WD Repeat and HMG Box DNA Binding Protein 1: An Oncoprotein at the Hub of Tumorigenesis and a Novel Therapeutic Target
Abstract
1. Introduction
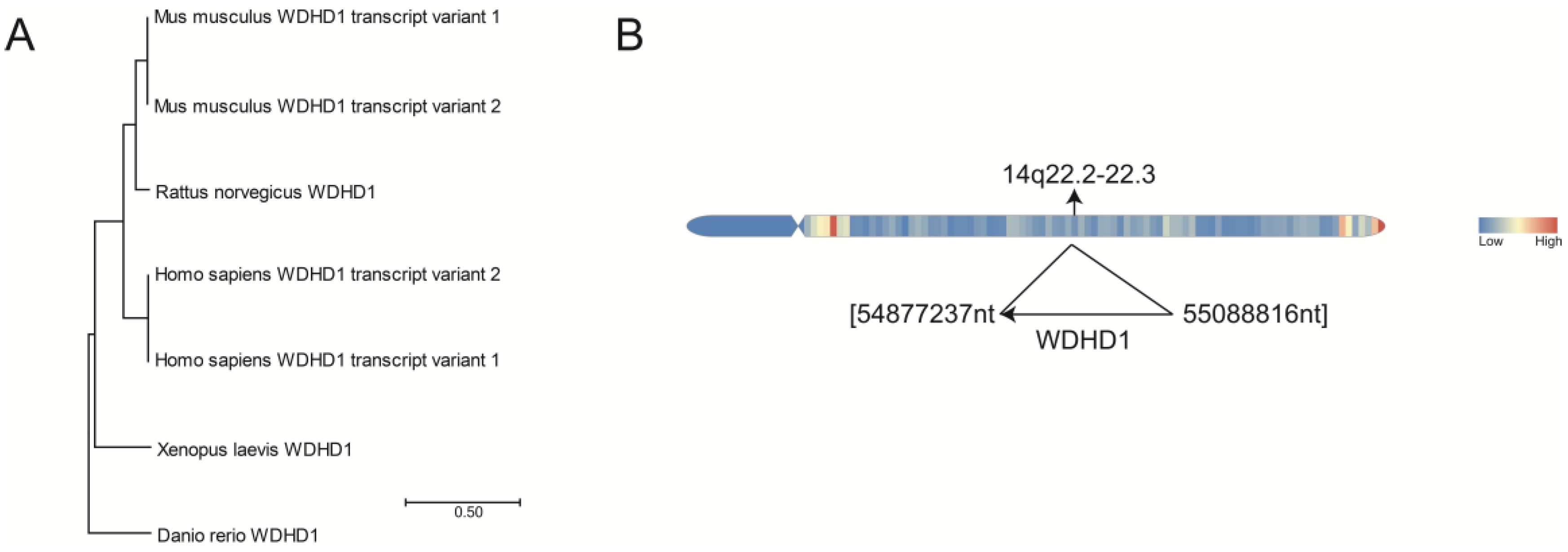
2. Evolutionary Conservation of WDHD1
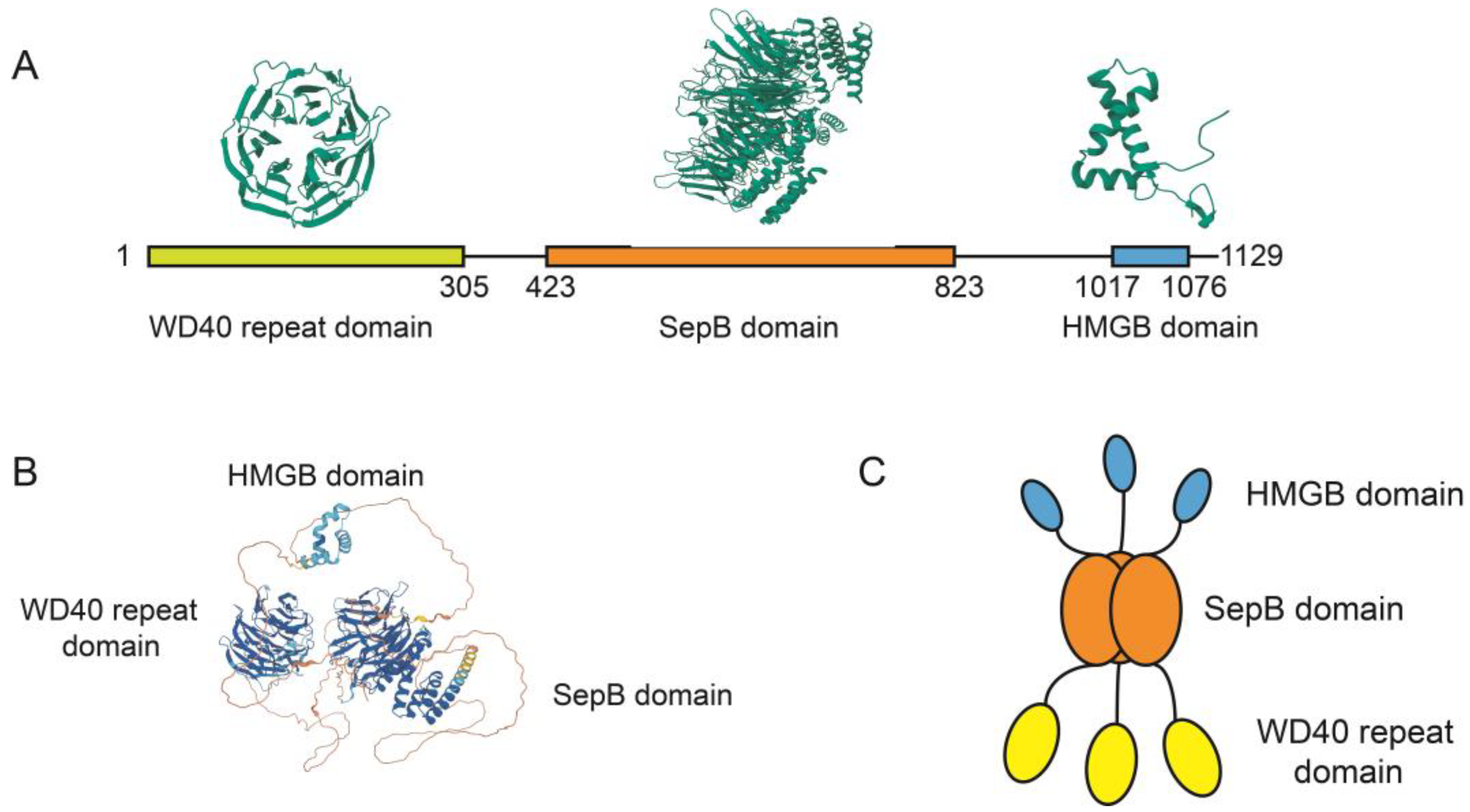
3. The Molecular Structure of WDHD1
4. Biological Functions of WDHD1
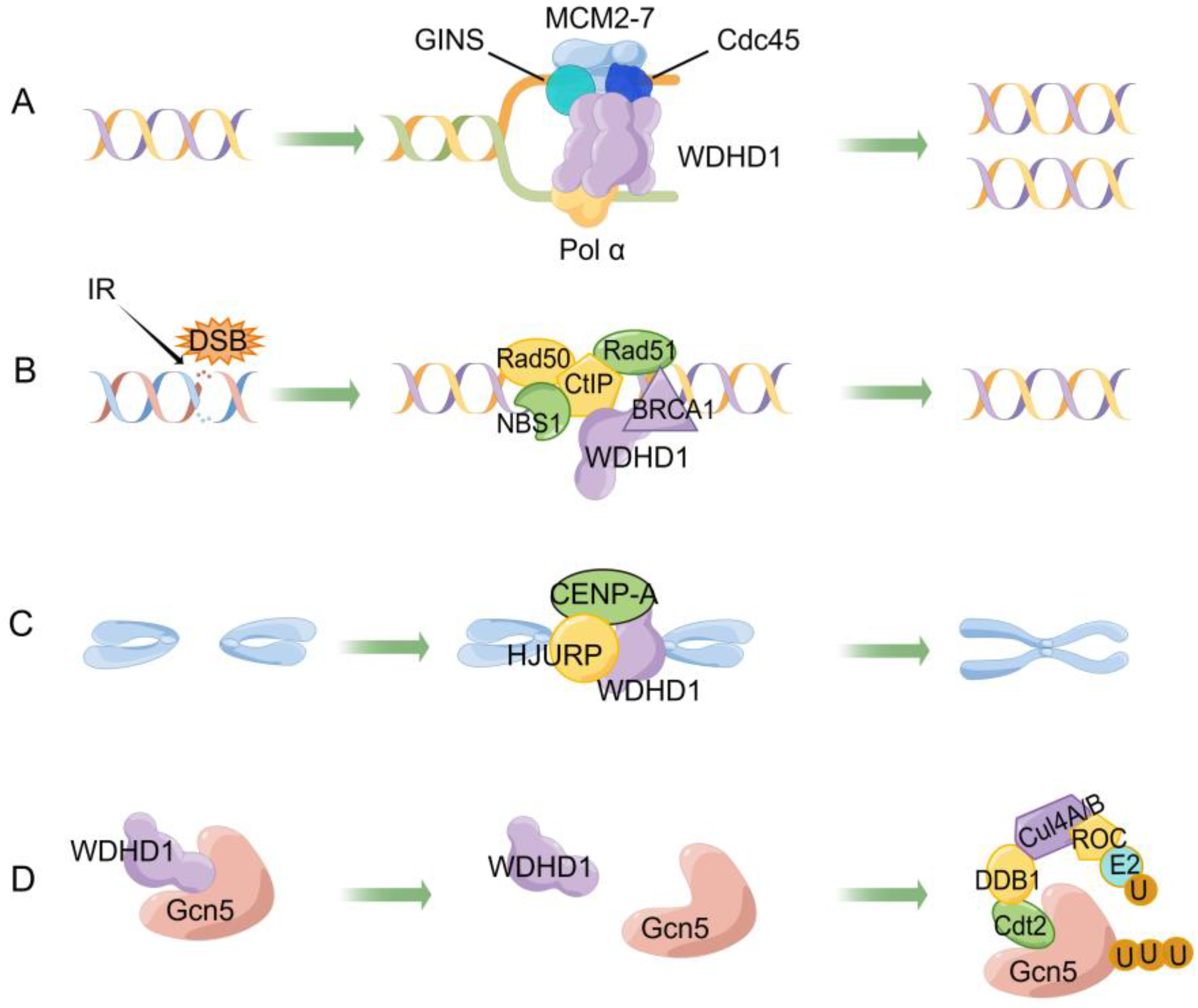
4.1. DNA Replication
4.2. Cell Cycle
4.3. DNA Damage Repair
4.4. Embryogenesis
4.5. Centromeric and Sister Chromatid
4.6. Histone Acetylation
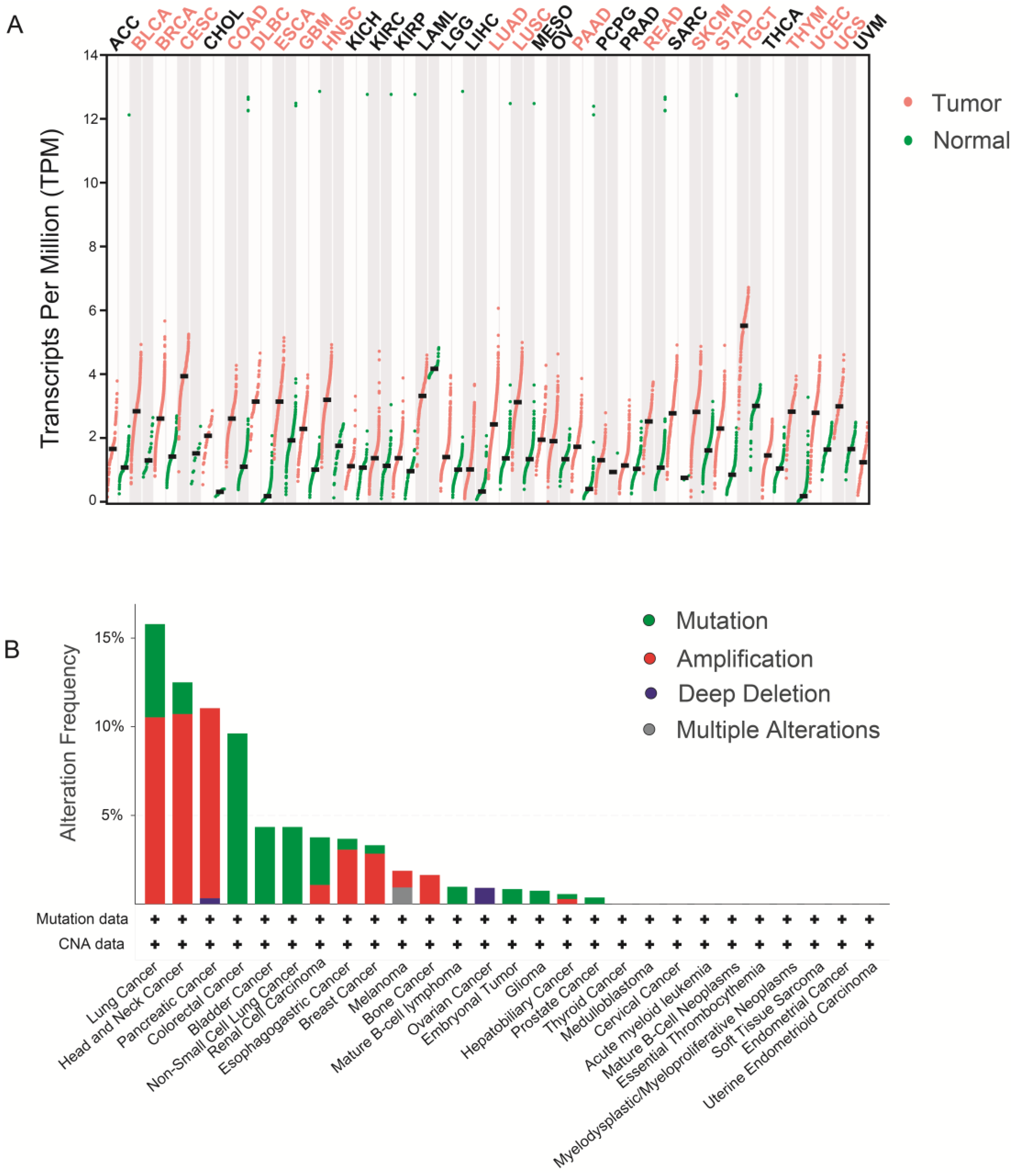
5. WDHD1 and Cancer
5.1. WDHD1 and High-Risk HPV
5.2. WDHD1 and Nasopharyngeal Carcinoma
5.3. WDHD1 and Laryngeal Cancer
5.4. WDHD1 and Esophageal Cancer
5.5. WDHD1 and Lung Cancer
5.6. WDHD1 and Breast Cancer
5.7. WDHD1 and Hepatocellular Carcinoma
5.8. WDHD1 and Cholangiocarcinoma
5.9. WDHD1 and Pancreatic Adenocarcinoma
5.10. WDHD1 and Cervical Cancer
5.11. WDHD1 and Ovarian Cancer
5.12. WDHD1 and Acute Myeloid Leukemia
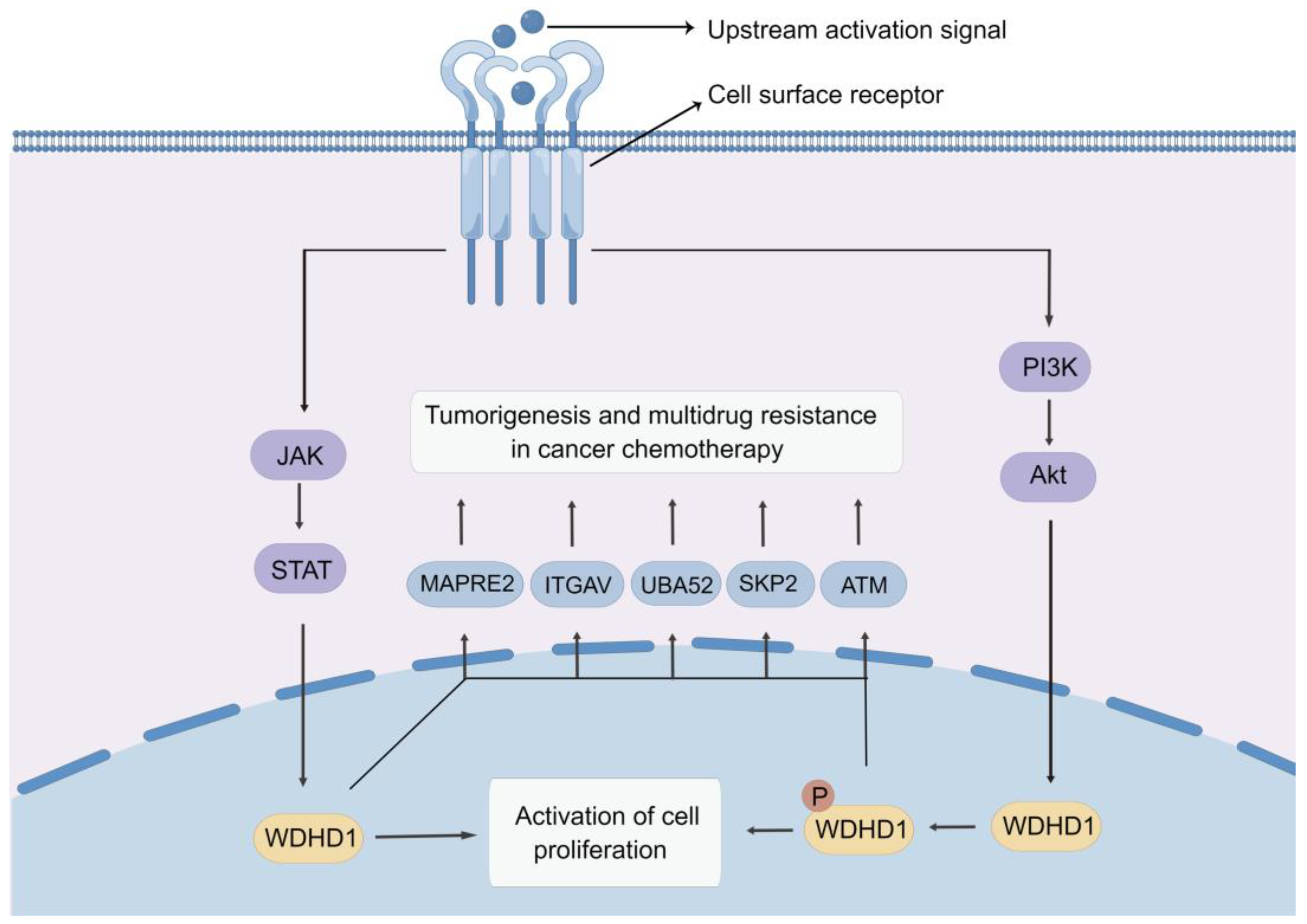
6. Molecular Mechanisms of Action of WDHD1
6.1. PI3K/AKT Pathway
6.2. JAK-STAT Signaling Pathway
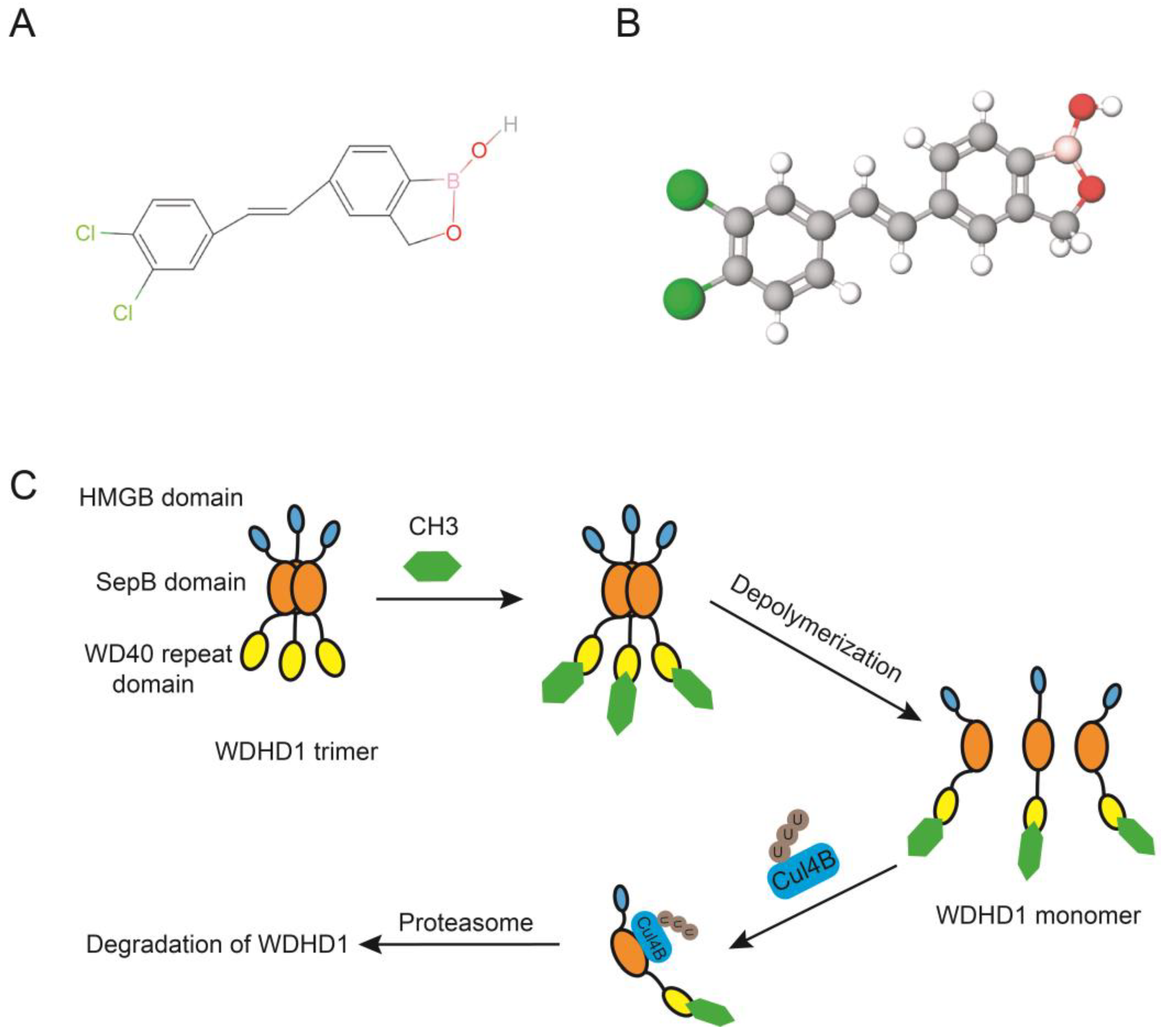
7. Inhibitors of WDHD1
8. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kouprina, N.; Kroll, E.; Bannikov, V.; Bliskovsky, V.; Gizatullin, R.; Kirillov, A.; Shestopalov, B.; Zakharyev, V.; Hieter, P.; Spencer, F.; et al. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1992, 12, 5736–5747. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Schmidt-Zachmann, M.S.; Franke, W.W. AND-1, a natural chimeric DNA-binding protein, combines an HMG-box with regulatory WD-repeats. J. Cell Sci. 1997, 110 Pt 9, 1051–1062. [Google Scholar] [CrossRef]
- Zhu, W.; Ukomadu, C.; Jha, S.; Senga, T.; Dhar, S.K.; Wohlschlegel, J.A.; Nutt, L.K.; Kornbluth, S.; Dutta, A. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007, 21, 2288–2299. [Google Scholar] [CrossRef]
- Guan, C.; Li, J.; Sun, D.; Liu, Y.; Liang, H. The structure and polymerase-recognition mechanism of the crucial adaptor protein AND-1 in the human replisome. J. Biol. Chem. 2017, 292, 9627–9636. [Google Scholar] [CrossRef]
- Xu, C.; Min, J. Structure and function of WD40 domain proteins. Protein Cell 2011, 2, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Stros, M.; Launholt, D.; Grasser, K.D. The HMG-box: A versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell. Mol. Life Sci. 2007, 64, 2590–2606. [Google Scholar] [CrossRef] [PubMed]
- Gambus, A.; van Deursen, F.; Polychronopoulos, D.; Foltman, M.; Jones, R.C.; Edmondson, R.D.; Calzada, A.; Labib, K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009, 28, 2992–3004. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.S.; Kroll, E.S.; Lundblad, V.; Spencer, F.A. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 2001, 21, 3144–3158. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.R.; McIntosh, J.R. mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryot. Cell 2002, 1, 758–773. [Google Scholar] [CrossRef]
