Molecular Changes in the Brain of the Wintering Calidris pusilla in the Mangroves of the Amazon River Estuary
Abstract
1. Introduction
2. Results
2.1. Sequencing Assembly of Semipalmated Sandpiper Transcriptome
2.2. Gene Expression between Experimental Groups
2.3. Gene Ontology and Functional Analysis
3. Discussion
3.1. GABBR2 and ARHGEF9 Differential Expressed Genes in Wintering C. pusilla
3.2. Gene Differential Expressions Related with Metabolic Pathways, Glial Changes, Neurogenesis, and Anti-Virus Response in Recently Arrived and Premigratory C. pusilla
Concluding Remarks
4. Materials and Methods
4.1. Bird Sampling and Ethics Recommendations for the Use of Animals in Research
4.2. Transcardiac Perfusion with RNA Later and RNA-Sequencing
4.3. Filtering, Trimming, and Transcriptome Assembly
4.4. Differential Expression Discovery and Functional Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RA | Recent Arrived Birds |
| PM | Premigratory |
| DEGs | Differentially Expressed Genes |
| FDR | False Discovery Rate |
| Log2FC | Log2 Fold Change |
| GO | Gene Ontology |
| CC | Cellular Component |
| MF | Molecular Function |
| BP | Biological Process |
| IDs | Identifications |
| TPM | Transcript Per Million |
| TMM | Trimmed Mean of M values |
References
- Kimmitt, A.A. Females as the Gatekeepers to Seasonal Breeding: What We Can Learn by Studying Reproductive Mechanisms in Both Sexes. Integr. Comp. Biol. 2020, 60, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Lifjeld, J.T.; Kleven, O.; Fossoy, F.; Jacobsen, F.; Laskemoen, T.; Rudolfsen, G.; Robertson, R.J. When Older Males Sire More Offspring-Increased Attractiveness or Higher Fertility? Behav. Ecol. Sociobiol. 2022, 76, 61. [Google Scholar] [CrossRef]
- Yoshimura, T. Thyroid hormone and seasonal regulation of reproduction. Front. Neuroendocrinol. 2013, 34, 157–166. [Google Scholar] [CrossRef]
- Sharma, A.; Das, S.; Komal, R.; Malik, S.; Rani, S.; Kumar, V. Seasonal reproductive state determines gene expression in the hypothalamus of a latitudinal migratory songbird during the spring and autumn migration. Mol. Cell Endocrinol. 2020, 508, 110794. [Google Scholar] [CrossRef]
- Costa, J.S.; Hahn, S.; Araujo, P.M.; Dhanjal-Adams, K.L.; Rocha, A.D.; Alves, J.A. Linking migratory performance to breeding phenology and productivity in an Afro-Palearctic long-distance migrant. Sci. Rep. 2021, 11, 23258. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, A.K.; Kumar, J.; Rani, S.; Kumar, V. Annual life history-dependent gene expression in the hypothalamus and liver of a migratory songbird: Insights into the molecular regulation of seasonal metabolism. J. Biol. Rhythms 2014, 29, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Singh, D.; Malik, S.; Gupta, N.J.; Rani, S.; Kumar, V. Difference in control between spring and autumn migration in birds: Insight from seasonal changes in hypothalamic gene expression in captive buntings. Proc. Biol. Sci. 2018, 285, 20181531. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kumar, V. Metabolic plasticity mediates differential responses to spring and autumn migrations: Evidence from gene expression patterns in migratory buntings. Exp. Physiol. 2019, 104, 1841–1857. [Google Scholar] [CrossRef]
- Sharma, A.; Singh, D.; Gupta, P.; Bhardwaj, S.K.; Kaur, I.; Kumar, V. Molecular changes associated with migratory departure from wintering areas in obligate songbird migrants. J. Exp. Biol. 2021, 224, jeb242153. [Google Scholar] [CrossRef]
- Frias-Soler, R.C.; Kelsey, N.A.; Villarin Pildain, L.; Wink, M.; Bairlein, F. Transcriptome signature changes in the liver of a migratory passerine. Genomics 2022, 114, 110283. [Google Scholar] [CrossRef]
- Frias-Soler, R.C.; Pildain, L.V.; Parau, L.G.; Wink, M.; Bairlein, F. Transcriptome signatures in the brain of a migratory songbird. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 34, 100681. [Google Scholar] [CrossRef] [PubMed]
- Henrique, E.P.; de Oliveira, M.A.; Paulo, D.C.; Pereira, P.D.C.; Dias, C.; de Siqueira, L.S.; de Lima, C.M.; Miranda, D.A.; do Rego, P.S.; Araripe, J.; et al. Contrasting migratory journeys and changes in hippocampal astrocyte morphology in shorebirds. Eur. J. Neurosci. 2021, 54, 5687–5704. [Google Scholar] [CrossRef]
- de Almeida Miranda, D.; Araripe, J.; de Morais Magalhaes, N.G.; de Siqueira, L.S.; de Abreu, C.C.; Pereira, P.D.C.; Henrique, E.P.; da Silva Chira, P.A.C.; de Melo, M.A.D.; do Rego, P.S.; et al. Shorebirds’ Longer Migratory Distances Are Associated With Larger ADCYAP1 Microsatellites and Greater Morphological Complexity of Hippocampal Astrocytes. Front. Psychol. 2021, 12, 784372. [Google Scholar] [CrossRef] [PubMed]
- Carvalho-Paulo, D.; de Morais Magalhaes, N.G.; de Almeida Miranda, D.; Diniz, D.G.; Henrique, E.P.; Moraes, I.A.M.; Pereira, P.D.C.; de Melo, M.A.D.; de Lima, C.M.; de Oliveira, M.A.; et al. Hippocampal Astrocytes in Migrating and Wintering Semipalmated Sandpiper Calidris pusilla. Front. Neuroanat. 2017, 11, 126. [Google Scholar] [CrossRef]
