Tomato Aphid (Aphis gossypii) Secreted Saliva Can Enhance Aphid Resistance by Upregulating Signaling Molecules in Tomato (Solanum lycopersicum)
Abstract
1. Introduction
2. Results
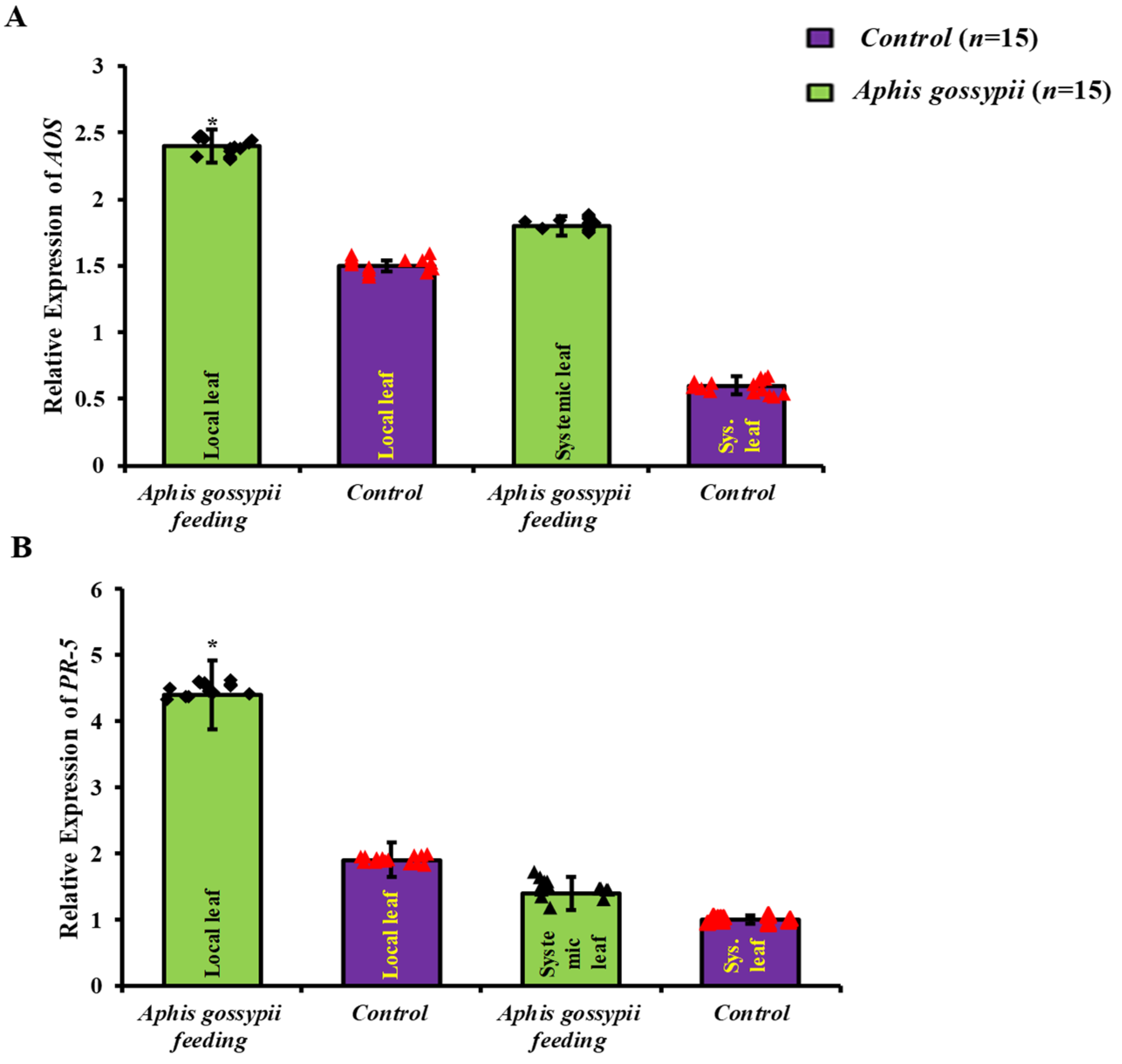
2.1. Local Defence Responses Were Triggered by A. gossypii Feeding
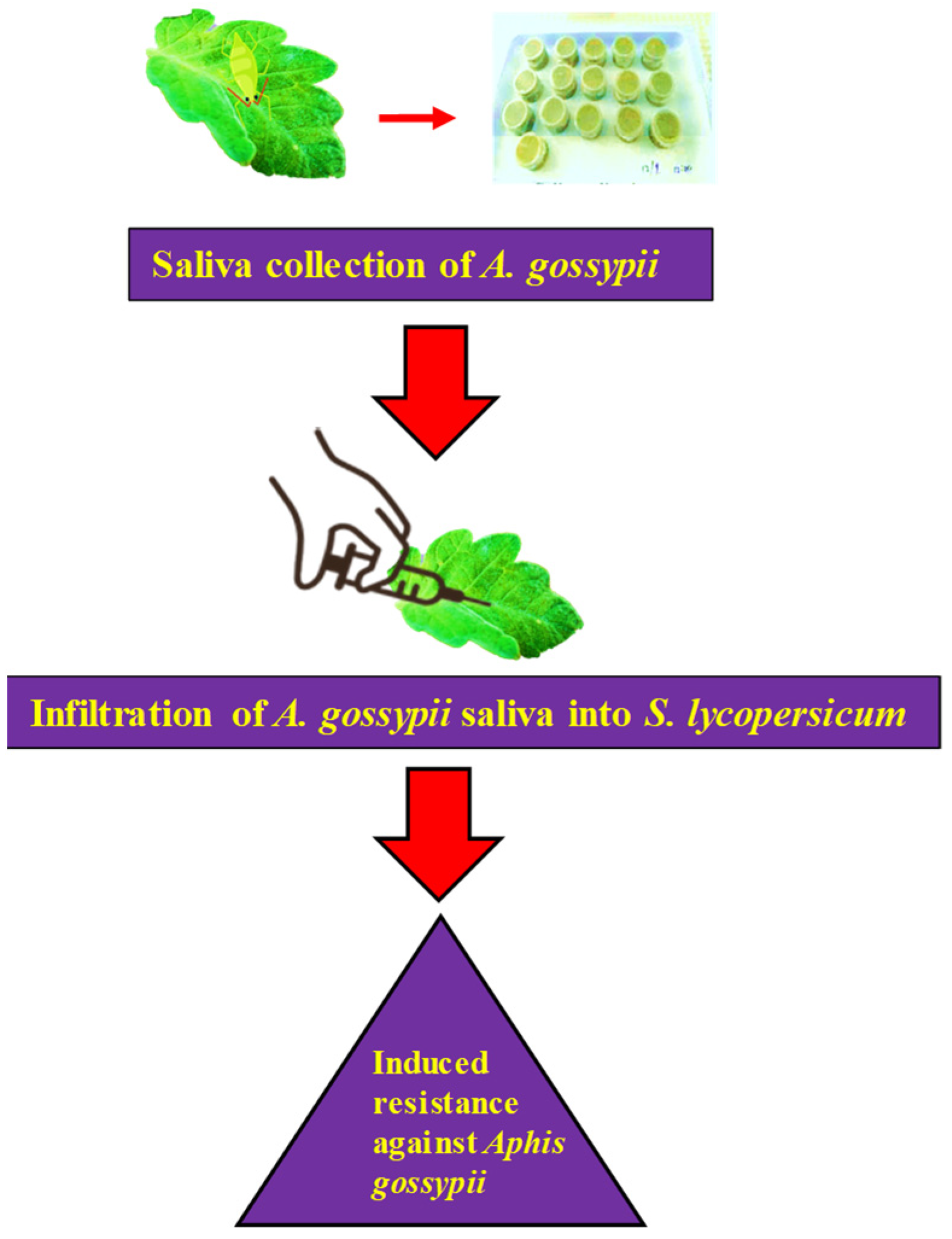
2.2. Assembly of A. gossypii Saliva
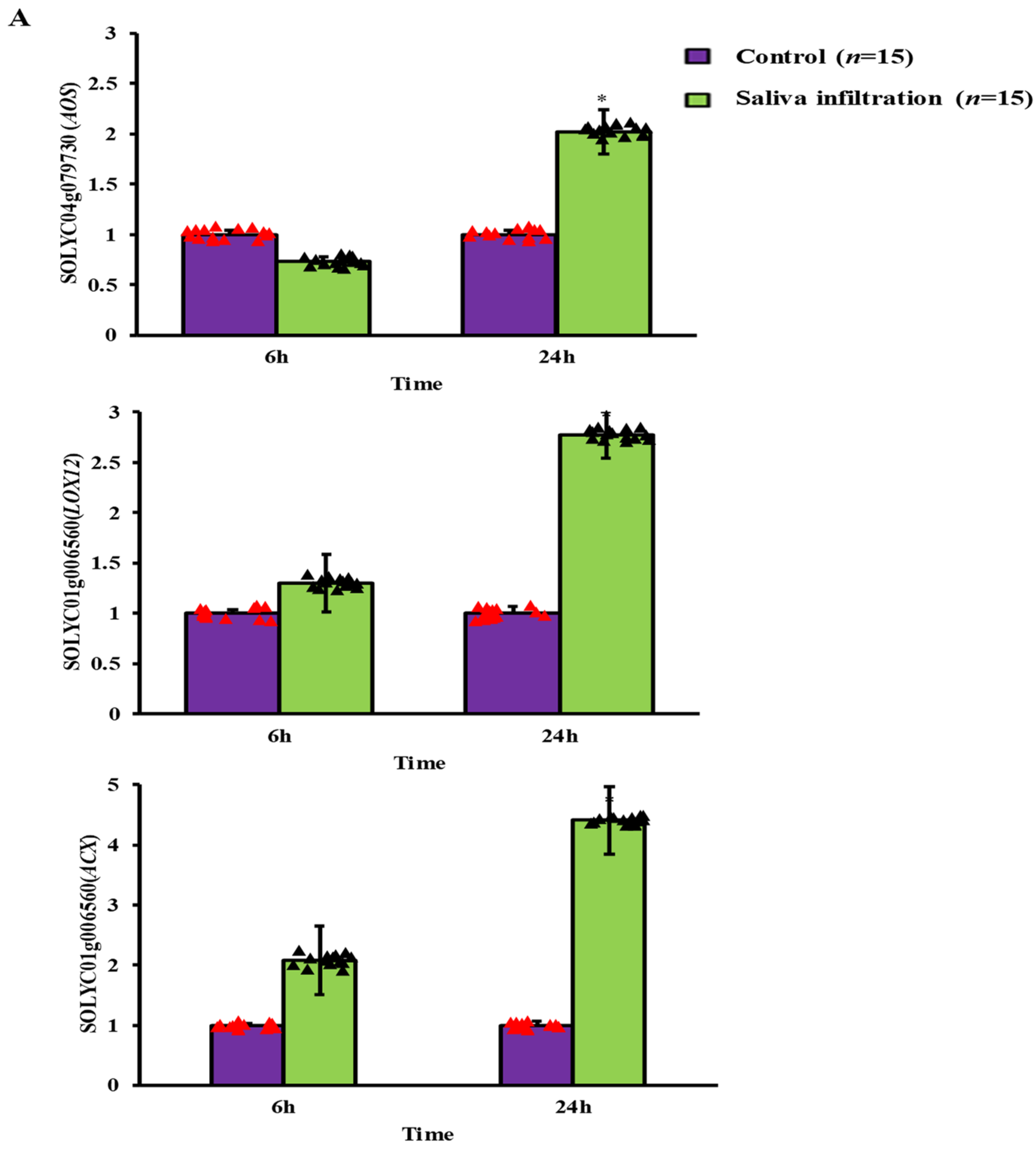
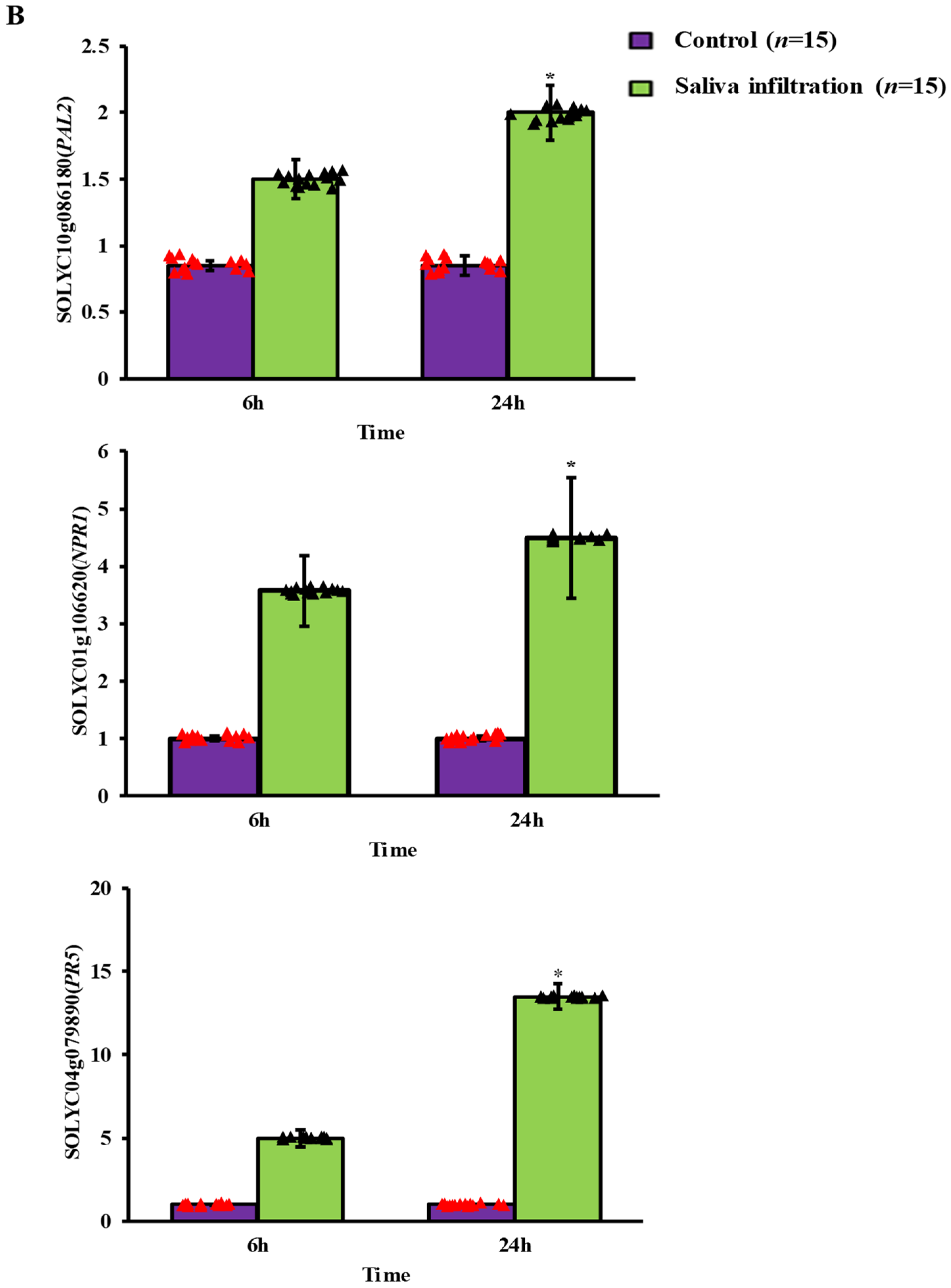
2.3. Defense-Related Gene Expression after A. gossypii Saliva Invasion
2.4. Performance of the A. gossypii
2.5. Aphis gossypii Feeding Activity by EPG
3. Discussion
4. Materials and Methods
4.1. Rearing of Insects and Prepration of Plant Colonies
4.2. Aphid Infestation Treatments
4.3. Collection of Aphid Saliva
4.4. Saliva Infiltration
4.5. Isolation of Total RNA and Synthesis of cDNA
4.6. RT-qPCR Analysis
4.7. Aphid Bioassay
4.7.1. Aphis gossypii Preference Assay
4.7.2. Intrinsic Population Growth Rate of A. gossypii
4.8. EPG Monitoring of A. gossypii Eating Activity
4.9. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mithöfer, A.; Boland, W. Plant defense against herbivores: Chemical aspects. Annu. Rev. Plant Biol. 2012, 63, 431–450. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, A.; Pieterse, C.M.J. Cross talk in defense signaling. Plant Physiol. 2008, 146, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Thaler, J.S.; Humphrey, P.T.; Whiteman, N.K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012, 17, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Jaouannet, M.; Rodriguez, P.A.; Thorpe, P.; Lenoir, C.J.G.; Macleod, R.; Escudero-Martinez, C.; Bos, J.I.B. Plant immunity in plant-aphid interactions. Front. Plant Sci. 2014, 5, 1–10. [Google Scholar] [CrossRef]
- Coppola, V.; Coppola, M.; Rocco, M.; Digilio, M.C.; D’Ambrosio, C.; Renzone, G.; Martinelli, R.; Scaloni, A.; Pennacchio, F.; Rao, R.; et al. Transcriptomic and proteomic analysis of a compatible tomato-aphid interaction reveals a predominant salicylic acid-dependent plant response. BMC Genom. 2013, 14, 1. [Google Scholar] [CrossRef]