- Yoshizawa-Sugata, N.; Masai, H. Roles of human AND-1 in chromosome transactions in S phase. J. Biol. Chem. 2009, 284, 20718–20728. [Google Scholar] [CrossRef]
- Li, Y.; Xiao, H.; de Renty, C.; Jaramillo-Lambert, A.; Han, Z.; DePamphilis, M.L.; Brown, K.J.; Zhu, W. The involvement of acidic nucleoplasmic DNA-binding protein (And-1) in the regulation of prereplicative complex (pre-RC) assembly in human cells. J. Biol. Chem. 2012, 287, 42469–42479. [Google Scholar] [CrossRef]
- Abe, T.; Kawasumi, R.; Giannattasio, M.; Dusi, S.; Yoshimoto, Y.; Miyata, K.; Umemura, K.; Hirota, K.; Branzei, D. AND-1 fork protection function prevents fork resection and is essential for proliferation. Nat. Commun. 2018, 9, 3091. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Im, J.S.; Park, S.R.; Kim, S.E.; Wang, H.J.; Lee, J.K. Mimosine arrests the cell cycle prior to the onset of DNA replication by preventing the binding of human Ctf4/And-1 to chromatin via Hif-1alpha activation in HeLa cells. Cell Cycle 2012, 11, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Pei, F.; Ji, M.; Zhang, F.; Sun, Y.; Zhao, Q.; Wang, X.; Hong, Y.; Tian, J.; Wang, Y.; et al. WDHD1 facilitates G1 checkpoint abrogation in HPV E7 expressing cells by modulating GCN5. BMC Cancer 2020, 20, 840. [Google Scholar] [CrossRef]
- Petronczki, M.; Chwalla, B.; Siomos, M.F.; Yokobayashi, S.; Helmhart, W.; Deutschbauer, A.M.; Davis, R.W.; Watanabe, Y.; Nasmyth, K. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J. Cell Sci. 2004, 117, 3547–3559. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Kubota, Y.; Tsujimura, T.; Kumano, M.; Masai, H.; Takisawa, H. Replisome progression complex links DNA replication to sister chromatid cohesion in Xenopus egg extracts. Genes Cells 2009, 14, 949–963. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Z.; Wu, R.; Han, Z.; Zhu, W. And-1 is required for homologous recombination repair by regulating DNA end resection. Nucleic Acids Res. 2017, 45, 2531–2545. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.H.; Lee, K.H.; Chen, Y.T.; Lin, L.J.; Yang, T.L.; Lin, S.W.; Yu, I.S. Wdhd1 is essential for early mouse embryogenesis. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119011. [Google Scholar] [CrossRef]
- Sato, N.; Koinuma, J.; Fujita, M.; Hosokawa, M.; Ito, T.; Tsuchiya, E.; Kondo, S.; Nakamura, Y.; Daigo, Y. Activation of WD repeat and high-mobility group box DNA binding protein 1 in pulmonary and esophageal carcinogenesis. Clin. Cancer Res. 2010, 16, 226–239. [Google Scholar] [CrossRef]
- Liu, B.; Hu, Y.; Qin, L.; Peng, X.B.; Huang, Y.X. MicroRNA-494-dependent WDHDI inhibition suppresses epithelial-mesenchymal transition, tumor growth and metastasis in cholangiocarcinoma. Dig. Liver Dis. 2019, 51, 397–411. [Google Scholar] [CrossRef]
- Chen, S.; Gao, C.; Wu, Y.; Huang, Z. Identification of Prognostic miRNA Signature and Lymph Node Metastasis-Related Key Genes in Cervical Cancer. Front. Pharmacol. 2020, 11, 544. [Google Scholar] [CrossRef]