- DeMoranville, K.J.; Corder, K.R.; Hamilton, A.; Russell, D.E.; Huss, J.M.; Schaeffer, P.J. PPAR expression, muscle size and metabolic rates across the gray catbird’s annual cycle are greatest in preparation for fall migration. J. Exp. Biol. 2019, 222 Pt 14, jeb198028. [Google Scholar] [CrossRef] [PubMed]
- Guglielmo, C.G. Obese super athletes: Fat-fueled migration in birds and bats. J. Exp. Biol. 2018, 221 (Suppl. S1), jeb165753. [Google Scholar] [CrossRef] [PubMed]
- Schmaljohann, H.; Eikenaar, C.; Sapir, N. Understanding the ecological and evolutionary function of stopover in migrating birds. Biol. Rev. Camb. Philos. Soc. 2022, 97, 1231–1252. [Google Scholar] [CrossRef]
- Catry, T.; Granadeiro, J.P.; Gutierrez, J.S.; Correia, E. Stopover use of a large estuarine wetland by dunlins during spring and autumn migrations: Linking local refuelling conditions to migratory strategies. PLoS ONE 2022, 17, e0263031. [Google Scholar] [CrossRef]
- Elowe, C.R.; Gerson, A.R. Migratory disposition alters lean mass dynamics and protein metabolism in migratory white-throated sparrows (Zonotrichia albicollis). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2022, 323, R98–R109. [Google Scholar] [CrossRef]
- Malik, S.; Singh, S.; Rani, S.; Kumar, V. Life at a different pace: Annual itineraries are conserved in seasonal songbirds. J. Biosci. 2014, 39, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Singh, D.; Das, S.; Kumar, V. Hypothalamic and liver transcriptome from two crucial life-history stages in a migratory songbird. Exp. Physiol. 2018, 103, 559–569. [Google Scholar] [CrossRef]
- Albert, S.; Wolfe, J.D.; Kellerman, J.; Sherry, T.; Stutchbury, B.J.M.; Bayly, N.J.; Ruiz-Sánchez, A. Habitat ecology of Nearctic–Neotropical migratory landbirds on the nonbreeding grounds. Condor 2020, 122, duaa055. [Google Scholar] [CrossRef]
- Gratto-Trevor, C.; Morrison, R.; Mizrahi, D.; Lank, D.; Hicklin, P.; Spaans, A. Migratory connectivity of Semipalmated Sandpipers: Winter distribution and migration routes of breeding populations. Waterbirds 2012, 35, 83–95. [Google Scholar] [CrossRef]
- Hicklin, P.; Gratto-Trevor, C.L.; Poole, A.F. Semipalmated Sandpiper (Calidris pusilla). The Birds of North. America Online. 2010. Available online: https://www.birds-of-north-america.net/Semipalmated_Sandpiper.html (accessed on 20 January 2023).
- Hicklin, P. The migration of shorebirds in the Bay of Fundy. Wilson Bull. 1987, 99, 540–570. [Google Scholar]
- Gwinner, E. Circadian and circannual programmes in avian migration. J. Exp. Biol. 1996, 199 Pt 1, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Haest, B.; Huppop, O.; Bairlein, F. The influence of weather on avian spring migration phenology: What, where and when? Glob. Chang. Biol. 2018, 24, 5769–5788. [Google Scholar] [CrossRef]
- Marra, P.P.; Francis, C.M.; Mulvihill, R.S.; Moore, F.R. The influence of climate on the timing and rate of spring bird migration. Oecologia 2005, 142, 307–315. [Google Scholar] [CrossRef]
- Horton, K.G.; Van Doren, B.M.; La Sorte, F.A.; Cohen, E.B.; Clipp, H.L.; Buler, J.J.; Fink, D.; Kelly, J.F.; Farnsworth, A. Holding steady: Little change in intensity or timing of bird migration over the Gulf of Mexico. Glob. Chang. Biol. 2019, 25, 1106–1118. [Google Scholar] [CrossRef]
- Huppop, O.; Huppop, K. North Atlantic Oscillation and timing of spring migration in birds. Proc. Biol. Sci. 2003, 270, 233–240. [Google Scholar] [CrossRef]
- Haest, B.; Huppop, O.; Bairlein, F. Weather at the winter and stopover areas determines spring migration onset, progress, and advancements in Afro-Palearctic migrant birds. Proc. Natl. Acad. Sci. USA 2020, 117, 17056–17062. [Google Scholar] [CrossRef]
- Brown, S. The Remarkable Odyssey of a Semipalmated Sandpiper. In Shorebird Science; Manomet Soaring Solutions Grounded Science: Coats Island, NU, Canada, 2014. [Google Scholar]
- Mata, A.; Marin, G.; Rodriguez, J.P.; Yurai Guerrero, H.; Cardillo, E. Plasma corticosterone levels of semipalmated sandpiper Calidris pusilla overwintering in a tropical coastal lagoon of northeastern Venezuela: Effect of capture and handling. Ann. N. Y. Acad. Sci. 2009, 1163, 460–463. [Google Scholar] [CrossRef]
- Larrazábal, M.; Azevedo-Júnior, S.d.; Pena, O. Monitoramento de aves limícolas na Salina diamante Branco, galinhos, Rio grande do Norte, Brasil. Rev. Bras. Zool. 2002, 19, 1081–1089. [Google Scholar] [CrossRef]
- Linhart, R.; Hamilton, D.; Paquet, J.; Monteiro, J.; Ramires, G.; Mobley, J. Movement and habitat use of non-breeding Semipalmated Sandpiper (Calidris pusilla) at the Banco dos Cajuais in Northeast Brazil. In Conservation Science and Practice; Wiley: Hoboken, NJ, USA, 2022; p. e12683. Available online: wileyonlinelibrary.com/journal/csp2 (accessed on 3 February 2022).