- Mohase, L.; Van Der Westhuizen, A.J. Salicylic acid is involved in resistance responses in the Russian wheat aphid-wheat interaction. Plant Physiol. 2002, 159, 585–590. [Google Scholar] [CrossRef]
- Martinez De Ilarduya, O.; Xie, Q.G.; Kaloshian, I. Aphid-induced defense responses in Mi-1-mediated compatible and incompatible tomato interactions. Mol. Plant-Microbe Interact. 2003, 16, 699–708. [Google Scholar] [CrossRef]
- Zhu-Salzman, K.; Salzman, R.A.; Ahn, J.E.; Koiwa, H. Transcriptional Regulation of Sorghum Defense Determinants against a Phloem-Feeding Aphid. Plant Physiol. 2004, 134, 420–431. [Google Scholar] [CrossRef]
- Moran, P.J.; Thompson, G.A. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001, 125, 1074–1085. [Google Scholar] [CrossRef]
- Zhu, F.; Poelman, E.H.; Dicke, M. Insect herbivore-associated organisms affect plant responses to herbivory. New Phytol. 2014, 204, 315–321. [Google Scholar] [CrossRef]
- Alborn, H.; Turlings, T.; Jones, T.; Stenhagen, G.; Loughrin, J.; Tumlinson, J. An elicitor of plant volatiles from beet armyworm oral secretion. Science 1997, 276, 945–949. [Google Scholar] [CrossRef]
- Miles, P.W. Plant-sucking bugs can remove the contents of cells without mechanical damage. Experientia 1987, 43, 937–939. [Google Scholar] [CrossRef]
- Miles, P.W. Aphid saliva. Biol. Rev. 1999, 74, 41–85. [Google Scholar] [CrossRef]
- Botha, A.M.; Lacock, L.; Van Niekerk, C.; Matsioloko, M.T.; Du Preez, F.B.; Loots, S.; Venter, E.; Kunert, K.J.; Cullis, C.A. Is photosynthetic transcriptional regulation in Triticum aestivum L. cv. “TugelaDN” a contributing factor for tolerance to Diuraphis noxia (Homoptera: Aphididae)? Plant Cell Rep. 2006, 25, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Prince, D.C.; Drurey, C.; Zipfel, C.; Hogenhout, S.A. The leucine-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 and the Cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in arabidopsis. Plant Physiol. 2014, 164, 2207–2219. [Google Scholar] [CrossRef]
