Robust ParB Binding to Half-parS Sites in Pseudomonas aeruginosa—A Mechanism for Retaining ParB on the Nucleoid?
Abstract
1. Introduction
2. Results
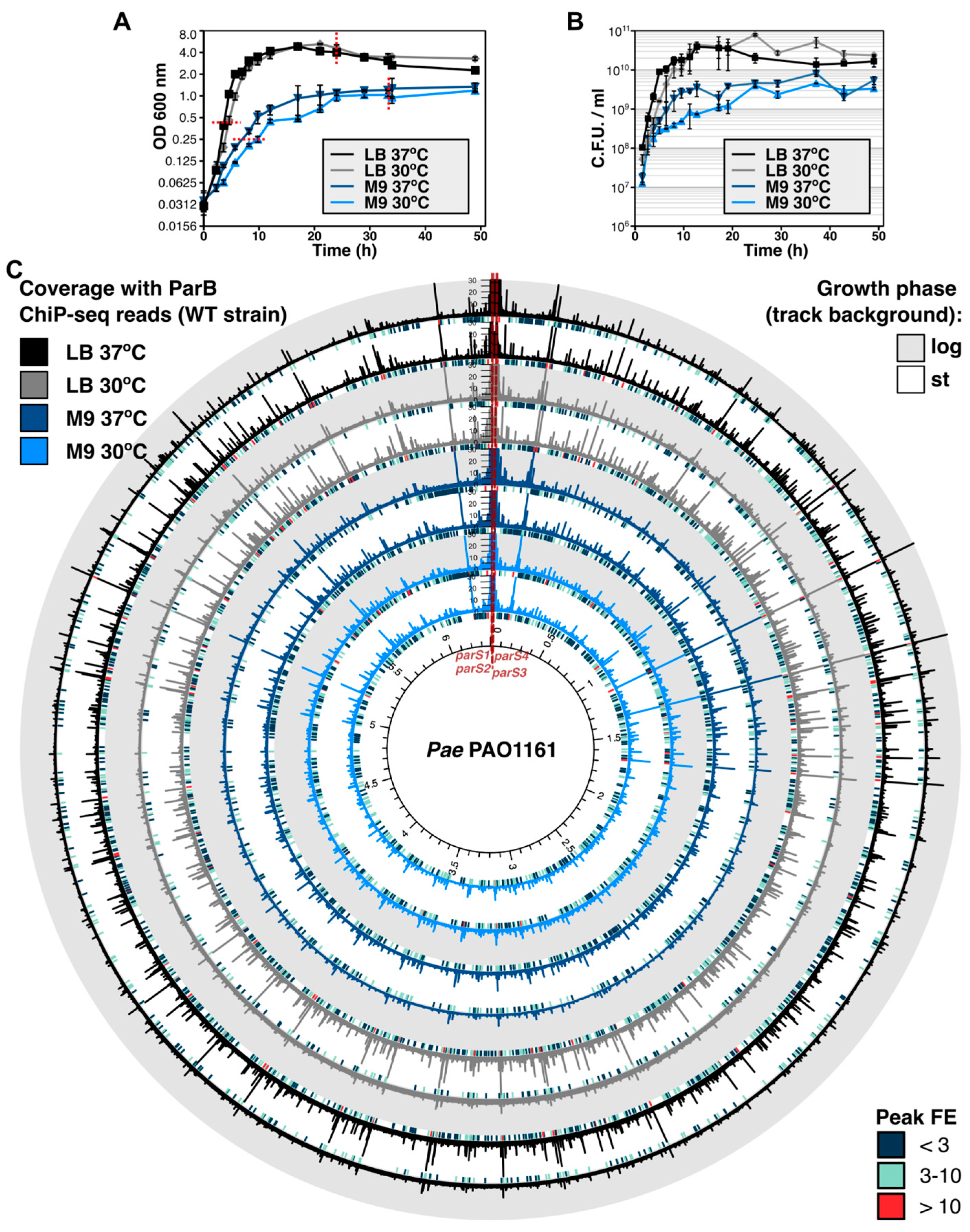
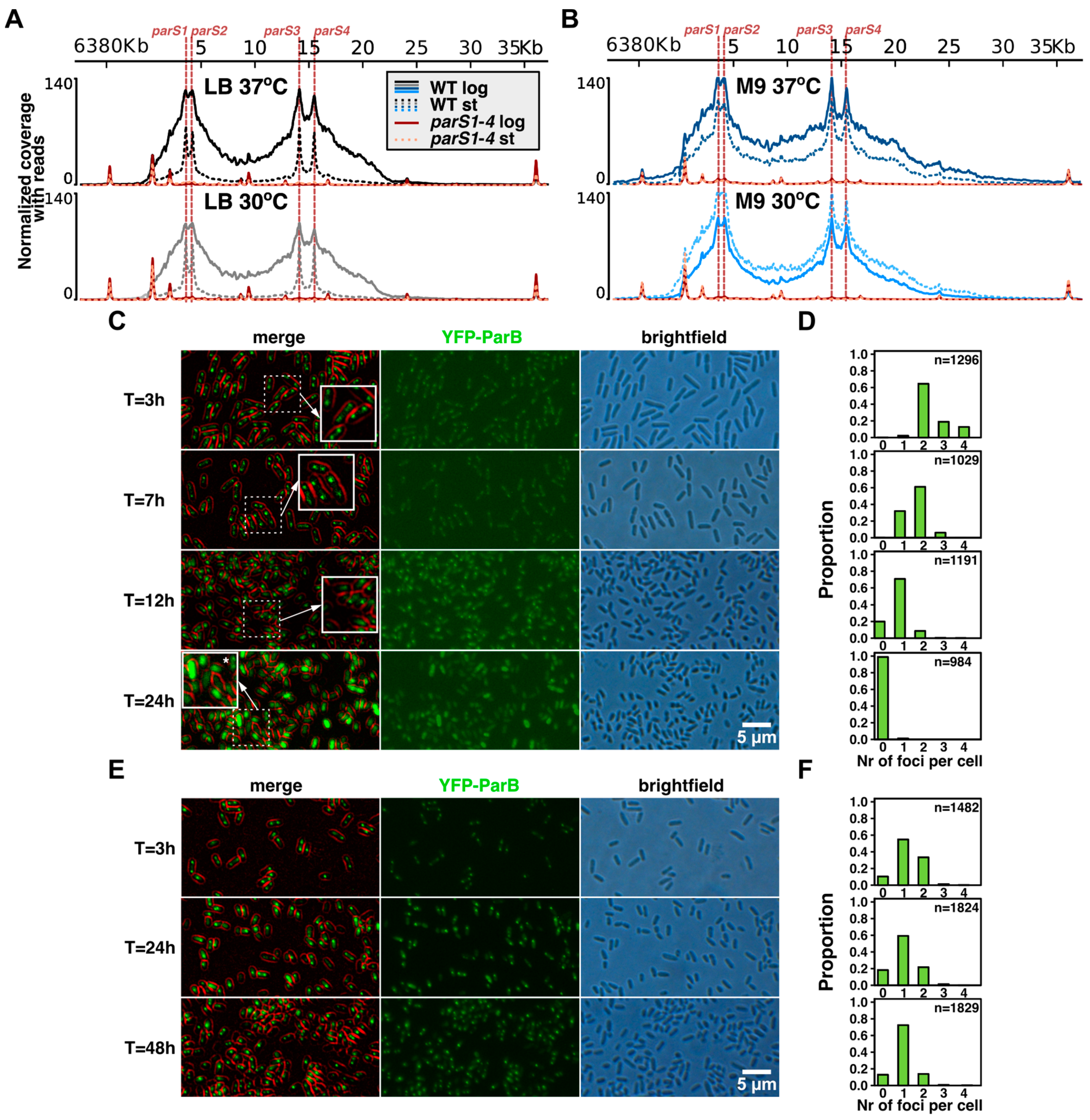
2.1. Distribution of ParB on Genome in Cells from Exponential and Stationary Phases of Cultures under Conditions of Varying Cell Division Time
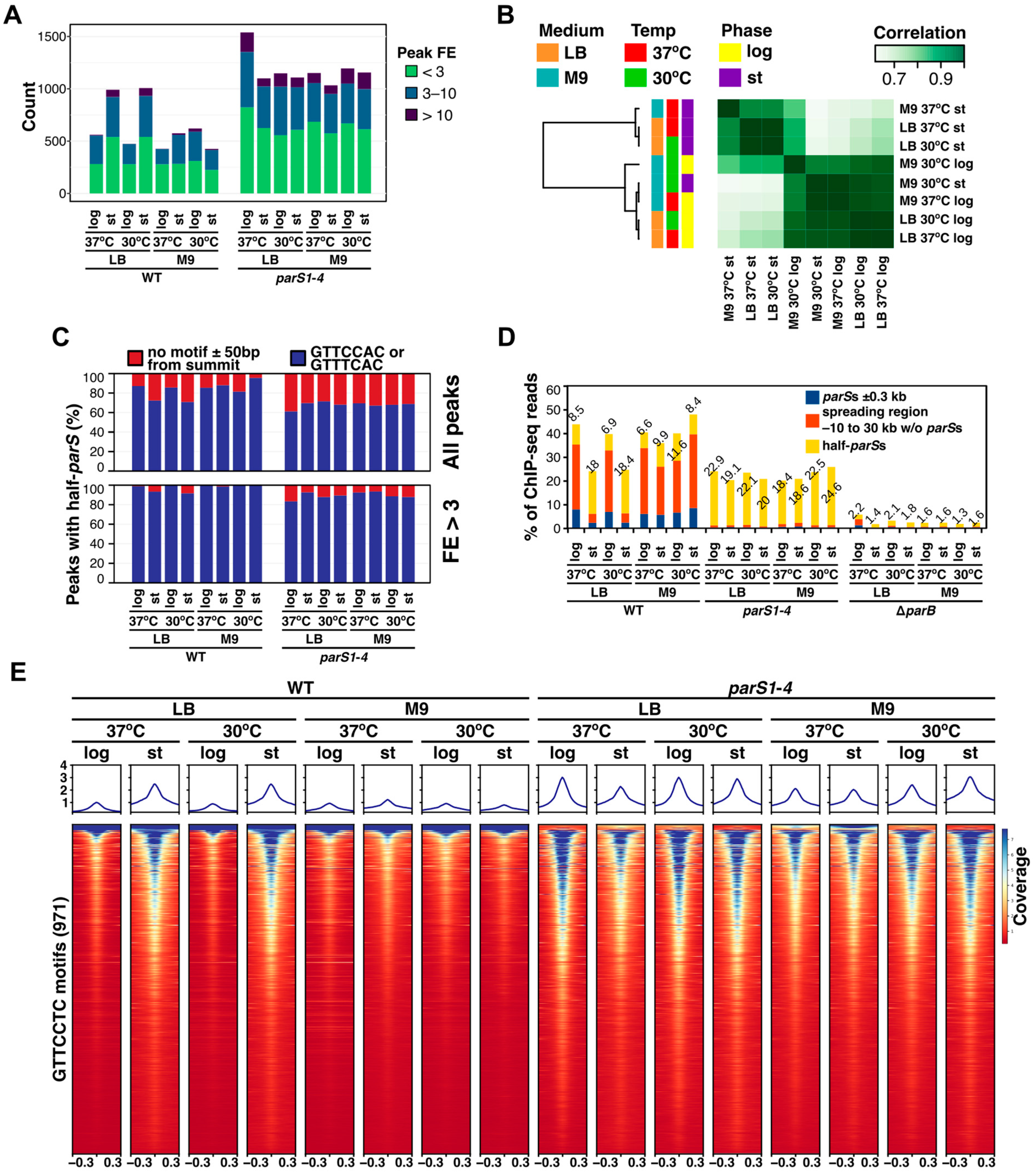
2.2. Growth Conditions Affect the Extent of ParB Binding to Half-parSs but Not the Individual Half-parS Preference
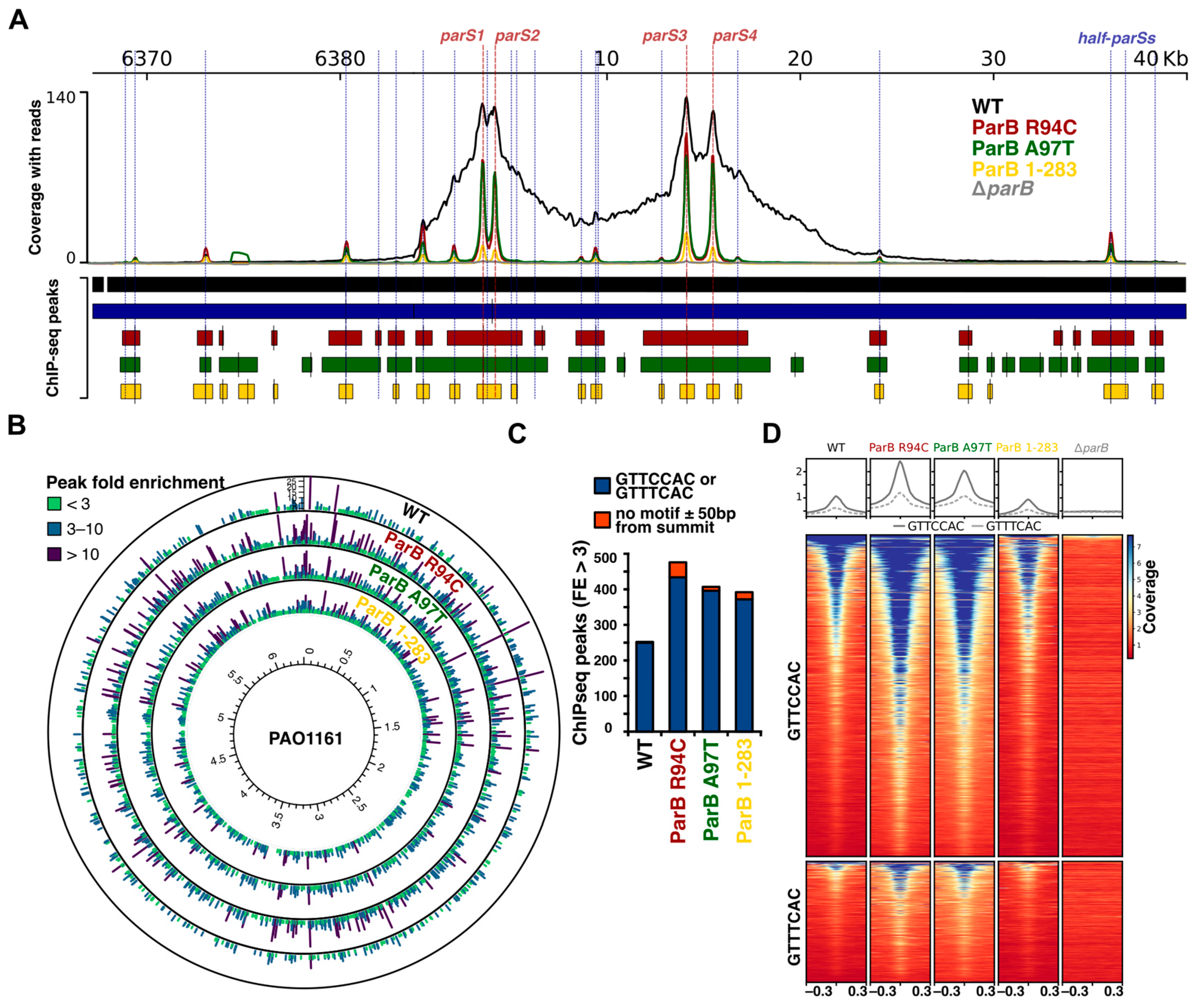
2.3. Mutations in the Conserved Arginine Patch and C-Terminal Dimerization Domain of Pae ParB Impair Its Spreading Ability but Not Binding to Half-parSs
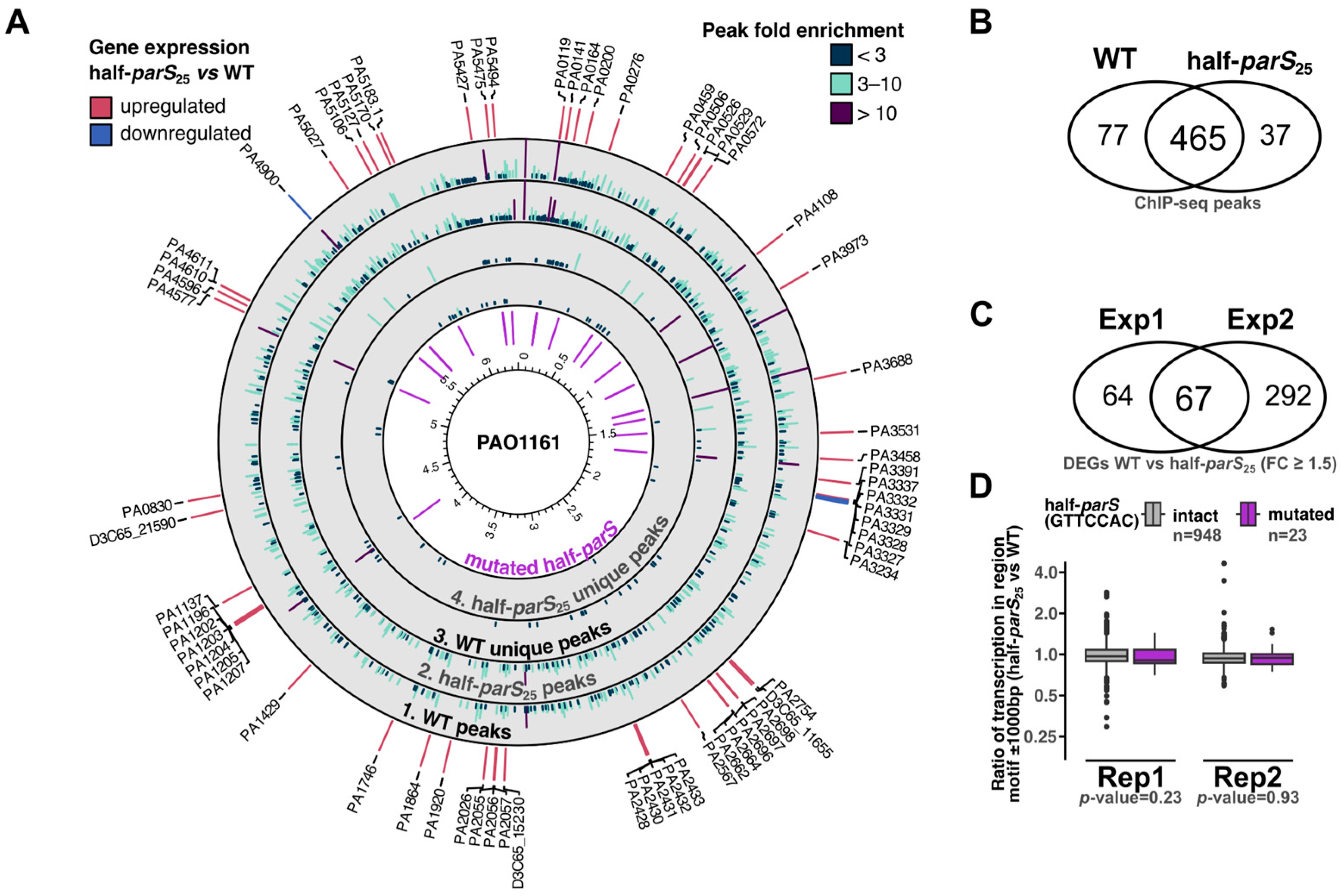
2.4. Effect of Half-parSs Removal on ParB Genome Distribution and Gene Expression
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. ChIP-seq
4.3. RNA-seq
4.4. Fluorescence Microscopy and Image Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reyes-Lamothe, R.; Sherratt, D.J. The bacterial cell cycle, chromosome inheritance and cell growth. Nat. Rev. Microbiol. 2019, 17, 467–478. [Google Scholar] [CrossRef]
- Badrinarayanan, A.; Le, T.B.K.; Laub, M.T. Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 2015, 31, 171–199. [Google Scholar] [CrossRef]
- den Blaauwen, T. Prokaryotic cell division: Flexible and diverse. Curr. Opin. Microbiol. 2013, 16, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Bouet, J.-Y.; Stouf, M.; Lebailly, E.; Cornet, F. Mechanisms for chromosome segregation. Curr. Opin. Microbiol. 2014, 22, 60–65. [Google Scholar] [CrossRef]
- Kawalek, A.; Wawrzyniak, P.; Bartosik, A.A.; Jagura-Burdzy, G. Rules and exceptions: The role of chromosomal ParB in DNA segregation and other cellular processes. Microorganisms 2020, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Marczynski, G.T.; Petit, K.; Patel, P. Crosstalk regulation between bacterial chromosome replication and chromosome partitioning. Front. Microbiol. 2019, 10, 279. [Google Scholar] [CrossRef]