- Ertay, A.; Liu, H.; Liu, D.; Peng, P.; Hill, C.; Xiong, H.; Hancock, D.; Yuan, X.; Przewloka, M.R.; Coldwell, M.; et al. WDHD1 is essential for the survival of PTEN-inactive triple-negative breast cancer. Cell Death Dis. 2020, 11, 1001. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Y.; Sun, J.; Chen, L.; Gou, W.; Chen, C.W.; Zhou, Y.; Li, Z.; Chan, D.W.; Huang, R.; et al. Discovery and characterization of potent And-1 inhibitors for cancer treatment. Clin. Transl. Med. 2021, 11, e627. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Xiao, M.; He, D.; Hu, Y.; Zhu, Y.; Xiang, L.; Bao, Y.; Liu, X.; Zeng, Q.; Liu, J.; et al. WDHD1 Leads to Cisplatin Resistance by Promoting MAPRE2 Ubiquitination in Lung Adenocarcinoma. Front. Oncol. 2020, 10, 461. [Google Scholar] [CrossRef] [PubMed]
- Gou, W.; Yu, X.; Wu, S.; Wu, H.; Chang, H.; Chen, L.; Wei, H.; Bi, C.; Ning, H.; Wu, Y.; et al. Targeted inhibition of acidic nucleoplasmic DNA-binding protein 1 enhances radiosensitivity of non-small cell lung cancer. Cancer Lett. 2022, 530, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, V.P.; Farina, A.; Tappin, I.; Hurwitz, J. Influence of the human cohesion establishment factor Ctf4/AND-1 on DNA replication. J. Biol. Chem. 2010, 285, 9493–9505. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Stirnimann, C.U.; Petsalaki, E.; Russell, R.B.; Muller, C.W. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010, 35, 565–574. [Google Scholar] [CrossRef]
- Fülöp, V.; Jones, D. Beta propellers: Structural rigidity and functional diversity. Curr. Opin. Struct. Biol. 1999, 9, 715–721. [Google Scholar] [CrossRef]
- Schapira, M.; Tyers, M.; Torrent, M.; Arrowsmith, C.H. WD40 repeat domain proteins: A novel target class? Nat. Rev. Drug Discov. 2017, 16, 773–786. [Google Scholar] [CrossRef]
- Gygax, S.E.; Semighini, C.P.; Goldman, G.H.; Harris, S.D. SepBCTF4 is required for the formation of DNA-damage-induced UvsCRAD51 foci in Aspergillus nidulans. Genetics 2005, 169, 1391–1402. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kilkenny, M.L.; Simon, A.C.; Mainwaring, J.; Wirthensohn, D.; Holzer, S.; Pellegrini, L. The human CTF4-orthologue AND-1 interacts with DNA polymerase alpha/primase via its unique C-terminal HMG box. Open Biol. 2017, 7, 170217. [Google Scholar] [CrossRef]
- Bustin, M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 2001, 26, 152–153. [Google Scholar] [CrossRef]
- Simon, A.C.; Zhou, J.C.; Perera, R.L.; van Deursen, F.; Evrin, C.; Ivanova, M.E.; Kilkenny, M.L.; Renault, L.; Kjaer, S.; Matak-Vinkovic, D.; et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature 2014, 510, 293–297. [Google Scholar] [CrossRef] [PubMed]
- O'Donnell, M.; Langston, L.; Stillman, B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 2013, 5, a010108. [Google Scholar] [CrossRef]
- Bell, S.P.; Labib, K. Chromosome Duplication in Saccharomyces cerevisiae. Genetics 2016, 203, 1027–1067. [Google Scholar] [CrossRef] [PubMed]