- Ma, R.; Chen, J.L.; Cheng, D.F.; Sun, J.R. Activation of defense mechanism in wheat by polyphenol oxidase from aphid saliva. J. Agric. Food Chem. 2010, 58, 2410–2418. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.A.; Stam, R.; Warbroek, T.; Bos, J.I.B. Mp10 and Mp42 from the aphid species Myzus persicae trigger plant defenses in Nicotiana benthamiana through different activities. Mol. Plant-Microbe Interact. 2014, 27, 30–39. [Google Scholar] [CrossRef]
- Blackman, R.L.; Eastop, V.F. Additions and amendments to “aphids on the World’s Plants”. Zootaxa 2011, 68, 57–68. [Google Scholar] [CrossRef]
- Zhao, L.Y.; Chen, J.L.; Cheng, D.F.; Sun, J.R.; Liu, Y.; Tian, Z. Biochemical and molecular characterizations of Sitobion avenae-induced wheat defense responses. Crop Prot. 2009, 28, 435–442. [Google Scholar] [CrossRef]
- Rao, S.A.K.; Carolan, J.C.; Wilkinson, T.L. Proteomic Profiling of Cereal Aphid Saliva Reveals Both Ubiquitous and Adaptive Secreted Proteins. PLoS ONE 2013, 8, e57413. [Google Scholar] [CrossRef]
- Chaudhary, R.; Peng, H.C.; He, J.; MacWilliams, J.; Teixeira, M.; Tsuchiya, T.; Chesnais, Q.; Mudgett, M.B.; Kaloshian, I. Aphid effector Me10 interacts with tomato TFT7, a 14-3-3 isoform involved in aphid resistance. New Phytol. 2019, 221, 1518–1528. [Google Scholar] [CrossRef] [PubMed]
- Tjallingii, W.F. Electronic Recording of Penetration Behaviour by Aphids. Entomol. Exp. Appl. 1978, 24, 721–730. [Google Scholar] [CrossRef]
- Kerchev, P.I.; Karpińska, B.; Morris, J.A.; Hussain, A.; Verrall, S.R.; Hedley, P.E.; Fenton, B.; Foyer, C.H.; Hancock, R.D. Vitamin C and the abscisic acid-insensitive 4 transcription factor are important determinants of aphid resistance in arabidopsis. Antioxid. Redox Signal. 2013, 18, 2091–2105. [Google Scholar] [CrossRef]