- Surovtsev, I.V.; Jacobs-Wagner, C. Subcellular organization: A critical feature of bacterial cell replication. Cell 2018, 172, 1271–1293. [Google Scholar] [CrossRef]
- Lioy, V.S.; Junier, I.; Boccard, F. Multiscale dynamic structuring of bacterial chromosomes. Annu. Rev. Microbiol. 2021, 75, 541–561. [Google Scholar] [CrossRef]
- Livny, J.; Yamaichi, Y.; Waldor, M.K. Distribution of centromere-like parS sites in bacteria: Insights from comparative genomics. J. Bacteriol. 2007, 189, 8693–8703. [Google Scholar] [CrossRef]
- Funnell, B.E. ParB partition proteins: Complex formation and spreading at bacterial and plasmid centromeres. Front. Mol. Biosci. 2016, 3, 44. [Google Scholar] [CrossRef]
- Jalal, A.S.B.; Le, T.B.K. Bacterial chromosome segregation by the ParABS system. Open Biol. 2021, 10, 200097. [Google Scholar] [CrossRef] [PubMed]
- Soh, Y.-M.; Davidson, I.F.; Zamuner, S.; Basquin, J.; Bock, F.P.; Taschner, M.; Veening, J.-W.; Rios, P.D.L.; Peters, J.-M.; Gruber, S. Self-organization of parS centromeres by the ParB CTP hydrolase. Science 2019, 366, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Osorio-Valeriano, M.; Altegoer, F.; Steinchen, W.; Urban, S.; Liu, Y.; Bange, G.; Thanbichler, M. ParB-type DNA segregation proteins are CTP-dependent molecular switches. Cell 2019, 179, 1512–1524.e15. [Google Scholar] [CrossRef] [PubMed]
- Osorio-Valeriano, M.; Altegoer, F.; Das, C.K.; Steinchen, W.; Panis, G.; Connolley, L.; Giacomelli, G.; Feddersen, H.; Corrales-Guerrero, L.; Giammarinaro, P.I.; et al. The CTPase activity of ParB determines the size and dynamics of prokaryotic DNA partition complexes. Mol. Cell 2021, 81, 3992–4007.e10. [Google Scholar] [CrossRef] [PubMed]
- Jalal, A.S.; Tran, N.T.; Stevenson, C.E.; Chimthanawala, A.; Badrinarayanan, A.; Lawson, D.M.; Le, T.B. A CTP-dependent gating mechanism enables ParB spreading on DNA. eLife 2021, 10, e69676. [Google Scholar] [CrossRef] [PubMed]
- McLean, T.C.; Le, T.B. CTP switches in ParABS-mediated bacterial chromosome segregation and beyond. Curr. Opin. Microbiol. 2023, 73, 102289. [Google Scholar] [CrossRef]