- Gambus, A.; Jones, R.C.; Sanchez-Diaz, A.; Kanemaki, M.; van Deursen, F.; Edmondson, R.D.; Labib, K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006, 8, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Gerbi, S.A.; Bielinsky, A.K. DNA replication and chromatin. Curr. Opin. Genet. Dev. 2002, 12, 243–248. [Google Scholar] [CrossRef]
- Branzei, D.; Foiani, M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 2010, 11, 208–219. [Google Scholar] [CrossRef]
- Takaya, J.; Kusunoki, S.; Ishimi, Y. Protein interaction and cellular localization of human CDC45. J. Biochem. 2013, 153, 381–388. [Google Scholar] [CrossRef]
- Fragkos, M.; Ganier, O.; Coulombe, P.; Méchali, M. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 2015, 16, 360–374. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dong, J.; Wang, W.; Yu, D.; Fan, X.; Hui, Y.; Lee, C.; Lam, W.; Alary, N.; Yang, Y.; et al. The human pre-replication complex is an open complex. Cell 2023, 186, 98–111e21. [Google Scholar] [CrossRef]
- Kang, Y.-H.; Farina, A.; Bermudez, V.P.; Tappin, I.; Du, F.; Galal, W.C.; Hurwitz, J. Interaction between human Ctf4 and the Cdc45/Mcm2-7/GINS (CMG) replicative helicase. Proc. Natl. Acad. Sci. USA 2013, 110, 19760–19765. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Simon, A.C.; Ortiz Bazan, M.A.; Kilkenny, M.L.; Wirthensohn, D.; Wightman, M.; Matak-Vinkovic, D.; Pellegrini, L.; Labib, K. Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase. Mol. Cell 2016, 63, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Errico, A.; Cosentino, C.; Rivera, T.; Losada, A.; Schwob, E.; Hunt, T.; Costanzo, V. Tipin/Tim1/And1 protein complex promotes Pol alpha chromatin binding and sister chromatid cohesion. EMBO J. 2009, 28, 3681–3692. [Google Scholar] [CrossRef]
- Hao, J.; de Renty, C.; Li, Y.; Xiao, H.; Kemp, M.G.; Han, Z.; DePamphilis, M.L.; Zhu, W. And-1 coordinates with Claspin for efficient Chk1 activation in response to replication stress. EMBO J. 2015, 34, 2096–2110. [Google Scholar] [CrossRef]
- Jones, M.L.; Baris, Y.; Taylor, M.R.G.; Yeeles, J.T.P. Structure of a human replisome shows the organisation and interactions of a DNA replication machine. EMBO J. 2021, 40, e108819. [Google Scholar] [CrossRef]
- Gao, X.; Leone, G.; Wang, H. Cyclin D-CDK4/6 functions in cancer. Adv. Cancer Res. 2020, 148, 147–169. [Google Scholar] [CrossRef]
- Lim, S.; Kaldis, P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013, 140, 3079–3093. [Google Scholar] [CrossRef]
- Koshiji, M.; Kageyama, Y.; Pete, E.; Horikawa, I.; Barrett, J.; Huang, L. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004, 23, 1949–1956. [Google Scholar] [CrossRef]
- Qiu, Z.; Oleinick, N.L.; Zhang, J. ATR/CHK1 inhibitors and cancer therapy. Radiother. Oncol. 2018, 126, 450–464. [Google Scholar] [CrossRef]
- Altieri, F.; Grillo, C.; Maceroni, M.; Chichiarelli, S. DNA damage and repair: From molecular mechanisms to health implications. Antioxid. Redox Signal. 2008, 10, 891–937. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A.; Lindsey-Boltz, L.; Unsal-Kaçmaz, K.; Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004, 73, 39–85. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Heyer, W.D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef]