- Stewart, S.A.; Hodge, S.; Bennett, M.; Mansfield, J.W.; Powell, G. Aphid induction of phytohormones in Medicago truncatula is dependent upon time post-infestation, aphid density and the genotypes of both plant and insect. Arthropod-Plant Interact. 2016, 10, 41–53. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, H.J.; Yang, Y.T.; Zhang, Y.; Guo, J.Y.; Liu, W.X.; Wan, F.H. Plant defense responses induced by Bemisia tabaci Middle East—Asia Minor 1 salivary components. Entomol. Exp. Appl. 2016, 159, 287–297. [Google Scholar] [CrossRef]
- Rodriguez-Saona, C.R.; Musser, R.O.; Vogel, H.; Hum-Musser, S.M.; Thaler, J.S. Molecular, Biochemical, and Organismal Analyses of Tomato Plants Simultaneously Attacked by Herbivores from Two Feeding Guilds. J. Chem. Ecol. 2010, 36, 1043–1057. [Google Scholar] [CrossRef]
- Boughton, A.J.; Hoover, K.; Felton, G.W. Impact of chemical elicitor applications on greenhouse tomato plants and population growth of the green peach aphid, Myzus persicae. Entomol. Exp. Appl. 2006, 120, 175–188. [Google Scholar] [CrossRef]
- Ellis, C.; Karafyllidis, I.; Turner, J.G. Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol. Plant-Microbe Interact. 2002, 15, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Züst, T.; Agrawal, A.A. Mechanisms and evolution of plant resistance to aphids. Nat. Plants 2016, 2, 15206. [Google Scholar] [CrossRef] [PubMed]
- Zarate, S.I.; Kempema, L.A.; Walling, L.L. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 2007, 143, 866–875. [Google Scholar] [CrossRef]
- Zehnder, G.W.; Nichols, A.J.; Edwards, O.R.; Ridsdill-Smith, T.J. Electronically monitored cowpea aphid feeding behavior on resistant and susceptible lupins. Entomol. Exp. Appl. 2001, 98, 259–269. [Google Scholar] [CrossRef]