- Mishra, D.; Srinivasan, R. Catching a Walker in the act—DNA partitioning by ParA family of proteins. Front. Microbiol. 2022, 13, 856547. [Google Scholar] [CrossRef]
- Lutkenhaus, J. The ParA/MinD family puts things in their place. Trends Microbiol. 2012, 20, 411–418. [Google Scholar] [CrossRef]
- Errington, J. Septation and chromosome segregation during sporulation in Bacillus subtilis. Curr. Opin. Microbiol. 2001, 4, 660–666. [Google Scholar] [CrossRef]
- Flärdh, K.; Buttner, M.J. Streptomyces morphogenetics: Dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 2009, 7, 36–49. [Google Scholar] [CrossRef]
- Wang, X.; Llopis, P.M.; Rudner, D.Z. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc. Natl. Acad. Sci. USA 2014, 111, 12877–12882. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Llopis, P.M.; Rudner, D.Z. Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 2013, 14, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Szafran, M.J.; Jakimowicz, D.; Elliot, M.A. Compaction and control—The role of chromosome-organizing proteins in Streptomyces. FEMS Microbiol. Rev. 2020, 44, 725–739. [Google Scholar] [CrossRef]
- Pióro, M.; Jakimowicz, D. Chromosome segregation proteins as coordinators of cell cycle in response to environmental conditions. Front. Microbiol. 2020, 11, 588. [Google Scholar] [CrossRef] [PubMed]
- Lioy, V.S.; Junier, I.; Lagage, V.; Vallet, I.; Boccard, F. Distinct activities of bacterial condensins for chromosome management in Pseudomonas aeruginosa. Cell Rep. 2020, 33, 108344. [Google Scholar] [CrossRef] [PubMed]
- Vallet-Gely, I.; Boccard, F. Chromosomal organization and segregation in Pseudomonas aeruginosa. PLoS Genet. 2013, 9, e1003492. [Google Scholar] [CrossRef]
- Bartosik, A.A.; Lasocki, K.; Mierzejewska, J.; Thomas, C.M.; Jagura-Burdzy, G. ParB of Pseudomonas aeruginosa: Interactions with its partner ParA and its target parS and specific effects on bacterial growth. J. Bacteriol. 2004, 186, 6983–6998. [Google Scholar] [CrossRef]