- Sartori, A.A.; Lukas, C.; Coates, J.; Mistrik, M.; Fu, S.; Bartek, J.; Baer, R.; Lukas, J.; Jackson, S.P. Human CtIP promotes DNA end resection. Nature 2007, 450, 509–514. [Google Scholar] [CrossRef]
- Kousholt, A.N.; Fugger, K.; Hoffmann, S.; Larsen, B.D.; Menzel, T.; Sartori, A.A.; Sorensen, C.S. CtIP-dependent DNA resection is required for DNA damage checkpoint maintenance but not initiation. J. Cell Biol. 2012, 197, 869–876. [Google Scholar] [CrossRef]
- Li, Y.; Jaramillo-Lambert, A.; Hao, J.; Yang, Y.; Zhu, W. The stability of histone acetyltransferase general control non-derepressible (Gcn) 5 is regulated by Cullin4-RING E3 ubiquitin ligase. J. Biol. Chem. 2011, 286, 41344–41352. [Google Scholar] [CrossRef]
- Li, Y.; Jaramillo-Lambert, A.N.; Yang, Y.; Williams, R.; Lee, N.H.; Zhu, W. And-1 is required for the stability of histone acetyltransferase Gcn5. Oncogene 2012, 31, 643–652. [Google Scholar] [CrossRef]
- Jaramillo-Lambert, A.; Hao, J.; Xiao, H.; Li, Y.; Han, Z.; Zhu, W. Acidic nucleoplasmic DNA-binding protein (And-1) controls chromosome congression by regulating the assembly of centromere protein A (CENP-A) at centromeres. J. Biol. Chem. 2013, 288, 1480–1488. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, H.; Zhang, H.; Sun, C.; Hu, Z.; Tian, Q.; Peng, C.; Jiang, P.; Hua, H.; Li, X.; et al. And-1 coordinates with CtIP for efficient homologous recombination and DNA damage checkpoint maintenance. Nucleic Acids Res. 2017, 45, 2516–2530. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Nievera, C.J.; Lee, A.Y.; Wu, X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008, 283, 7713–7720. [Google Scholar] [CrossRef]
- Chen, Z.; Tran, M.; Tang, M.; Wang, W.; Gong, Z.; Chen, J. Proteomic Analysis Reveals a Novel Mutator S (MutS) Partner Involved in Mismatch Repair Pathway. Mol. Cell Proteom. 2016, 15, 1299–1308. [Google Scholar] [CrossRef]
- Kim, J.M.; Nakao, K.; Nakamura, K.; Saito, I.; Katsuki, M.; Arai, K.; Masai, H. Inactivation of Cdc7 kinase in mouse ES cells results in S-phase arrest and p53-dependent cell death. EMBO J. 2002, 21, 2168–2179. [Google Scholar] [CrossRef]
- Prakash Yadav, R.; Leskinen, S.; Ma, L.; Mäkelä, J.A.; Kotaja, N. Chromatin remodelers HELLS, WDHD1 and BAZ1A are dynamically expressed during mouse spermatogenesis. Reproduction 2023, 165, 49–63. [Google Scholar] [CrossRef]
- Wang, J.; Chen, H.; Zeng, X. Identification of hub genes associated with follicle development in multiple births sheep by WGCNA. Front. Vet.-Sci. 2022, 9, 1057282. [Google Scholar] [CrossRef]