- Fedrizzi, C.E.; Azevedo Júnior, S.M.d.; Larrazábal, M.E.L.d. Body mass and acquisition of breeding plumage of wintering Calidris pusilla (Linnaeus) (Aves, Scolopacidae) in the coast of Pernambuco, north-eastern Brazil. Rev. Bras. Zool. 2004, 21, 249–252. [Google Scholar] [CrossRef]
- Buccitelli, C.; Selbach, M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 2020, 21, 630–644. [Google Scholar] [CrossRef]
- de Morais Magalhaes, N.G.; Guerreiro Diniz, C.; Guerreiro Diniz, D.; Pereira Henrique, E.; Correa Pereira, P.D.; Matos Moraes, I.A.; Damasceno de Melo, M.A.; Sherry, D.F.; Wanderley Picanco Diniz, C. Hippocampal neurogenesis and volume in migrating and wintering semipalmated sandpipers (Calidris pusilla). PLoS ONE 2017, 12, e0179134. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, V.; Kraft, L.; Mesny, F.; Rigerte, L. A simple guide to de novo transcriptome assembly and annotation. Brief. Bioinform. 2022, 23, bbab563. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- McDermaid, A.; Monier, B.; Zhao, J.; Liu, B.; Ma, Q. Interpretation of differential gene expression results of RNA-seq data: Review and integration. Brief. Bioinform. 2019, 20, 2044–2054. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, M.C.; Konate, M.M.; Chen, L.; Das, B.; Karlovich, C.; Williams, P.M.; Evrard, Y.A.; Doroshow, J.H.; McShane, L.M. TPM, FPKM, or Normalized Counts? A Comparative Study of Quantification Measures for the Analysis of RNA-seq Data from the NCI Patient-Derived Models Repository. J. Transl. Med. 2021, 19, 269. [Google Scholar] [CrossRef]
- Barkan, S.; Yom-Tov, Y.; Barnea, A. Exploring the Relationship between Brain Plasticity, Migratory Lifestyle, and Social Structure in Birds. Front. Neurosci. 2017, 11, 139. [Google Scholar] [CrossRef]
- Rytova, V.; Ganella, D.E.; Hawkes, D.; Bathgate, R.A.D.; Ma, S.; Gundlach, A.L. Chronic activation of the relaxin-3 receptor on GABA neurons in rat ventral hippocampus promotes anxiety and social avoidance. Hippocampus 2019, 29, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; McLvor, G.E.; van der Vaart, K.; Vaughan, R.T.; Thornton, A.; Ouellette, N.T. Costs and benefits of social relationships in the collective motion of bird flocks. Nat. Ecol. Evol. 2019, 3, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, L.C.F.; Henrique, E.P.; da Silva Rosa, J.B.; Pereira, P.D.C.; de Abreu, C.C.; Fernandes, T.N.; de Morais Magalhaes, N.G.; de Jesus Falcao da Silva, A.; da Costa, E.R.; Guerreiro-Diniz, C.; et al. Plasticity in the hippocampal formation of shorebirds during the wintering period: Stereological analysis of parvalbumin neurons in Actitis macularius. Learn. Behav. 2022, 50, 45–54. [Google Scholar] [CrossRef]