- Lasocki, K.; Bartosik, A.A.; Mierzejewska, J.; Thomas, C.M.; Jagura-Burdzy, G. Deletion of the parA (soj) homologue in Pseudomonas aeruginosa causes ParB instability and affects growth rate, chromosome segregation, and motility. J. Bacteriol. 2007, 189, 5762–5772. [Google Scholar] [CrossRef]
- Bartosik, A.A.; Mierzejewska, J.; Thomas, C.M.; Jagura-Burdzy, G. ParB deficiency in Pseudomonas aeruginosa destabilizes the partner protein ParA and affects a variety of physiological parameters. Microbiol. Read. Engl. 2009, 155, 1080–1092. [Google Scholar] [CrossRef]
- Jecz, P.; Bartosik, A.A.; Glabski, K.; Jagura-Burdzy, G. A single parS sequence from the cluster of four sites closest to oriC is necessary and sufficient for proper chromosome segregation in Pseudomonas aeruginosa. PLoS ONE 2015, 10, e0120867. [Google Scholar] [CrossRef]
- Lagage, V.; Boccard, F.; Vallet-Gely, I. Regional control of chromosome segregation in Pseudomonas aeruginosa. PLoS Genet. 2016, 12, e1006428. [Google Scholar] [CrossRef] [PubMed]
- Kawalek, A.; Bartosik, A.A.; Glabski, K.; Jagura-Burdzy, G. Pseudomonas aeruginosa partitioning protein ParB acts as a nucleoid-associated protein binding to multiple copies of a parS-related motif. Nucleic Acids Res. 2018, 46, 4592–4606. [Google Scholar] [CrossRef] [PubMed]
- Bartosik, A.A.; Glabski, K.; Jecz, P.; Mikulska, S.; Fogtman, A.; Koblowska, M.; Jagura-Burdzy, G. Transcriptional profiling of parA and parB mutants in actively dividing cells of an opportunistic human pathogen Pseudomonas aeruginosa. PLoS ONE 2014, 9, e87276. [Google Scholar] [CrossRef] [PubMed]
- Kawalek, A.; Glabski, K.; Bartosik, A.A.; Wozniak, D.; Kusiak, M.; Gawor, J.; Zuchniewicz, K.; Jagura-Burdzy, G. Diverse partners of the partitioning ParB protein in Pseudomonas aeruginosa. Microbiol. Spectr. 2023, 11, e0428922. [Google Scholar] [CrossRef]
- Wang, J.D.; Levin, P.A. Metabolism, cell growth and the bacterial cell cycle. Nat. Rev. Microbiol. 2009, 7, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Alpers, K.; Vatareck, E.; Gröbe, L.; Müsken, M.; Scharfe, M.; Häussler, S.; Tomasch, J. Transcriptome dynamics of Pseudomonas aeruginosa during transition from overlapping to non-overlapping cell cycles. mSystems 2023, 8, e0113022. [Google Scholar] [CrossRef]