- Sullivan, K. A solid foundation: Functional specialization of centromeric chromatin. Curr. Opin. Genet. Dev. 2001, 11, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Foltz, D.R.; Jansen, L.E.; Bailey, A.O.; Yates, J.R., 3rd; Bassett, E.A.; Wood, S.; Black, B.E.; Cleveland, D.W. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 2009, 137, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Samora, C.P.; Saksouk, J.; Goswami, P.; Wade, B.O.; Singleton, M.R.; Bates, P.A.; Lengronne, A.; Costa, A.; Uhlmann, F. Ctf4 Links DNA Replication with Sister Chromatid Cohesion Establishment by Recruiting the Chl1 Helicase to the Replisome. Mol. Cell 2016, 63, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Zhang, K.; Nicolas, E.; Cam, H.; Zofall, M.; Grewal, S. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 2008, 451, 734–737. [Google Scholar] [CrossRef]
- Moazed, D. Small RNAs in transcriptional gene silencing and genome defence. Nature 2009, 457, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.; Moazed, D. Centromere assembly and propagation. Cell 2007, 128, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.L.; Lin, C.L.; Liu, H.; Chang, Y.J.; Shih, C.J.; Zhong, C.Z.; Lee, S.C.; Tan, B.C. WDHD1 modulates the post-transcriptional step of the centromeric silencing pathway. Nucleic Acids Res. 2011, 39, 4048–4062. [Google Scholar] [CrossRef]
- Shahbazian, M.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.H.; Brownell, J.E.; Sobel, R.E.; Ranalli, T.A.; Cook, R.G.; Edmondson, D.G.; Roth, S.Y.; Allis, C.D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 1996, 383, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Jakaria, M.; Akther, M.; Cho, D.; Kim, I.; Choi, D. The GCN5: Its biological functions and therapeutic potentials. Clin. Sci. 2021, 135, 231–257. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, Y.; Liu, J.; Zhang, J.; Xu, M.; You, Z.; Peng, C.; Gong, Z.; Liu, W. Acetyltransferase GCN5 regulates autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep. 2020, 21, e48335. [Google Scholar] [CrossRef]
- Wu, J.Y.; Niu, Y.T.; Huang, S.N.; Tan, Y.M.; Yang, Z.D.; Fang, Y.Y.; Jiang, L.; Zhang, T.T.; Zeng, X.F.; Peng, Y.X.; et al. WDHD1 is over-expressed in nasopharyngeal carcinoma and may control the expression of ITGAV. FEBS Open Bio 2022, 13, 102–117. [Google Scholar] [CrossRef]
- Wu, J.Y.; Lan, X.L.; Yan, D.M.; Fang, Y.Y.; Peng, Y.X.; Liang, F.F.; Jiang, L.; Huang, S.N.; Mo, M.; Lin, C.X.; et al. The clinical significance of transcription factor WD repeat and HMG-box DNA binding protein 1 in laryngeal squamous cell carcinoma and its potential molecular mechanism. Pathol. Res. Pract. 2022, 230, 153751. [Google Scholar] [CrossRef]
- Xian, Q.; Zhu, D. The Involvement of WDHD1 in the Occurrence of Esophageal Cancer as a Downstream Target of PI3K/AKT Pathway. J. Oncol. 2022, 2022, 5871188. [Google Scholar] [CrossRef]
- Chen, N.; Zhang, G.; Fu, J.; Wu, Q. Identification of Key Modules and Hub Genes Involved in Esophageal Squamous Cell Carcinoma Tumorigenesis Using WCGNA. Cancer Control 2020, 27, 107327482097881. [Google Scholar] [CrossRef] [PubMed]
- He, R.-Q.; Li, J.-D.; He, W.-Y.; Chen, G.; Huang, Z.-G.; Li, M.-F.; Wu, W.-Z.; Chen, J.-T.; Pan, Y.-Q.; Jiang, H.; et al. Prognosis prediction ability and prospective biological mechanisms of WDHD1 in hepatocellular carcinoma tissues. Electron. J. Biotechnol. 2022, 55, 78–90. [Google Scholar] [CrossRef]