- Donato, F.; Rompani, S.B.; Caroni, P. Parvalbumin-expressing basket-cell network plasticity induced by experience regulates adult learning. Nature 2013, 504, 272–276. [Google Scholar] [CrossRef]
- Caroni, P. Regulation of Parvalbumin Basket cell plasticity in rule learning. Biochem. Biophys. Res. Commun. 2015, 460, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Caroni, P. Inhibitory microcircuit modules in hippocampal learning. Curr. Opin. Neurobiol. 2015, 35, 66–73. [Google Scholar] [CrossRef]
- Patenaude, C.; Chapman, C.A.; Bertrand, S.; Congar, P.; Lacaille, J.C. GABAB receptor- and metabotropic glutamate receptor-dependent cooperative long-term potentiation of rat hippocampal GABAA synaptic transmission. J. Physiol. 2003, 553 Pt 1, 155–167. [Google Scholar] [CrossRef]
- Patenaude, C.; Massicotte, G.; Lacaille, J.C. Cell-type specific GABA synaptic transmission and activity-dependent plasticity in rat hippocampal stratum radiatum interneurons. Eur. J. Neurosci. 2005, 22, 179–188. [Google Scholar] [CrossRef]
- Honore, E.; Khlaifia, A.; Bosson, A.; Lacaille, J.C. Hippocampal Somatostatin Interneurons, Long-Term Synaptic Plasticity and Memory. Front. Neural Circuits 2021, 15, 687558. [Google Scholar] [CrossRef]
- Topolnik, L.; Tamboli, S. The role of inhibitory circuits in hippocampal memory processing. Nat. Rev. Neurosci. 2022, 23, 476–492. [Google Scholar] [CrossRef]
- Chamberland, S.; Topolnik, L. Inhibitory control of hippocampal inhibitory neurons. Front. Neurosci. 2012, 6, 165. [Google Scholar] [CrossRef]
- Mouritsen, H.; Heyers, D.; Gunturkun, O. The Neural Basis of Long-Distance Navigation in Birds. Annu. Rev. Physiol. 2016, 78, 133–154. [Google Scholar] [CrossRef]
- Tyagi, T.; Bhardwaj, S.K. Magnetic Compass Orientation in a Palaearctic-Indian Night Migrant, the Red-Headed Bunting. Animals 2021, 11, 1541. [Google Scholar] [CrossRef] [PubMed]
- Gronroos, J.; Muheim, R.; Akesson, S. Orientation and autumn migration routes of juvenile sharp-tailed sandpipers at a staging site in Alaska. J. Exp. Biol. 2010, 213, 1829–1835. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, T.; Eulenburg, V.; Reddy-Alla, S.; Mansuy, I.M.; Li, Y.; Betz, H. Collybistin is required for both the formation and maintenance of GABAergic postsynapses in the hippocampus. Mol. Cell Neurosci. 2008, 39, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Ibaraki, K.; Mizuno, M.; Aoki, H.; Niwa, A.; Iwamoto, I.; Hara, A.; Tabata, H.; Ito, H.; Nagata, K.I. Biochemical and Morphological Characterization of a Guanine Nucleotide Exchange Factor ARHGEF9 in Mouse Tissues. Acta Histochem. Cytochem. 2018, 51, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Chiou, T.T.; Bonhomme, B.; Jin, H.; Miralles, C.P.; Xiao, H.; Fu, Z.; Harvey, R.J.; Harvey, K.; Vicini, S.; De Blas, A.L. Differential regulation of the postsynaptic clustering of gamma-aminobutyric acid type A (GABAA) receptors by collybistin isoforms. J. Biol. Chem. 2011, 286, 22456–22468. [Google Scholar] [CrossRef] [PubMed]
