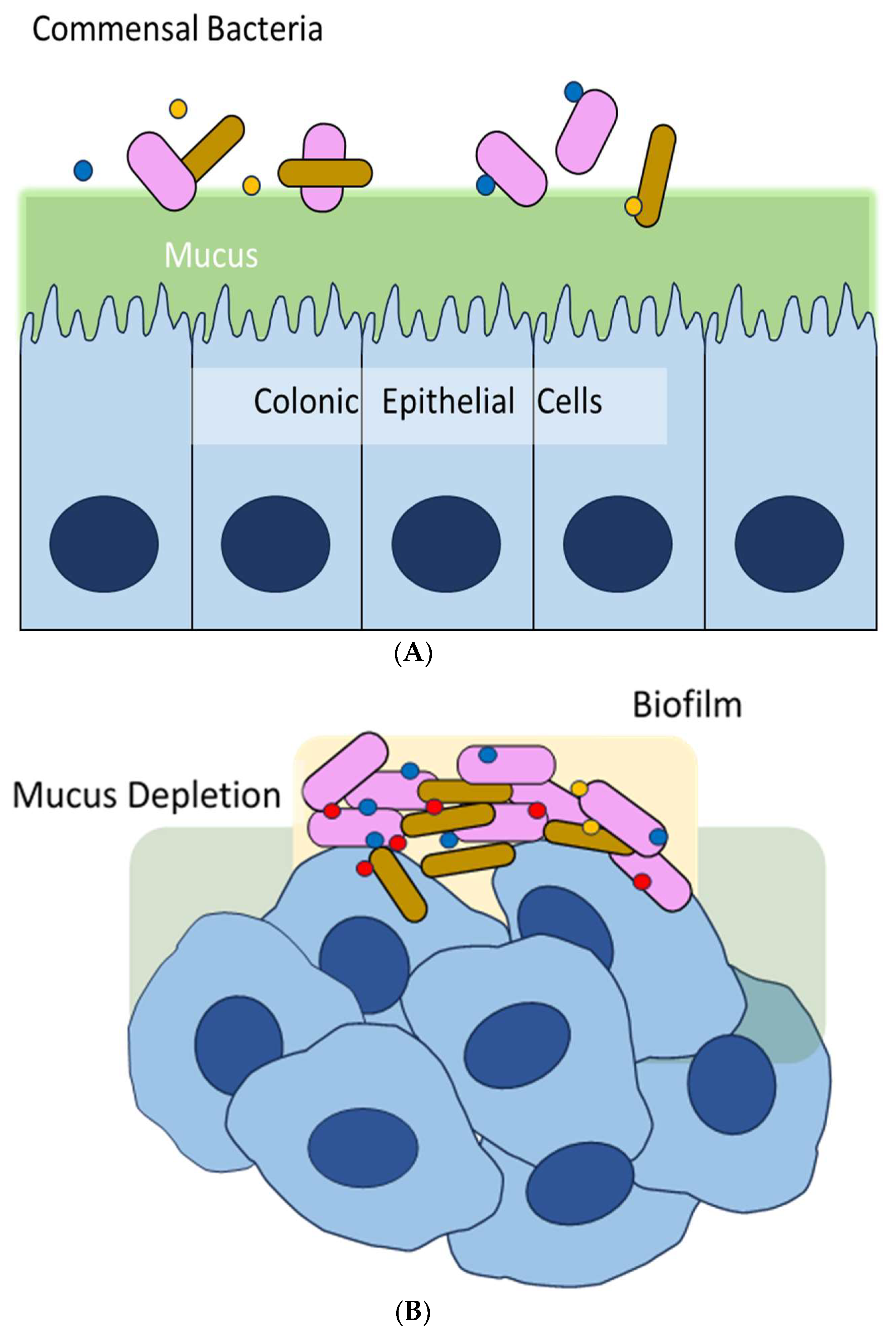
1. Introduction
2. Biofilm and Colorectal Cancer (CRC)
3. Biofilm-Associated Helicobacter Pylori and Gastric Cancer
4. Tumor Microbiome and Other Cancers
5. Therapeutics and Their Clinical Implications
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vestby, L.K.; Gronseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Jayaraman, N.; Chatterji, D. Small-Molecule Inhibition of Bacterial Biofilm. ACS Omega 2020, 5, 3108–3115. [Google Scholar] [CrossRef]
- Sonderholm, M.; Kragh, K.N.; Koren, K.; Jakobsen, T.H.; Darch, S.E.; Alhede, M.; Jensen, P.O.; Whiteley, M.; Kuhl, M.; Bjarnsholt, T. Pseudomonas aeruginosa Aggregate Formation in an Alginate Bead Model System Exhibits In Vivo-Like Characteristics. Appl. Environ. Microbiol. 2017, 83, e00113-17. [Google Scholar] [CrossRef]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Fey, P.D.; Olson, M.E. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 2010, 5, 917–933. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef]
- Elgundi, Z.; Papanicolaou, M.; Major, G.; Cox, T.R.; Melrose, J.; Whitelock, J.M.; Farrugia, B.L. Cancer Metastasis: The Role of the Extracellular Matrix and the Heparan Sulfate Proteoglycan Perlecan. Front. Oncol. 2019, 9, 1482. [Google Scholar] [CrossRef]
- Cluntun, A.A.; Lukey, M.J.; Cerione, R.A.; Locasale, J.W. Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer 2017, 3, 169–180. [Google Scholar] [CrossRef]
- Wan, N.; Wang, H.; Ng, C.K.; Mukherjee, M.; Ren, D.; Cao, B.; Tang, Y.J. Bacterial Metabolism during Biofilm Growth Investigated by (13)C Tracing. Front. Microbiol. 2018, 9, 2657. [Google Scholar] [CrossRef]
- Gong, T.; Zheng, C.; Ou, X.; Zheng, J.; Yu, J.; Chen, S.; Duan, Y.; Liu, W. Glutamine metabolism in cancers: Targeting the oxidative homeostasis. Front. Oncol. 2022, 12, 994672. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis—The ‘accidental’ pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef]
- Ellermann, M.; Sartor, R.B. Intestinal bacterial biofilms modulate mucosal immune responses. J. Immunol. Sci. 2018, 2, 13–18. [Google Scholar] [PubMed]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Ciernikova, S.; Sevcikova, A.; Stevurkova, V.; Mego, M. Tumor microbiome—An integral part of the tumor microenvironment. Front. Oncol. 2022, 12, 1063100. [Google Scholar] [CrossRef]
- Alipour, M. Molecular Mechanism of Helicobacter pylori-Induced Gastric Cancer. J. Gastrointest. Cancer 2021, 52, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Nougayrede, J.P.; Homburg, S.; Taieb, F.; Boury, M.; Brzuszkiewicz, E.; Gottschalk, G.; Buchrieser, C.; Hacker, J.; Dobrindt, U.; Oswald, E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 2006, 313, 848–851. [Google Scholar] [CrossRef]
- Putze, J.; Hennequin, C.; Nougayrede, J.P.; Zhang, W.; Homburg, S.; Karch, H.; Bringer, M.A.; Fayolle, C.; Carniel, E.; Rabsch, W.; et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect. Immun. 2009, 77, 4696–4703. [Google Scholar] [CrossRef]
- Garrett, W.S. Cancer and the microbiota. Science 2015, 348, 80–86. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Abreu, M.T.; Peek, R.M., Jr. Gastrointestinal malignancy and the microbiome. Gastroenterology 2014, 146, 1534–1546.e3. [Google Scholar] [CrossRef]
- Lopes, A.; Billard, E.; Casse, A.H.; Villeger, R.; Veziant, J.; Roche, G.; Carrier, G.; Sauvanet, P.; Briat, A.; Pages, F.; et al. Colibactin-positive Escherichia coli induce a procarcinogenic immune environment leading to immunotherapy resistance in colorectal cancer. Int. J. Cancer 2020, 146, 3147–3159. [Google Scholar] [CrossRef]