- Kawalek, A.; Kotecka, K.; Modrzejewska, M.; Gawor, J.; Jagura-Burdzy, G.; Bartosik, A.A. Genome sequence of Pseudomonas aeruginosa PAO1161, a PAO1 derivative with the ICEPae1161 integrative and conjugative element. BMC Genomics 2020, 21, 14. [Google Scholar] [CrossRef]
- Meisner, J.; Goldberg, J.B. The Escherichia coli rhaSR-prhaBAD Inducible promoter system allows tightly controlled gene expression over a wide range in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2016, 82, 6715–6727. [Google Scholar] [CrossRef]
- Lagendijk, E.L.; Validov, S.; Lamers, G.E.M.; De Weert, S.; Bloemberg, G.V. Genetic tools for tagging Gram-negative bacteria with mCherry for visualization in vitro and in natural habitats, biofilm and pathogenicity studies. FEMS Microbiol. Lett. 2010, 305, 81–90. [Google Scholar] [CrossRef]
- Modrzejewska, M.; Kawalek, A.; Bartosik, A.A. The LysR-Type transcriptional regulator BsrA (PA2121) controls vital metabolic pathways in Pseudomonas aeruginosa. mSystems 2021, 6, e0001521. [Google Scholar] [CrossRef]
- Kotecka, K.; Kawalek, A.; Kobylecki, K.; Bartosik, A.A. The MarR-type regulator PA3458 is involved in osmoadaptation control in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2021, 22, 3982. [Google Scholar] [CrossRef]
- Kusiak, M.; Gapczyńska, A.; Płochocka, D.; Thomas, C.M.; Jagura-Burdzy, G. Binding and spreading of ParB on DNA determine its biological function in Pseudomonas aeruginosa. J. Bacteriol. 2011, 193, 3342–3355. [Google Scholar] [CrossRef]
- Mierzejewska, J.; Bartosik, A.A.; Macioszek, M.; Płochocka, D.; Thomas, C.M.; Jagura-Burdzy, G. Identification of C-terminal hydrophobic residues important for dimerization and all known functions of ParB of Pseudomonas aeruginosa. Microbiol. Read. Engl. 2012, 158, 1183–1195. [Google Scholar] [CrossRef]
- Turner, K.H.; Vallet-Gely, I.; Dove, S.L. Epigenetic control of virulence gene expression in Pseudomonas aeruginosa by a LysR-type transcription regulator. PLoS Genet. 2009, 5, e1000779. [Google Scholar] [CrossRef] [PubMed]