- Smart, T.G.; Stephenson, F.A. A half century of gamma-aminobutyric acid. Brain Neurosci. Adv. 2019, 3, 2398212819858249. [Google Scholar] [CrossRef]
- Agrawal, J.; Dwivedi, Y. GABA(A) Receptor Subunit Transcriptional Regulation, Expression Organization, and Mediated Calmodulin Signaling in Prefrontal Cortex of Rats Showing Testosterone-Mediated Impulsive Behavior. Front. Neurosci. 2020, 14, 600099. [Google Scholar] [CrossRef]
- Blank, M.H.; de Oliveira, M.J.; Cubas, Z.S.; de Morae, W.; Moreira, N.; Pereira, R.J.G. Fecal sex steroids and reproductive behaviors in harpy eagles (Harpia harpyja). Zoo Biol. 2020, 39, 315–324. [Google Scholar] [CrossRef]
- Landys, M.M.; Piersma, T.; Guglielmo, C.G.; Jukema, J.; Ramenofsky, M.; Wingfield, J.C. Metabolic profile of long–distance migratory flight and stopover in a shorebird. Proc. R. Soc. B Biol. Sci. 2005, 272, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Guglielmo, C.G. Move that fatty acid: Fuel selection and transport in migratory birds and bats. Integr. Comp. Biol. 2010, 50, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, G.M.; Jiang, L.; Rothman, D.L.; Behar, K.L. The contribution of ketone bodies to basal and activity-dependent neuronal oxidation in vivo. J. Cereb. Blood Flow. Metab. 2014, 34, 1233–1242. [Google Scholar] [CrossRef] [PubMed]
- Achanta, L.B.; Rae, C.D. β-Hydroxybutyrate in the Brain: One Molecule, Multiple Mechanisms. Neurochem. Res. 2017, 42, 35–49. [Google Scholar] [CrossRef]
- Brister, J.R.; Ako-adjei, D.; Bao, Y.; Blinkova, O. NCBI Viral Genomes Resource. Nucleic Acids Res. 2015, 43, D571–D577. [Google Scholar] [CrossRef]
- Pereira, P.D.C. Genes inflammatory response, tolerance, and resistance to virus infections in migratory birds, bats, and rodents. Front. Immunol. 2023; under review. [Google Scholar]
- Eikenaar, C.; Hegemann, A.; Packmor, F.; Kleudgen, I.; Isaksson, C. Not just fuel: Energy stores are correlated with immune function and oxidative damage in a long-distance migrant. Curr. Zool. 2020, 66, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Schneider, D.S.; Soares, M.P. Disease tolerance as a defense strategy. Science 2012, 335, 936–941. [Google Scholar] [CrossRef]
- Rivas, F.V.; Chervonsky, A.V.; Medzhitov, R. ART and immunology. Trends Immunol. 2014, 35, 451. [Google Scholar] [CrossRef]