- Barth, H.; Fischer, S.; Moglich, A.; Fortsch, C. Clostridial C3 Toxins Target Monocytes/Macrophages and Modulate Their Functions. Front. Immunol. 2015, 6, 339. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhou, R.; Wang, H.; Li, W.; Pan, M.; Yao, X.; Zhan, W.; Yang, S.; Xu, L.; Ding, Y.; et al. Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ. 2019, 26, 2447–2463. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Kang, M.; Martin, A. Microbiome and colorectal cancer: Unraveling host-microbiota interactions in colitis-associated colorectal cancer development. Semin. Immunol. 2017, 32, 3–13. [Google Scholar] [CrossRef]
- Burns, M.B.; Lynch, J.; Starr, T.K.; Knights, D.; Blekhman, R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med. 2015, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef]
- Genua, F.; Raghunathan, V.; Jenab, M.; Gallagher, W.M.; Hughes, D.J. The Role of Gut Barrier Dysfunction and Microbiome Dysbiosis in Colorectal Cancer Development. Front. Oncol. 2021, 11, 626349. [Google Scholar] [CrossRef]
- Dejea, C.M.; Wick, E.C.; Hechenbleikner, E.M.; White, J.R.; Mark Welch, J.L.; Rossetti, B.J.; Peterson, S.N.; Snesrud, E.C.; Borisy, G.G.; Lazarev, M.; et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 18321–18326. [Google Scholar] [CrossRef] [PubMed]
- Ye, P.; Xi, Y.; Huang, Z.; Xu, P. Linking Obesity with Colorectal Cancer: Epidemiology and Mechanistic Insights. Cancers 2020, 12, 1408. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Dejea, C.M.; Edler, D.; Hoang, L.T.; Santidrian, A.F.; Felding, B.H.; Ivanisevic, J.; Cho, K.; Wick, E.C.; Hechenbleikner, E.M.; et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015, 21, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Meng, Y.; Wu, X.; Sun, Y. Polyamines and related signaling pathways in cancer. Cancer Cell Int. 2020, 20, 539. [Google Scholar] [CrossRef]
- Bernstein, C.; Holubec, H.; Bhattacharyya, A.K.; Nguyen, H.; Payne, C.M.; Zaitlin, B.; Bernstein, H. Carcinogenicity of deoxycholate, a secondary bile acid. Arch. Toxicol. 2011, 85, 863–871. [Google Scholar] [CrossRef]
- Cao, H.; Luo, S.; Xu, M.; Zhang, Y.; Song, S.; Wang, S.; Kong, X.; He, N.; Cao, X.; Yan, F.; et al. The secondary bile acid, deoxycholate accelerates intestinal adenoma-adenocarcinoma sequence in Apc (min/+) mice through enhancing Wnt signaling. Fam. Cancer 2014, 13, 563–571. [Google Scholar] [CrossRef]
- Guzior, D.V.; Quinn, R.A. Review: Microbial transformations of human bile acids. Microbiome 2021, 9, 140. [Google Scholar] [CrossRef]
- Tomkovich, S.; Dejea, C.M.; Winglee, K.; Drewes, J.L.; Chung, L.; Housseau, F.; Pope, J.L.; Gauthier, J.; Sun, X.; Muhlbauer, M.; et al. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J. Clin. Investig. 2019, 129, 1699–1712. [Google Scholar] [CrossRef] [PubMed]
- Allen-Vercoe, E.; Strauss, J.; Chadee, K. Fusobacterium nucleatum: An emerging gut pathogen? Gut Microbes 2011, 2, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.L.; Freeman, D.J.; Pleasance, S.; Watson, P.; Moore, R.A.; Cochrane, K.; Allen-Vercoe, E.; Holt, R.A. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 2013, 1, 16. [Google Scholar] [CrossRef]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef]
- Kinder, S.A.; Holt, S.C. Localization of the Fusobacterium nucleatum T18 adhesin activity mediating coaggregation with Porphyromonas gingivalis T22. J. Bacteriol. 1993, 175, 840–850. [Google Scholar] [CrossRef]