- Goggin, F.L. Plant-aphid interactions: Molecular and ecological perspectives. Curr. Opin. Plant Biol. 2007, 10, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Park, K.C. Methyl salicylate, a soybean aphid-induced plant volatile attractive to the predator Coccinella septempunctata. J. Chem. Ecol. 2005, 31, 1733–1746. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, S.Y.; Pickett, J.A. Production of semiochemical and allelobiotic agents as a consequence of aphid feeding. Chemoecology 2010, 20, 89–96. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, W.L.; Guo, G.X.; Ji, X.L. Volatile emission in wheat and parasitism by Aphidius avenae after exogenous application of salivary enzymes of Sitobion avenae. Entomol. Exp. Appl. 2009, 130, 215–221. [Google Scholar] [CrossRef]
- Botha, C.E.J.; Matsiliza, B. Reduction in transport in wheat (Triticum aestivum) is caused by sustained phloem feeding by the Russian wheat aphid (Diuraphis noxia). S. Afr. J. Bot. 2004, 70, 249–254. [Google Scholar] [CrossRef]
- Kehr, J. Phloem sap proteins: Their identities and potential roles in the interaction between plants and phloem-feeding insects. J. Exp. Bot. 2006, 57, 767–774. [Google Scholar] [CrossRef]
- Atamian, H.S.; Chaudhary, R.; Cin, V.D.; Bao, E.; Girke, T.; Kaloshian, I. In planta expression or delivery of potato aphid Macrosiphum euphorbiae effectors Me10 and Me23 enhances aphid fecundity. Mol. Plant-Microbe Interact. 2013, 26, 67–74. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, Y.; Tong, J.; Ge, P.; Wang, Q.; Zhao, Z.; Zhu-Salzman, K.; Hogenhout, S.A.; Ge, F.; Sun, Y. An Aphid-Secreted Salivary Protease Activates Plant Defense in Phloem. Curr. Biol. 2020, 30, 4826–4836.e7. [Google Scholar] [CrossRef]
- Yin, C.; Hulbert, S. Prospects for functional analysis of effectors from cereal rust fungi. Euphytica 2011, 179, 57–67. [Google Scholar] [CrossRef]