- Huang, L.; Ye, T.; Wang, J.; Gu, X.; Ma, R.; Sheng, L.; Ma, B. Identification of Survival-Associated Hub Genes in Pancreatic Adenocarcinoma Based on WGCNA. Front. Genet. 2021, 12, 814798. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, J.; Zhou, Y.; Li, Z.; Peng, C.; Pei, H.; Zhu, W. And-1 Coordinates with the FANCM Complex to Regulate Fanconi Anemia Signaling and Cisplatin Resistance. Cancer Res. 2022, 82, 3249–3262. [Google Scholar] [CrossRef] [PubMed]
- Wermke, M.; Camgoz, A.; Paszkowski-Rogacz, M.; Thieme, S.; von Bonin, M.; Dahl, A.; Platzbecker, U.; Theis, M.; Ehninger, G.; Brenner, S.; et al. RNAi profiling of primary human AML cells identifies ROCK1 as a therapeutic target and nominates fasudil as an antileukemic drug. Blood 2015, 125, 3760–3768. [Google Scholar] [CrossRef] [PubMed]
- Shillitoe, E.J. Papillomaviruses as targets for cancer gene therapy. Cancer Gene Ther. 2006, 13, 445–450. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cortes-Malagon, E.M.; Bonilla-Delgado, J.; Diaz-Chavez, J.; Hidalgo-Miranda, A.; Romero-Cordoba, S.; Uren, A.; Celik, H.; McCormick, M.; Munguia-Moreno, J.A.; Ibarra-Sierra, E.; et al. Gene expression profile regulated by the HPV16 E7 oncoprotein and estradiol in cervical tissue. Virology 2013, 447, 155–165. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Q.; Gao, G.; Zhang, X.; Liu, Y.; Yuan, S.; Wang, X.; Chen, J.J. Role of WDHD1 in Human Papillomavirus-Mediated Oncogenesis Identified by Transcriptional Profiling of E7-Expressing Cells. J. Virol. 2016, 90, 6071–6084. [Google Scholar] [CrossRef]
- Mao, Y.P.; Xie, F.Y.; Liu, L.Z.; Sun, Y.; Li, L.; Tang, L.L.; Liao, X.B.; Xu, H.Y.; Chen, L.; Lai, S.Z.; et al. Re-evaluation of 6th edition of AJCC staging system for nasopharyngeal carcinoma and proposed improvement based on magnetic resonance imaging. Int. J. Radiat. Oncol. Biol. Phys. 2009, 73, 1326–1334. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Steuer, C.E.; El-Deiry, M.; Parks, J.R.; Higgins, K.A.; Saba, N.F. An update on larynx cancer. CA Cancer J. Clin. 2017, 67, 31–50. [Google Scholar] [CrossRef]
- Reichenbach, Z.W.; Murray, M.G.; Saxena, R.; Farkas, D.; Karassik, E.G.; Klochkova, A.; Patel, K.; Tice, C.; Hall, T.M.; Gang, J.; et al. Clinical and translational advances in esophageal squamous cell carcinoma. Adv. Cancer Res. 2019, 144, 95–135. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Otake, R.; Kozuki, R.; Toihata, T.; Takahashi, K.; Okamura, A.; Imamura, Y. Recent progress in multidisciplinary treatment for patients with esophageal cancer. Surg. Today 2020, 50, 12–20. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- O'Reilly, E.A.; Gubbins, L.; Sharma, S.; Tully, R.; Guang, M.H.; Weiner-Gorzel, K.; McCaffrey, J.; Harrison, M.; Furlong, F.; Kell, M.; et al. The fate of chemoresistance in triple negative breast cancer (TNBC). BBA Clin. 2015, 3, 257–275. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.; Stroup, A.M.; Paddock, L.E.; Hill, S.M.; Plascak, J.J.; Llanos, A.A.M. Breast Cancer Incidence and Mortality by Molecular Subtype: Statewide Age and Racial/Ethnic Disparities in New Jersey. Cancer Health Disparities 2019, 3, e1–e17. [Google Scholar] [CrossRef]