- Edwards, A.M.; Grossman, T.J.; Rudney, J.D. Fusobacterium nucleatum transports noninvasive Streptococcus cristatus into human epithelial cells. Infect. Immun. 2006, 74, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Kunzmann, A.T.; Proenca, M.A.; Jordao, H.W.; Jiraskova, K.; Schneiderova, M.; Levy, M.; Liska, V.; Buchler, T.; Vodickova, L.; Vymetalkova, V.; et al. Fusobacterium nucleatum tumor DNA levels are associated with survival in colorectal cancer patients. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1891–1899. [Google Scholar] [CrossRef] [PubMed]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Baik, J.E.; Lagana, S.M.; Han, R.P.; Raab, W.J.; Sahoo, D.; Dalerba, P.; Wang, T.C.; Han, Y.W. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/beta-catenin modulator Annexin A1. EMBO Rep. 2019, 20, e47638. [Google Scholar] [CrossRef]
- Dejea, C.M.; Sears, C.L. Do biofilms confer a pro-carcinogenic state? Gut Microbes 2016, 7, 54–57. [Google Scholar] [CrossRef]
- World Cancer Research Fund International. Stomach Cancer Statistics. (0228). Available online: https://www.wcrf.org/cancer-trends/stomach-cancer-statistics (accessed on 2 February 2023).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Han, Z.; Liu, J.; Zhang, W.; Kong, Q.; Wan, M.; Lin, M.; Lin, B.; Ding, Y.; Duan, M.; Li, Y.; et al. Cardia and non-cardia gastric cancer risk associated with Helicobacter pylori in East Asia and the West: A systematic review, meta-analysis, and estimation of population attributable fraction. Helicobacter 2023, 28, e12950. [Google Scholar] [CrossRef]
- Parsonnet, J.; Hansen, S.; Rodriguez, L.; Gelb, A.B.; Warnke, R.A.; Jellum, E.; Orentreich, N.; Vogelman, J.H.; Friedman, G.D. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 1994, 330, 1267–1271. [Google Scholar] [CrossRef]
- Bugaytsova, J.A.; Bjornham, O.; Chernov, Y.A.; Gideonsson, P.; Henriksson, S.; Mendez, M.; Sjostrom, R.; Mahdavi, J.; Shevtsova, A.; Ilver, D.; et al. Helicobacter pylori Adapts to Chronic Infection and Gastric Disease via pH-Responsive BabA-Mediated Adherence. Cell Host Microbe 2017, 21, 376–389. [Google Scholar] [CrossRef]
- Aitken, A. 14-3-3 proteins: A historic overview. Semin. Cancer Biol. 2006, 16, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.J.; Jan, Y.J.; Ko, B.S.; Liang, S.M.; Liou, J.Y. Involvement of 14-3-3 Proteins in Regulating Tumor Progression of Hepatocellular Carcinoma. Cancers 2015, 7, 1022–1036. [Google Scholar] [CrossRef] [PubMed]
- Sougleri, I.S.; Papadakos, K.S.; Zadik, M.P.; Mavri-Vavagianni, M.; Mentis, A.F.; Sgouras, D.N. Helicobacter pylori CagA protein induces factors involved in the epithelial to mesenchymal transition (EMT) in infected gastric epithelial cells in an EPIYA-phosphorylation-dependent manner. FEBS J. 2016, 283, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.; Zhang, H.; Meng, F.; He, L.; Zhang, J.; Xiao, D. Proteomic and transcriptomic studies of BGC823 cells stimulated with Helicobacter pylori isolates from gastric MALT lymphoma. PLoS ONE 2020, 15, e0238379. [Google Scholar] [CrossRef]
- Hathroubi, S.Z.J.; Ottemann, K.M. Helicobacter pylori Biofilm Cells are Metabolically Distinct, Express Flagella, and Antibiotic Tolerant. bioRxiv 2019, 728766. [Google Scholar] [CrossRef]
- Krzyzek, P.; Grande, R.; Migdal, P.; Paluch, E.; Gosciniak, G. Biofilm Formation as a Complex Result of Virulence and Adaptive Responses of Helicobacter pylori. Pathogens 2020, 9, 1062. [Google Scholar] [CrossRef]