- Rajput, A.; Tsunemoto, H.; Sastry, A.V.; Szubin, R.; Rychel, K.; Chauhan, S.M.; Pogliano, J.; Palsson, B.O. Advanced transcriptomic analysis reveals the role of efflux pumps and media composition in antibiotic responses of Pseudomonas aeruginosa. Nucleic Acids Res. 2022, 50, 9675–9688. [Google Scholar] [CrossRef]
- Kawalek, A.; Glabski, K.; Bartosik, A.A.; Fogtman, A.; Jagura-Burdzy, G. Increased ParB level affects expression of stress response, adaptation and virulence operons and potentiates repression of promoters adjacent to the high affinity binding sites parS3 and parS4 in Pseudomonas aeruginosa. PLoS ONE 2017, 12, e0181726. [Google Scholar] [CrossRef] [PubMed]
- Kotecka, K.; Kawalek, A.; Modrzejewska-Balcerek, M.; Gawor, J.; Zuchniewicz, K.; Gromadka, R.; Bartosik, A.A. Functional characterization of TetR-like transcriptional regulator PA3973 from Pseudomonas aeruginosa. Int. J. Mol. Sci. 2022, 23, 14584. [Google Scholar] [CrossRef]
- Lundgren, B.R.; Bailey, F.J.; Moley, G.; Nomura, C.T. DdaR (PA1196) regulates expression of dimethylarginine dimethylaminohydrolase for the metabolism of methylarginines in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2017, 199, e0000117. [Google Scholar] [CrossRef] [PubMed]
- Baxter, J.C.; Funnell, B.E. Plasmid partition mechanisms. Microbiol. Spec. 2014, 2, 6. [Google Scholar] [CrossRef]
- Taylor, J.A.; Pastrana, C.L.; Butterer, A.; Pernstich, C.; Gwynn, E.J.; Sobott, F.; Moreno-Herrero, F.; Dillingham, M.S. Specific and non-specific interactions of ParB with DNA: Implications for chromosome segregation. Nucleic Acids Res. 2015, 43, 719–731. [Google Scholar] [CrossRef]
- Madariaga-Marcos, J.; Pastrana, C.L.; Fisher, G.L.; Dillingham, M.S.; Moreno-Herrero, F. ParB dynamics and the critical role of the CTD in DNA condensation unveiled by combined force-fluorescence measurements. eLife 2019, 8, e43812. [Google Scholar] [CrossRef] [PubMed]