- Raberg, L.; Graham, A.L.; Read, A.F. Decomposing health: Tolerance and resistance to parasites in animals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 37–49. [Google Scholar] [CrossRef]
- Zhong, C.; Liu, F.; Hajnik, R.J.; Yao, L.; Chen, K.; Wang, M.; Liang, Y.; Sun, J.; Soong, L.; Hou, W.; et al. Type I Interferon Promotes Humoral Immunity in Viral Vector Vaccination. J. Virol. 2021, 95, e0092521. [Google Scholar] [CrossRef] [PubMed]
- de Weerd, N.A.; Nguyen, T. The interferons and their receptors--distribution and regulation. Immunol. Cell Biol. 2012, 90, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Tachedjian, M.; Wynne, J.W.; Boyd, V.; Cui, J.; Smith, I.; Cowled, C.; Ng, J.H.; Mok, L.; Michalski, W.P.; et al. Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc. Natl. Acad. Sci. USA 2016, 113, 2696–2701. [Google Scholar] [CrossRef]
- Carty, M.; Guy, C.; Bowie, A.G. Detection of Viral Infections by Innate Immunity. Biochem. Pharmacol. 2021, 183, 114316. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.; Anderson, D.E.; Zhang, Q.; Tan, C.W.; Lim, B.L.; Luko, K.; Wen, M.; Chia, W.N.; Mani, S.; Wang, L.C.; et al. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat. Microbiol. 2019, 4, 789–799. [Google Scholar] [CrossRef]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef]
- Vrentas, C.E.; Schaut, R.G.; Boggiatto, P.M.; Olsen, S.C.; Sutterwala, F.S.; Moayeri, M. Inflammasomes in livestock and wildlife: Insights into the intersection of pathogens and natural host species. Vet. Immunol. Immunopathol. 2018, 201, 49–56. [Google Scholar] [CrossRef] [PubMed]
- FastQC: A Quality Control Tool for High Throughput Sequence Data. 2016. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 30 June 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium, T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, D61–D65. [Google Scholar] [CrossRef]
- Suzek, B.E.; Huang, H.; McGarvey, P.; Mazumder, R.; Wu, C.H. UniRef: Comprehensive and non-redundant UniProt reference clusters. Bioinformatics 2007, 23, 1282–1288. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2019, 47, D94–D99. [Google Scholar] [CrossRef] [PubMed]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Frias-Soler, R.C.; Villarin Pildain, L.; Hotz-Wagenblatt, A.; Kolibius, J.; Bairlein, F.; Wink, M. De novo annotation of the transcriptome of the Northern Wheatear (Oenanthe oenanthe). PeerJ 2018, 6, e5860. [Google Scholar] [CrossRef]
- Guigo, R.; de Hoon, M. Recent advances in functional genome analysis. F1000Research 2018, 7, 1968. [Google Scholar] [CrossRef] [PubMed]
- Colquitt, B.M. Organizational Conservation and Flexibility in the Evolution of Birdsong and Avian Motor Control. Brain Behav. Evol. 2022, 97, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Prather, J.F.; Okanoya, K.; Bolhuis, J.J. Brains for birds and babies: Neural parallels between birdsong and speech acquisition. Neurosci. Biobehav. Rev. 2017, 81 Pt B, 225–237. [Google Scholar] [CrossRef]