- Choi, I.J.; Kim, C.G.; Lee, J.Y.; Kim, Y.I.; Kook, M.C.; Park, B.; Joo, J. Family History of Gastric Cancer and Helicobacter pylori Treatment. N. Engl. J. Med. 2020, 382, 427–436. [Google Scholar] [CrossRef]
- Stathis, A.; Chini, C.; Bertoni, F.; Proserpio, I.; Capella, C.; Mazzucchelli, L.; Pedrinis, E.; Cavalli, F.; Pinotti, G.; Zucca, E. Long-term outcome following Helicobacter pylori eradication in a retrospective study of 105 patients with localized gastric marginal zone B-cell lymphoma of MALT type. Ann. Oncol. 2009, 20, 1086–1093. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Hanawa, T.; Kurata, S.; Ochiai, K.; Kamiya, S. Impact of Helicobacter pylori biofilm formation on clarithromycin susceptibility and generation of resistance mutations. PLoS ONE 2013, 8, e73301. [Google Scholar] [CrossRef]
- Waskito, L.A.; Salama, N.R.; Yamaoka, Y. Pathogenesis of Helicobacter pylori infection. Helicobacter 2018, 23 (Suppl. 1), e12516. [Google Scholar] [CrossRef] [PubMed]
- Dascalu, R.I.; Bolocan, A.; Paduaru, D.N.; Constantinescu, A.; Mitache, M.M.; Stoica, A.D.; Andronic, O. Multidrug resistance in Helicobacter pylori infection. Front. Microbiol. 2023, 14, 1128497. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, M.; El-Zataari, M.; Owyang, S.Y.; Eaton, K.A.; Liu, M.; Chang, Y.M.; Zou, W.; Kao, J.Y. TLR2 mediates Helicobacter pylori-induced tolerogenic immune response in mice. PLoS ONE 2013, 8, e74595. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xuan, C.; Shamonki, J.M.; Chung, A.; Dinome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial dysbiosis is associated with human breast cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef] [PubMed]
- Greathouse, K.L.; White, J.R.; Vargas, A.J.; Bliskovsky, V.V.; Beck, J.A.; von Muhlinen, N.; Polley, E.C.; Bowman, E.D.; Khan, M.A.; Robles, A.I.; et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018, 19, 123. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef]
- Fu, A.; Yao, B.; Dong, T.; Chen, Y.; Yao, J.; Liu, Y.; Li, H.; Bai, H.; Liu, X.; Zhang, Y.; et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022, 185, 1356–1372.e26. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 2017, 152, 851–866.e24. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Huffnagle, G.B. The role of the bacterial microbiome in lung disease. Expert. Rev. Respir. Med. 2013, 7, 245–257. [Google Scholar] [CrossRef]
- Yu, G.; Gail, M.H.; Consonni, D.; Carugno, M.; Humphrys, M.; Pesatori, A.C.; Caporaso, N.E.; Goedert, J.J.; Ravel, J.; Landi, M.T. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016, 17, 163. [Google Scholar] [CrossRef]
- Lee, S.H.; Sung, J.Y.; Yong, D.; Chun, J.; Kim, S.Y.; Song, J.H.; Chung, K.S.; Kim, E.Y.; Jung, J.Y.; Kang, Y.A.; et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer 2016, 102, 89–95. [Google Scholar] [CrossRef] [PubMed]
- O’Dwyer, D.N.; Dickson, R.P.; Moore, B.B. The Lung Microbiome, Immunity, and the Pathogenesis of Chronic Lung Disease. J. Immunol. 2016, 196, 4839–4847. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S.; et al. Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell 2019, 176, 998–1013.e16. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef]
- Shannon, B.; Yi, T.J.; Perusini, S.; Gajer, P.; Ma, B.; Humphrys, M.S.; Thomas-Pavanel, J.; Chieza, L.; Janakiram, P.; Saunders, M.; et al. Association of HPV infection and clearance with cervicovaginal immunology and the vaginal microbiota. Mucosal Immunol. 2017, 10, 1310–1319. [Google Scholar] [CrossRef]