- Guilhas, B.; Walter, J.C.; Rech, J.; David, G.; Walliser, N.O.; Palmeri, J.; Mathieu-Demaziere, C.; Parmeggiani, A.; Bouet, J.-Y.; Le Gall, A.; et al. ATP-driven separation of liquid phase condensates in bacteria. Mol. Cell 2020, 79, 293–303.e4. [Google Scholar] [CrossRef] [PubMed]
- Antar, H.; Soh, Y.-M.; Zamuner, S.; Bock, F.P.; Anchimiuk, A.; Rios, P.D.L.; Gruber, S. Relief of ParB autoinhibition by parS DNA catalysis and recycling of ParB by CTP hydrolysis promote bacterial centromere assembly. Sci. Adv. 2022, 7, eabj2854. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbour, NY, USA, 1989. [Google Scholar]
- El-Sayed, A.K.; Hothersall, J.; Thomas, C.M. Quorum-sensing-dependent regulation of biosynthesis of the polyketide antibiotic mupirocin in Pseudomonas fluorescens NCIMB 10586. Microbiol Read Engl. 2001, 147, 2127–2139. [Google Scholar] [CrossRef]
- Deatherage, D.E.; Barrick, J.E. Identification of mutations in laboratory evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014, 1151, 165–188. [Google Scholar]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Schulz, S.; Häussler, S. Chromatin immunoprecipitation for ChIP-chip and ChIP-seq. Methods Mol. Biol. 2014, 1149, 591–605. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Ramírez, F.; Ryan, D.P.; Grüning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dündar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based Analysis of ChIP-Seq [MACS]. Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Delisle, L.; Rabbani, L.; Wolff, J.; Bhardwaj, V.; Backofen, R.; Grüning, B.; Ramírez, F.; Manke, T. pyGenomeTracks: Reproducible plots for multivariate genomic datasets. Bioinformatics 2021, 37, 422–423. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.J.; Gazin, C.; Lawson, N.D.; Pagès, H.; Lin, S.M.; Lapointe, D.S.; Green, M.R. ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 2010, 11, 237. [Google Scholar] [CrossRef]
- Ross-Innes, C.S.; Stark, R.; Teschendorff, A.E.; Holmes, K.A.; Ali, H.R.; Dunning, M.J.; Brown, G.D.; Gojis, O.; Ellis, I.O.; Green, A.R.; et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012, 481, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Ducret, A.; Quardokus, E.M.; Brun, Y.V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 2016, 1, 16077. [Google Scholar] [CrossRef]