- Upadhyaya, N.M.; Mago, R.; Staskawicz, B.J.; Ayliffe, M.A.; Ellis, J.G.; Dodds, P.N. A bacterial type III secretion assay for delivery of fungal effector proteins into wheat. Mol. Plant-Microbe Interact. 2014, 27, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wu, K.; Yao, J.; Li, S.; Wang, X.; Huang, L.; Kang, Z. PSTha5a23, a candidate effector from the obligate biotrophic pathogen Puccinia striiformis f. sp. tritici, is involved in plant defense suppression and rust pathogenicity. Environ. Microbiol. 2017, 19, 1717–1729. [Google Scholar] [CrossRef] [PubMed]
- Ashford, D.A.; Smith, W.A.; Douglas, A.E. Living on a high sugar diet: The fate of sucrose ingested by a phloem- feeding insect, the pea aphid Acyrthosiphon pisum. J. Insect Physiol. 2000, 46, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Chincinska, I.A. Leaf infiltration in plant science: Old method, new possibilities. Plant Methods 2021, 17, 1–21. [Google Scholar] [CrossRef]
- Liu, X.; Meng, J.; Starkey, S.; Smith, C.M. Wheat Gene Expression is Differentially Affected by a Virulent Russian Wheat Aphid Biotype. J. Chem. Ecol. 2011, 37, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zheng, Z.; Huang, J.; Lai, Z.; Fan, B. Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 2009, 4, 493–496. [Google Scholar] [CrossRef]
- Javed, K.; Javed, H.; Qiu, D. Biocontrol Potential of Purified Elicitor Protein PeBL1 Extracted from Brevibacillus laterosporus Strain A60 and Its Capacity in the Induction of Defense Process against Cucumber Aphid (Myzus persicae ) in Cucumber (Cucumis sativus). Biology 2020, 9, 179. [Google Scholar] [CrossRef]
- Javed, K.; Talha, H.; Ayesha, H.; Wang, Y.; Javed, H. PeaT1 and PeBC1 Microbial Protein Elicitors Enhanced Resistance against Myzus persicae Sulzer in Chili. Microorganisms 2021, 9, 2197. [Google Scholar] [CrossRef]
- Javed, K.; Javed, H.; Qiu, D. PeBL1 of Brevibacillus laterosporus a new biocontrol tool for wheat aphid management (Sitobion avenae) in Triticum aestivum. Int. J. Trop. Insect. Sci. 2022, 42, 535–544. [Google Scholar]