- Frank, D.N.; Qiu, Y.; Cao, Y.; Zhang, S.; Lu, L.; Kofonow, J.M.; Robertson, C.E.; Liu, Y.; Wang, H.; Levens, C.L.; et al. A dysbiotic microbiome promotes head and neck squamous cell carcinoma. Oncogene 2022, 41, 1269–1280. [Google Scholar] [CrossRef] [PubMed]
- Galeano Nino, J.L.; Wu, H.; LaCourse, K.D.; Kempchinsky, A.G.; Baryiames, A.; Barber, B.; Futran, N.; Houlton, J.; Sather, C.; Sicinska, E.; et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 2022, 611, 810–817. [Google Scholar] [CrossRef]
- LaCourse, K.D.; Zepeda-Rivera, M.; Kempchinsky, A.G.; Baryiames, A.; Minot, S.S.; Johnston, C.D.; Bullman, S. The cancer chemotherapeutic 5-fluorouracil is a potent Fusobacterium nucleatum inhibitor and its activity is modified by intratumoral microbiota. Cell Rep. 2022, 41, 111625. [Google Scholar] [CrossRef]
- Inamura, K. Beyond Bacteria: Fungi in the Tumor Microbiome. Cancers 2023, 15, 572. [Google Scholar] [CrossRef]
- Broecker, F.; Moelling, K. The Roles of the Virome in Cancer. Microorganisms 2021, 9, 2538. [Google Scholar] [CrossRef]
- Nakkarach, A.; Foo, H.L.; Song, A.A.; Mutalib, N.E.A.; Nitisinprasert, S.; Withayagiat, U. Anti-cancer and anti-inflammatory effects elicited by short chain fatty acids produced by Escherichia coli isolated from healthy human gut microbiota. Microb. Cell Fact. 2021, 20, 36. [Google Scholar] [CrossRef]
- Ryu, T.Y.; Kim, K.; Han, T.S.; Lee, M.O.; Lee, J.; Choi, J.; Jung, K.B.; Jeong, E.J.; An, D.M.; Jung, C.R.; et al. Human gut-microbiome-derived propionate coordinates proteasomal degradation via HECTD2 upregulation to target EHMT2 in colorectal cancer. ISME J. 2022, 16, 1205–1221. [Google Scholar] [CrossRef]
- Gutierrez-Merino, J.; Isla, B.; Combes, T.; Martinez-Estrada, F.; Maluquer De Motes, C. Beneficial bacteria activate type-I interferon production via the intracellular cytosolic sensors STING and MAVS. Gut Microbes 2020, 11, 771–788. [Google Scholar] [CrossRef]
- Cammarota, G.; Branca, G.; Ardito, F.; Sanguinetti, M.; Ianiro, G.; Cianci, R.; Torelli, R.; Masala, G.; Gasbarrini, A.; Fadda, G.; et al. Biofilm demolition and antibiotic treatment to eradicate resistant Helicobacter pylori: A clinical trial. Clin. Gastroenterol. Hepatol. 2010, 8, 817–820.e3. [Google Scholar] [CrossRef]
- Gurbuz, A.K.; Ozel, A.M.; Ozturk, R.; Yildirim, S.; Yazgan, Y.; Demirturk, L. Effect of N-acetyl cysteine on Helicobacter pylori. South. Med. J. 2005, 98, 1095–1097. [Google Scholar] [CrossRef][Green Version]
- Sankar, S.; Yuwanati, M.; Ganesh, P.S. Pitchaipillai Sankar Ganesh, Acyl homoserine lactone inhibitors for oral squamous cell carcinoma–Novel insights and therapeutic perspective. Med. Hypotheses 2023, 170, 111003. [Google Scholar] [CrossRef]
- Chai, H.; Hazawa, M.; Shirai, N.; Igarashi, J.; Takahashi, K.; Hosokawa, Y.; Suga, H.; Kashiwakura, I. Functional properties of synthetic N-acyl-L-homoserine lactone analogs of quorum-sensing gram-negative bacteria on the growth of human oral squamous carcinoma cells. Investig. New Drugs 2012, 30, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Chai, H.; Hazawa, M.; Hosokawa, Y.; Igarashi, J.; Suga, H.; Kashiwakura, I. Novel acridine-based N-acyl-homoserine lactone analogs induce endoreduplication in the human oral squamous carcinoma cell line SAS. Biol. Pharm. Bull. 2012, 35, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Liu, S.Y.; Chua, S.L.; Khoo, B.L. The effects of biofilms on tumor progression in a 3D cancer-biofilm microfluidic model. Biosens. Bioelectron. 2021, 180, 113113. [Google Scholar] [CrossRef]