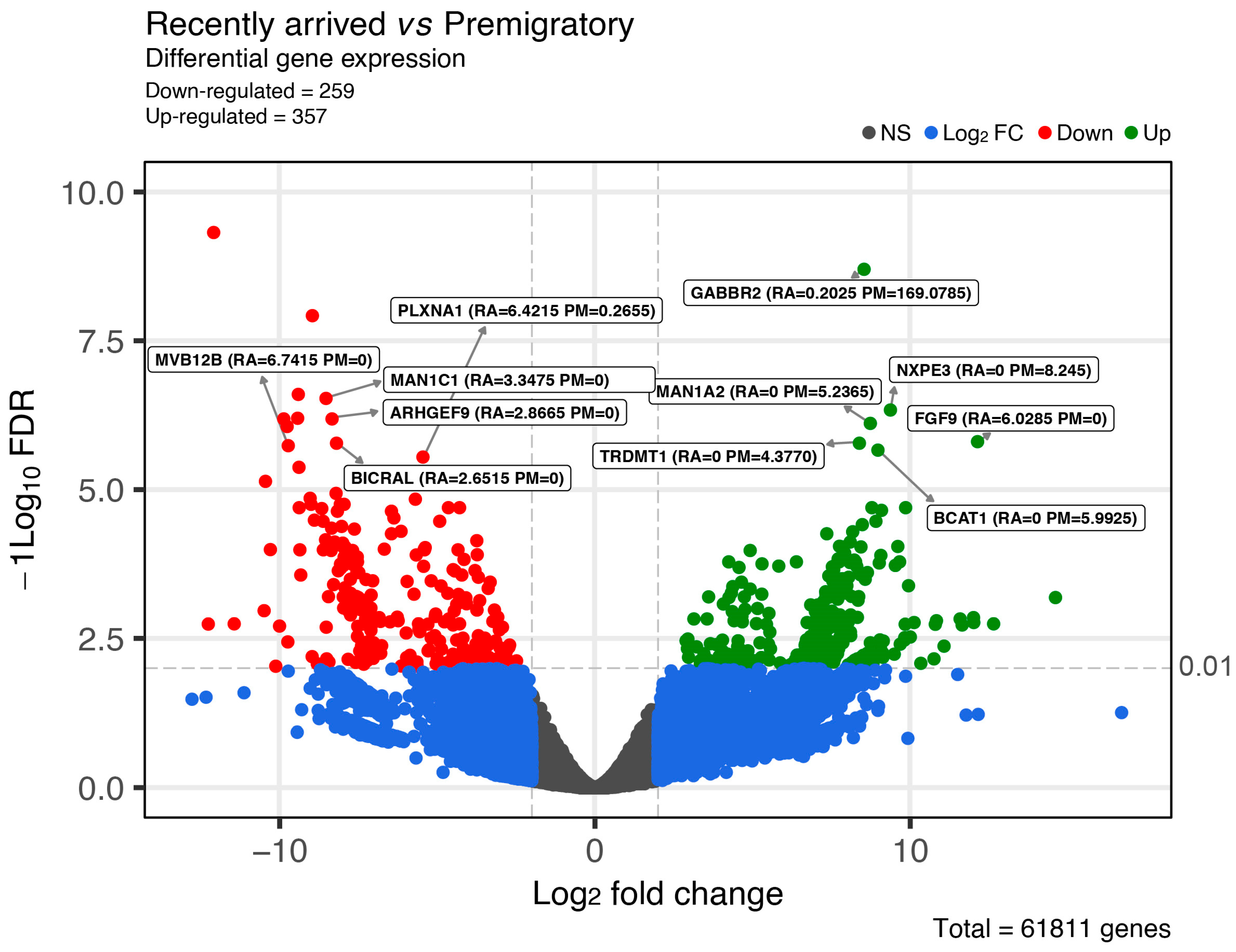
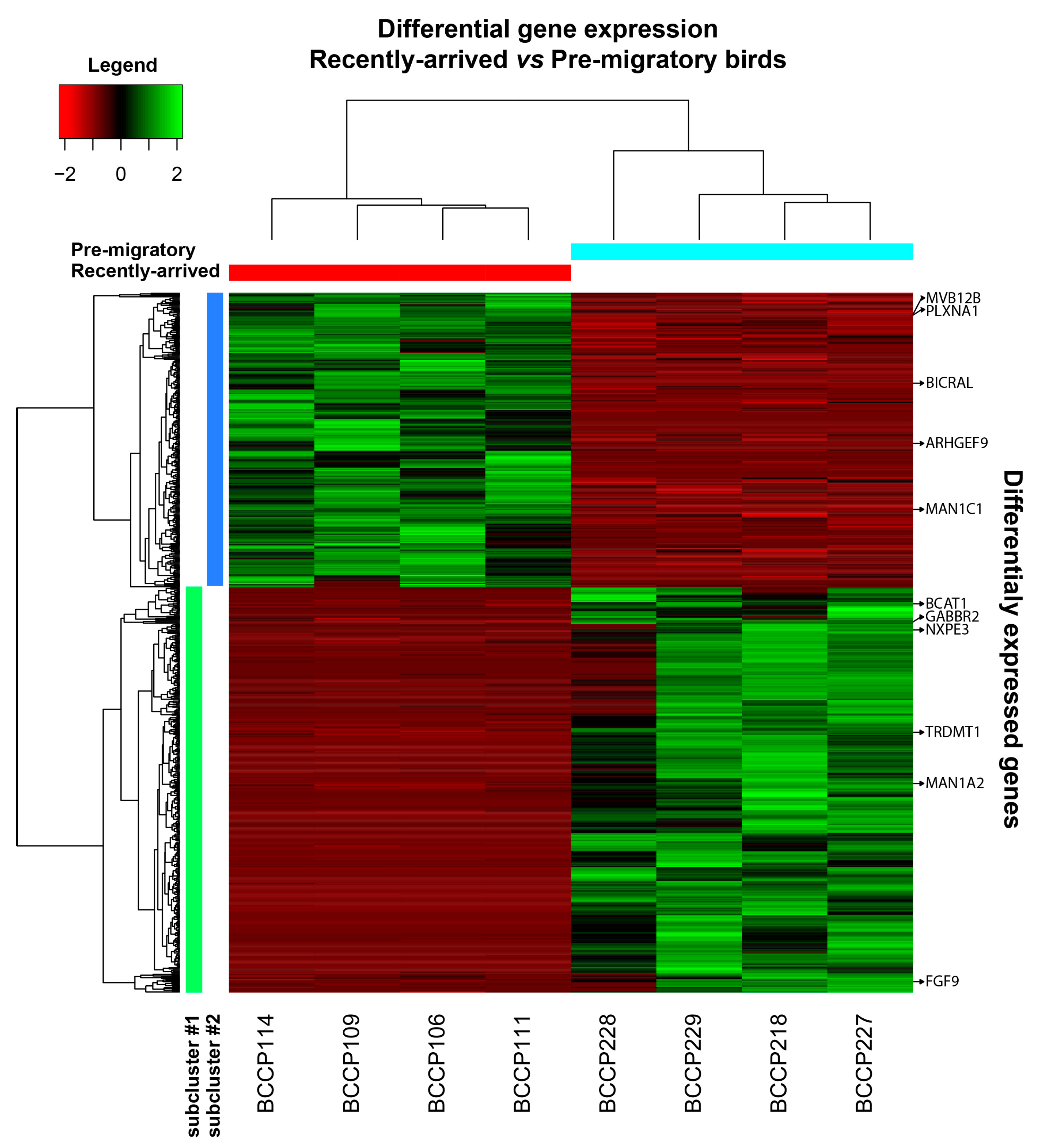
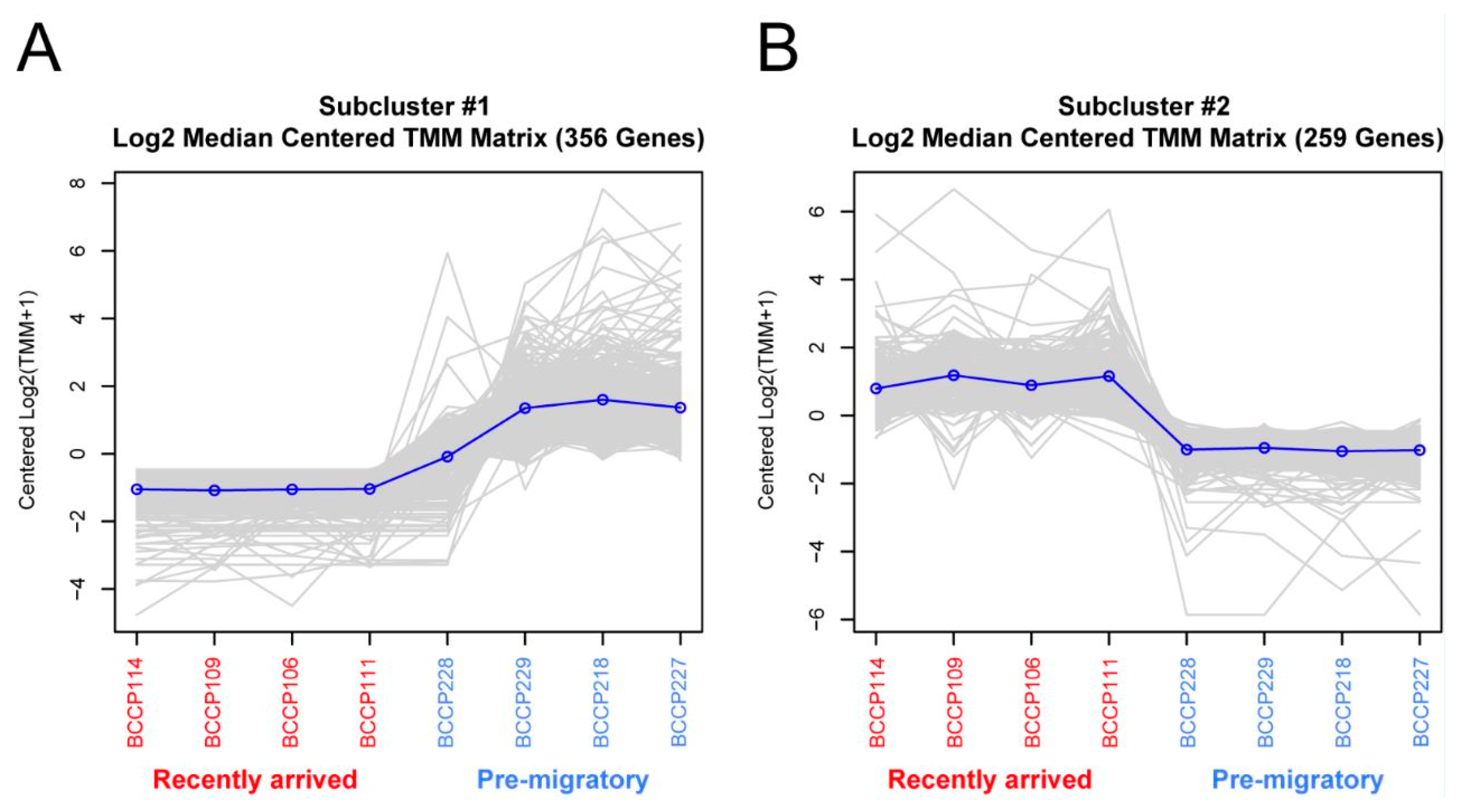
| Sequence ID | Gene Symbol | GO IDs | GO Names |
|---|---|---|---|
| Up-Regulated Genes | |||
| TRINITY_DN40544_c0_g1 | GABBR2 | GO:0007186; GO:0007214; | G protein-coupled receptor signaling pathway; Gamma-aminobutyric acid signaling pathway; |
| TRINITY_DN3467_c1_g1 | MAN1A2 | GO:0005975; GO:0006491; | Carbohydrate metabolic process; N-glycan processing; |
| TRINITY_DN41739_c0_g1 | BCAT1 | GO:0009082; GO:0009098; GO:0009099; | Branched-chain amino acid biosynthetic process; Leucine biosynthetic process; Valine biosynthetic process; |
| TRINITY_DN56325_c0_g1 | NXPE3 | GO:0008150; | Encode a member of neurexophilin family of neuropeptide-like glycoproteins |
| TRINITY_DN62709_c0_g1 | FGF9 | GO:0000122; GO:0001525; GO:0001649; GO:0001654; GO:0001934; GO:0002053; GO:0002062; GO:0006606; GO:0008543; GO:0008584; GO:0010628; GO:0030178; GO:0030238; GO:0030326; GO:0030334; GO:0030949; GO:0032927; GO:0042472; GO:0045880; GO:0048505; GO:0048566; GO:0048706; GO:0050679; GO:0051781; GO:0060045; GO:0060484; GO:0090263; GO:1904707; | Negative regulation of transcription by RNA polymerase II; Angiogenesis; Osteoblast differentiation; Eye development; Positive regulation of protein phosphorylation; Positive regulation of mesenchymal cell proliferation; Chondrocyte differentiation; Protein import into nucleus; Fibroblast growth factor receptor signaling pathway; Male gonad development; Positive regulation of gene expression; Negative regulation of Wnt signaling pathway; Male sex determination; Embryonic limb morphogenesis; Regulation of cell migration; Positive regulation of vascular endothelial growth factor receptor signaling pathway; Positive regulation of activin receptor signaling pathway; Inner ear morphogenesis; positive regulation of smoothened signaling pathway; Regulation of timing of cell differentiation; Embryonic digestive tract development; Embryonic skeletal system development; Positive regulation of epithelial cell proliferation; Positive regulation of cell division; Positive regulation of cardiac muscle cell proliferation; Lung-associated mesenchyme development; Positive regulation of canonical Wnt signaling pathway; Positive regulation of vascular associated smooth muscle cell proliferation; |
| TRINITY_DN147173_c0_g1 | TRDMT1 | GO:0030488; GO:0036416; | tRNA methylation; A stabilization; |
| Downregulated Genes | |||