- Sarria, E.; Cid, M.; Garzo, E.; Fereres, A. Excel Workbook for automatic parameter calculation of EPG data. Comput. Electron. Agric. 2009, 67, 35–42. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Generations of A. gossypii | Td (Day) (n = 30 for Each Aphid Generation) | No. of Nymphs per Day (n = 30 for Each Aphid Generation) | rm (n = 30 for Each Aphid Generation) | |||
|---|---|---|---|---|---|---|
| Saliva Infiltration | Control | Saliva Infiltration | Control | Saliva Infiltration | Control | |
| 1st | 8.63 ± 0.02 | 8.54 ± 0.01 | 3.73 ± 0.01 | 4.10 ± 0.02 | 0.32 ± 0.001 | 0.36 ± 0.001 |
| 2nd | 8.54 ± 0.02 | 7.44 ± 0.03 | 3.34 ± 0.012 | 3.53 ± 0.01 | 0.29 ± 0.01 * | 0.35 ± 0.01 * |
| 3rd | 8.89 ± 0.03 | 8.74 ± 0.007 | 2.42 ± 0.02 | 2.43 ± 0.018 | 0.19 ± 0.002 * | 0.21 ± 0.01 * |
| Time in (h) | Percentage of Aphids | |
|---|---|---|
| Saliva Infiltration (n = 20 for Each Time Period) | Control (n = 20 for Each Time Period) | |
| 6 h | 25.21 ± 0.01 b | 45.17 ± 0.02 *a |
| 24 h | 44.10 ± 0.02 *b | 54.10 ± 0.05 a |
| 48 h | 48.13 ± 0.01 b | 63.12 ± 0.04 a |
| Parameters of the EPG | Saliva-Infiltrated (n = 20 for Each Parameter) | Control (n = 20 for Each Parameter) |
|---|---|---|
| Total amount of time spent probing (h) | 4.22 ± 0.13 | 5.19 ± 0.07 * |
| Number of C | 18.12 ± 0.01 * | 10.94 ± 0.15 |
| Number of short probes (C < 3 min) | 8.41 ± 0.02 * | 4.15 ± 0.11 |
| Duration of non-probe period before the 1st E (h) | 2.71 ± 0.02 | 2.62 ± 0.05 |
| Mean duration of pd (s) | 7.52 ± 0.03 | 7.81 ± 0.02 |
| Number of pd | 131.21 ± 0.11 * | 91.25 ± 0.05 |
| Number of E1 | 7.51 ± 0.10 | 6.41 ± 0.02 |
| Mean duration of E1 (min) | 11.15 ± 1.01 | 9.81 ± 0.22 |
| Number of E2 | 4.61 ± 0.03 | 5.24 ± 0.11 |
| Mean duration of E2 (h) | 0.16 ± 0.02 | 1.51 ± 0.02 |
| Number of G | 0.16 ± 0.01 | 0.72 ± 0.02 |
| Mean Duration of G (min) | 12.13 ± 0.02 | 11.35 ± 0.01 |
| Number of F | 0.21 ± 0.02 * | 0.91 ± 0.02 |
| Mean duration of F (min) | 3.11 ± 0.03 * | 2.12 ± 0.03 |