- Vetizou, M.; Pitt, J.M.; Daillere, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillere, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Salminen, E.; Elomaa, I.; Minkkinen, J.; Vapaatalo, H.; Salminen, S. Preservation of intestinal integrity during radiotherapy using live Lactobacillus acidophilus cultures. Clin. Radiol. 1988, 39, 435–437. [Google Scholar] [CrossRef]
- Delia, P.; Sansotta, G.; Donato, V.; Messina, G.; Frosina, P.; Pergolizzi, S.; De Renzis, C.; Famularo, G. Prevention of radiation-induced diarrhea with the use of VSL#3, a new high-potency probiotic preparation. Am. J. Gastroenterol. 2002, 97, 2150–2152. [Google Scholar] [PubMed]
- Urbancsek, H.; Kazar, T.; Mezes, I.; Neumann, K. Results of a double-blind, randomized study to evaluate the efficacy and safety of Antibiophilus in patients with radiation-induced diarrhoea. Eur. J. Gastroenterol. Hepatol. 2001, 13, 391–396. [Google Scholar] [CrossRef]
- Kasinskas, R.W.; Forbes, N.S. Salmonella typhimurium specifically chemotax and proliferate in heterogeneous tumor tissue in vitro. Biotechnol. Bioeng. 2006, 94, 710–721. [Google Scholar] [CrossRef]
- Pawelek, J.M.; Low, K.B.; Bermudes, D. Tumor-targeted Salmonella as a novel anticancer vector. Cancer Res. 1997, 57, 4537–4544. [Google Scholar]
- Leschner, S.; Weiss, S. Salmonella-allies in the fight against cancer. J. Mol. Med. 2010, 88, 763–773. [Google Scholar] [CrossRef]
- Friedlos, F.; Lehouritis, P.; Ogilvie, L.; Hedley, D.; Davies, L.; Bermudes, D.; King, I.; Martin, J.; Marais, R.; Springer, C.J. Attenuated Salmonella targets prodrug activating enzyme carboxypeptidase G2 to mouse melanoma and human breast and colon carcinomas for effective suicide gene therapy. Clin. Cancer Res. 2008, 14, 4259–4266. [Google Scholar] [CrossRef]
- Saltzman, D.A.; Heise, C.P.; Hasz, D.E.; Zebede, M.; Kelly, S.M.; Curtiss, R., 3rd; Leonard, A.S.; Anderson, P.M. Attenuated Salmonella typhimurium containing interleukin-2 decreases MC-38 hepatic metastases: A novel anti-tumor agent. Cancer Biother. Radiopharm. 1996, 11, 145–153. [Google Scholar] [PubMed]
- Sorenson, B.S.; Banton, K.L.; Frykman, N.L.; Leonard, A.S.; Saltzman, D.A. Attenuated Salmonella typhimurium with IL-2 gene reduces pulmonary metastases in murine osteosarcoma. Clin. Orthop. Relat. Res. 2008, 466, 1285–1291. [Google Scholar] [CrossRef] [PubMed]
- Al-Ramadi, B.K.; Fernandez-Cabezudo, M.J.; El-Hasasna, H.; Al-Salam, S.; Bashir, G.; Chouaib, S. Potent anti-tumor activity of systemically-administered IL2-expressing Salmonella correlates with decreased angiogenesis and enhanced tumor apoptosis. Clin. Immunol. 2009, 130, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Gorjifard, S.; Goldszmid, R.S. Microbiota-myeloid cell crosstalk beyond the gut. J. Leukoc. Biol. 2016, 100, 865–879. [Google Scholar] [CrossRef]
- Liu, X.; Yin, S.; Chen, Y.; Wu, Y.; Zheng, W.; Dong, H.; Bai, Y.; Qin, Y.; Li, J.; Feng, S.; et al. LPS-induced proinflammatory cytokine expression in human airway epithelial cells and macrophages via NF-kappaB, STAT3 or AP-1 activation. Mol. Med. Rep. 2018, 17, 5484–5491. [Google Scholar]
- Dasari, S.; Kathera, C.; Janardhan, A.; Praveen Kumar, A.; Viswanath, B. Surfacing role of probiotics in cancer prophylaxis and therapy: A systematic review. Clin. Nutr. 2017, 36, 1465–1472. [Google Scholar] [CrossRef]
- Xu, J.; Saklatvala, R.; Mittal, S.; Deshmukh, S.; Procopio, A. Recent Progress of Potentiating Immune Checkpoint Blockade with External Stimuli-an Industry Perspective. Adv. Sci. 2020, 7, 1903394. [Google Scholar] [CrossRef]