| Strain Name | Description/Relevant Genotype | Origin |
|---|---|---|
| Escherichia coli | ||
| DH5α | F− Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoA supE44 λ thi-1 gyrA96 relA | [57] |
| S17-1 | pro ΔhsdR hsdM+ recA TpR SmR ΩRP4-Tc::Mu Kn::Tn7 | [58] |
| Pseudomonas aeruginosa | ||
| PAO1161 (WT) | RifR, leu-, PAO1 derivative | [28,37] |
| ∆parB | PAO1161 with parB deletion | [34] |
| parS1–4 | PAO1161 with mutations in parS1, parS2, parS3 and parS4 impairing ParB binding; termed parSmut15 in the original paper | [30] |
| parB94 | PAO1161 producing ParB R94C, non-spreading variant | [42] |
| parB97 | PAO1161 producing ParB T97A, non-spreading variant | [42] |
| parB Δ284-290 | PAO1161 producing truncated ParB 1-283 variant | [43] |
| half-parS25 | PAO1161 with 25 half-parS sites modified in 18 loci | this work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawalek, A.; Bartosik, A.A.; Jagura-Burdzy, G. Robust ParB Binding to Half-parS Sites in Pseudomonas aeruginosa—A Mechanism for Retaining ParB on the Nucleoid? Int. J. Mol. Sci. 2023, 24, 12517. https://doi.org/10.3390/ijms241512517
Kawalek A, Bartosik AA, Jagura-Burdzy G. Robust ParB Binding to Half-parS Sites in Pseudomonas aeruginosa—A Mechanism for Retaining ParB on the Nucleoid? International Journal of Molecular Sciences. 2023; 24(15):12517. https://doi.org/10.3390/ijms241512517
Chicago/Turabian StyleKawalek, Adam, Aneta Agnieszka Bartosik, and Grazyna Jagura-Burdzy. 2023. "Robust ParB Binding to Half-parS Sites in Pseudomonas aeruginosa—A Mechanism for Retaining ParB on the Nucleoid?" International Journal of Molecular Sciences 24, no. 15: 12517. https://doi.org/10.3390/ijms241512517
APA StyleKawalek, A., Bartosik, A. A., & Jagura-Burdzy, G. (2023). Robust ParB Binding to Half-parS Sites in Pseudomonas aeruginosa—A Mechanism for Retaining ParB on the Nucleoid? International Journal of Molecular Sciences, 24(15), 12517. https://doi.org/10.3390/ijms241512517