| Test Genes | Pathways | Forward Sequence (5′……3′) | Reverse Sequence (5′……3′) |
|---|---|---|---|
| SOLYC04g079730 (AOS) | JA | GCTACAATTCCCCTCGCATA | ACAGGTGGTGATGACGATGA |
| SOLYC01g006560 (LOX12) | JA | AGAGGCGGTTCAACAAAAGA | CAATCGCCAAAGGTCTCAAT |
| SOLYC04g054890 (ACX) | JA | GCAGCCTGCCTACAATTAGC | GCAAAATGGAATGCGTAGGT |
| SOLYC10g086180 (PAL2) | SA | GGATCCTTTGCAGAAACCAA | GCCACCATGTAAAGCCTTGT |
| SOLYC01g106620 (NPR1) | SA | TACTCAGGTGGTGTGGCGTA | TCAAAAGCCGGTTGATTTTC |
| SOLYC04g079890 (PR5) | SA | CTGGGCAATGAACAGGATTT | AAGCACTTTTGCAAGCCACT |
| Actin | GTGACGGGTGACGGAGAATT | GACACTAATGCGCCCGGTAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Javed, K.; Wang, Y.; Javed, H.; Wang, C.; Liu, C.; Huang, Y. Tomato Aphid (Aphis gossypii) Secreted Saliva Can Enhance Aphid Resistance by Upregulating Signaling Molecules in Tomato (Solanum lycopersicum). Int. J. Mol. Sci. 2023, 24, 12768. https://doi.org/10.3390/ijms241612768
Javed K, Wang Y, Javed H, Wang C, Liu C, Huang Y. Tomato Aphid (Aphis gossypii) Secreted Saliva Can Enhance Aphid Resistance by Upregulating Signaling Molecules in Tomato (Solanum lycopersicum). International Journal of Molecular Sciences. 2023; 24(16):12768. https://doi.org/10.3390/ijms241612768
Chicago/Turabian StyleJaved, Khadija, Yong Wang, Humayun Javed, Chen Wang, Chuang Liu, and Yuqian Huang. 2023. "Tomato Aphid (Aphis gossypii) Secreted Saliva Can Enhance Aphid Resistance by Upregulating Signaling Molecules in Tomato (Solanum lycopersicum)" International Journal of Molecular Sciences 24, no. 16: 12768. https://doi.org/10.3390/ijms241612768
APA StyleJaved, K., Wang, Y., Javed, H., Wang, C., Liu, C., & Huang, Y. (2023). Tomato Aphid (Aphis gossypii) Secreted Saliva Can Enhance Aphid Resistance by Upregulating Signaling Molecules in Tomato (Solanum lycopersicum). International Journal of Molecular Sciences, 24(16), 12768. https://doi.org/10.3390/ijms241612768