- Yu, N.; Li, J.; Singh, P.K.; Ding, D.; Sun, W.; Tang, Q.; Xu, H. The Superior Anticancer Effect of Reactive Oxygen Species-Responsive Paclitaxel Nanoparticles is Mediated Through Autophagic Cell Death. J. Biomed. Nanotechnol. 2019, 15, 2251–2261. [Google Scholar] [CrossRef]
- Kulshrestha, S.; Khan, S.; Meena, R.; Singh, B.R.; Khan, A.U. A graphene/zinc oxide nanocomposite film protects dental implant surfaces against cariogenic Streptococcus mutans. Biofouling 2014, 30, 1281–1294. [Google Scholar] [CrossRef]
- Kulshrestha, S.; Khan, S.; Hasan, S.; Khan, M.E.; Misba, L.; Khan, A.U. Calcium fluoride nanoparticles induced suppression of Streptococcus mutans biofilm: An in vitro and in vivo approach. Appl. Microbiol. Biotechnol. 2016, 100, 1901–1914. [Google Scholar] [CrossRef] [PubMed]
- Kulshrestha, S.; Khan, A.U. Nanomedicine for anticancer and antimicrobial treatment: An overview. IET Nanobiotechnol. 2018, 12, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.H.; Mao, L.P.; Cai, X.J.; Mo, X.Y.; Zhu, Q.Q.; Yang, F.T.; Wang, M.H. Zinc oxide nanoparticles reduce the chemoresistance of gastric cancer by inhibiting autophagy. World J. Gastroenterol. 2021, 27, 3851–3862. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Du, J.; Wu, X.; Abdelrehem, A.; Ren, Y.; Liu, C.; Zhou, X.; Wang, S. Crosstalk between autophagy and microbiota in cancer progression. Mol. Cancer 2021, 20, 163. [Google Scholar] [CrossRef] [PubMed]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef]
- Banerjee, S.; Alwine, J.C.; Wei, Z.; Tian, T.; Shih, N.; Sperling, C.; Guzzo, T.; Feldman, M.D.; Robertson, E.S. Microbiome signatures in prostate cancer. Carcinogenesis 2019, 40, 749–764. [Google Scholar] [CrossRef]
- Audirac-Chalifour, A.; Torres-Poveda, K.; Bahena-Roman, M.; Tellez-Sosa, J.; Martinez-Barnetche, J.; Cortina-Ceballos, B.; Lopez-Estrada, G.; Delgado-Romero, K.; Burguete-Garcia, A.I.; Cantu, D.; et al. Cervical Microbiome and Cytokine Profile at Various Stages of Cervical Cancer: A Pilot Study. PLoS ONE 2016, 11, e0153274. [Google Scholar] [CrossRef]
- Thyagarajan, S.; Zhang, Y.; Thapa, S.; Allen, M.S.; Phillips, N.; Chaudhary, P.; Kashyap, M.V.; Vishwanatha, J.K. Comparative analysis of racial differences in breast tumor microbiome. Sci. Rep. 2020, 10, 14116. [Google Scholar] [CrossRef]
- Yan, X.; Yang, M.; Liu, J.; Gao, R.; Hu, J.; Li, J.; Zhang, L.; Shi, Y.; Guo, H.; Cheng, J.; et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am. J. Cancer Res. 2015, 5, 3111–3122. [Google Scholar]
- Dickson, I. Microbiome promotes pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 328. [Google Scholar] [CrossRef]
- Yang, W.; Chen, C.H.; Jia, M.; Xing, X.; Gao, L.; Tsai, H.T.; Zhang, Z.; Liu, Z.; Zeng, B.; Yeung, S.J.; et al. Tumor-Associated Microbiota in Esophageal Squamous Cell Carcinoma. Front. Cell Dev. Biol. 2021, 9, 641270. [Google Scholar] [CrossRef]
- Schmidt, B.L.; Kuczynski, J.; Bhattacharya, A.; Huey, B.; Corby, P.M.; Queiroz, E.L.; Nightingale, K.; Kerr, A.R.; DeLacure, M.D.; Veeramachaneni, R.; et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS ONE 2014, 9, e98741. [Google Scholar] [CrossRef]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806.e12. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, K.; Baba, Y.; Nakagawa, S.; Mima, K.; Miyake, K.; Nakamura, K.; Sawayama, H.; Kinoshita, K.; Ishimoto, T.; Iwatsuki, M.; et al. Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated with Prognosis. Clin. Cancer Res. 2016, 22, 5574–5581. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Ren, Z.; Li, A.; Li, J.; Xu, S.; Zhang, H.; Jiang, J.; Yang, J.; Luo, Q.; Zhou, K.; et al. Tongue coating microbiome data distinguish patients with pancreatic head cancer from healthy controls. J. Oral. Microbiol. 2019, 11, 1563409. [Google Scholar] [CrossRef]