| TRINITY_DN1477_c0_g1 | PLXNA1 | GO:0007162; GO:0008360; GO:0030334; GO:0043087; GO:0050772; GO:1902287 | Negative regulation of cell adhesion; Regulation of cell shape; Regulation of cell migration; Regulation of GTPase activity; Positive regulation of axonogenesis; Semaphorin-plexin signaling pathway involved in axon guidance; |
| TRINITY_DN161848_c0_g1 | BICRAL | GO:0045893; | Positive regulation of DNA-templated transcription; |
| TRINITY_DN51404_c0_g1 | ARHGEF9 | GO:0050790; | Regulation of catalytic activity; |
| TRINITY_DN123969_c0_g1 | MVB12B | GO:0015031; GO:0019075; GO:0042058; GO:0046755; | Protein transport; Virus maturation; Regulation of epidermal growth factor receptor signaling pathway; Viral budding; |
| TRINITY_DN13053_c4_g1 | MAN1C1 | GO:0005975; GO:0006491; GO:0004571; GO:0005509; GO:0000139; GO:0005783; GO:0016020 | Carbohydrate metabolic process; N-glycan processing; Mannosyl-oligosaccharide 1,2-α-mannosidase activity; Calcium ion binding; Golgi membrane; Endoplasmic reticulum; membrane |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, P.D.C.; Henrique, E.P.; da Costa, E.R.; Falcão, A.d.J.; de Melo, M.A.D.; Schneider, M.P.C.; Burbano, R.M.R.; Diniz, D.G.; Magalhães, N.G.d.M.; Sherry, D.F.; et al. Molecular Changes in the Brain of the Wintering Calidris pusilla in the Mangroves of the Amazon River Estuary. Int. J. Mol. Sci. 2023, 24, 12712. https://doi.org/10.3390/ijms241612712
Pereira PDC, Henrique EP, da Costa ER, Falcão AdJ, de Melo MAD, Schneider MPC, Burbano RMR, Diniz DG, Magalhães NGdM, Sherry DF, et al. Molecular Changes in the Brain of the Wintering Calidris pusilla in the Mangroves of the Amazon River Estuary. International Journal of Molecular Sciences. 2023; 24(16):12712. https://doi.org/10.3390/ijms241612712
Chicago/Turabian StylePereira, Patrick Douglas Corrêa, Ediely Pereira Henrique, Emanuel Ramos da Costa, Anderson de Jesus Falcão, Mauro André Damasceno de Melo, Maria Paula Cruz Schneider, Rommel Mario Rodriguez Burbano, Daniel Guerreiro Diniz, Nara Gyzely de Morais Magalhães, David Francis Sherry, and et al. 2023. "Molecular Changes in the Brain of the Wintering Calidris pusilla in the Mangroves of the Amazon River Estuary" International Journal of Molecular Sciences 24, no. 16: 12712. https://doi.org/10.3390/ijms241612712
APA StylePereira, P. D. C., Henrique, E. P., da Costa, E. R., Falcão, A. d. J., de Melo, M. A. D., Schneider, M. P. C., Burbano, R. M. R., Diniz, D. G., Magalhães, N. G. d. M., Sherry, D. F., Diniz, C. W. P., & Guerreiro-Diniz, C. (2023). Molecular Changes in the Brain of the Wintering Calidris pusilla in the Mangroves of the Amazon River Estuary. International Journal of Molecular Sciences, 24(16), 12712. https://doi.org/10.3390/ijms241612712