- Peters, B.A.; Wu, J.; Pei, Z.; Yang, L.; Purdue, M.P.; Freedman, N.D.; Jacobs, E.J.; Gapstur, S.M.; Hayes, R.B.; Ahn, J. Oral Microbiome Composition Reflects Prospective Risk for Esophageal Cancers. Cancer Res. 2017, 77, 6777–6787. [Google Scholar] [CrossRef] [PubMed]
- Dohlman, A.B.; Arguijo Mendoza, D.; Ding, S.; Gao, M.; Dressman, H.; Iliev, I.D.; Lipkin, S.M.; Shen, X. The cancer microbiome atlas: A pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants. Cell Host Microbe 2021, 29, 281–298.e5. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Markowski, M.C.; Boorjian, S.A.; Burton, J.P.; Hahn, N.M.; Ingersoll, M.A.; Maleki Vareki, S.; Pal, S.K.; Sfanos, K.S. The Microbiome and Genitourinary Cancer: A Collaborative Review. Eur. Urol. 2019, 75, 637–646. [Google Scholar] [CrossRef]

| Stages of Carcinogenesis | Potential Mechanisms of Biofilm/Intratumoral Microbes | Name of the Microbes and Their Components |
|---|---|---|
| Tumor initiation | Promoting mutagenesis and epigenetic alterations by producing substances that cause DNA damage, cell cycle arrest, and instability of genetic materials. | Colibactin produced by some E. coli and other Enterobacteriaceae strains causes DNA damage [16,17]. It promotes tumorigenesis in CRC [18]. |
| Producing or releasing metabolites that induce carcinogenesis in host cells. | Polymicrobial bacteria (see text for details). | |
| Tumor promotion | Activating Wnt/β-catenin signaling pathway, leading to higher gene expression of c-MYC and Cyclin-D-1 (uncontrolled cell division). | Activation of β-catenin by FadA from Fusobacterium nucleatum is associated with CRC and gastric cancers [19,20] |
| Inducing a chronic pro-inflammatory response. | Fusobacterium nucleatum activates NF-κB, a critical factor for cancer, promoting inflammation [13,18]. | |
| Tumor progression/Metastases | Direct or indirect immunomodulation by suppressing anti-tumor immune response. | Colibactin-positive E. coli is associated with a reduction of CD3+CD8+ T cells in a CRC mouse model [21]. |
| Clostridial C3 toxins from Clostridium botulinum and Clostridium limosum disturb phagocytosis and migration of macrophages [22]. | ||
| Supporting tumor metastasis. | E. coli in the gut stimulates cathepsin K, a metastasis-associated secretory protein, which mediates M2 macrophage polarization in a TLR-4-dependent manner in CRC [23]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, E.; Murray, B.; Choi, S. Biofilm and Cancer: Interactions and Future Directions for Cancer Therapy. Int. J. Mol. Sci. 2023, 24, 12836. https://doi.org/10.3390/ijms241612836
Choi E, Murray B, Choi S. Biofilm and Cancer: Interactions and Future Directions for Cancer Therapy. International Journal of Molecular Sciences. 2023; 24(16):12836. https://doi.org/10.3390/ijms241612836
Chicago/Turabian StyleChoi, Euna, Ben Murray, and Sunga Choi. 2023. "Biofilm and Cancer: Interactions and Future Directions for Cancer Therapy" International Journal of Molecular Sciences 24, no. 16: 12836. https://doi.org/10.3390/ijms241612836
APA StyleChoi, E., Murray, B., & Choi, S. (2023). Biofilm and Cancer: Interactions and Future Directions for Cancer Therapy. International Journal of Molecular Sciences, 24(16), 12836. https://doi.org/10.3390/ijms241612836