- Lee, A.; Djamgoz, M.B.A. Triple negative breast cancer: Emerging therapeutic modalities and novel combination therapies. Cancer Treat. Rev. 2018, 62, 110–122. [Google Scholar] [CrossRef]
- Shi, J.F.; Cao, M.; Wang, Y.; Bai, F.Z.; Lei, L.; Peng, J.; Feletto, E.; Canfell, K.; Qu, C.; Chen, W. Is it possible to halve the incidence of liver cancer in China by 2050? Int. J. Cancer 2021, 148, 1051–1065. [Google Scholar] [CrossRef]
- Galle, P.R.; Foerster, F.; Kudo, M.; Chan, S.L.; Llovet, J.M.; Qin, S.; Schelman, W.R.; Chintharlapalli, S.; Abada, P.B.; Sherman, M.; et al. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019, 39, 2214–2229. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Cao, Q.; Wen, W.; Wang, H. Targeted therapy for hepatocellular carcinoma: Challenges and opportunities. Cancer Lett. 2019, 460, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Razumilava, N.; Gores, G.J. Cholangiocarcinoma. Lancet 2014, 383, 2168–2179. [Google Scholar] [CrossRef] [PubMed]
- Arai, Y.; Totoki, Y.; Hosoda, F.; Shirota, T.; Hama, N.; Nakamura, H.; Ojima, H.; Furuta, K.; Shimada, K.; Okusaka, T.; et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology 2014, 59, 1427–1434. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical cancer. Lancet 2019, 393, 169–182. [Google Scholar] [CrossRef]
- Doorbar, J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 2006, 110, 525–541. [Google Scholar] [CrossRef]
- Jayson, G.; Kohn, E.; Kitchener, H.; Ledermann, J. Ovarian cancer. Lancet 2014, 384, 1376–1388. [Google Scholar] [CrossRef]
- Rocha, C.; Silva, M.; Quinet, A.; Cabral-Neto, J.; Menck, C. DNA repair pathways and cisplatin resistance: An intimate relationship. Clinics 2018, 73, e478s. [Google Scholar] [CrossRef]
- Ley, T.; Miller, C.; Ding, L.; Raphael, B.; Mungall, A.; Robertson, A.; Hoadley, K.; Triche, T.; Laird, P.; Baty, J.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef]
- Tewari, D.; Patni, P.; Bishayee, A.; Sah, A.; Bishayee, A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: A novel therapeutic strategy. Semin. Cancer Biol. 2022, 80, 1–17. [Google Scholar] [CrossRef]
- Nepstad, I.; Hatfield, K.; Grønningsæter, I.; Reikvam, H. The PI3K-Akt-mTOR Signaling Pathway in Human Acute Myeloid Leukemia (AML) Cells. Int. J. Mol. Sci. 2020, 21, 2907. [Google Scholar] [CrossRef] [PubMed]
- Jafari, M.; Ghadami, E.; Dadkhah, T.; Akhavan-Niaki, H. PI3k/AKT signaling pathway: Erythropoiesis and beyond. J. Cell Physiol. 2019, 234, 2373–2385. [Google Scholar] [CrossRef] [PubMed]
- Noorolyai, S.; Shajari, N.; Baghbani, E.; Sadreddini, S.; Baradaran, B. The relation between PI3K/AKT signalling pathway and cancer. Gene 2019, 698, 120–128. [Google Scholar] [CrossRef]
- Matsuda, T.; Muromoto, R.; Sekine, Y.; Togi, S.; Kitai, Y.; Kon, S.; Oritani, K. Signal transducer and activator of transcription 3 regulation by novel binding partners. World J. Biol. Chem. 2015, 6, 324–332. [Google Scholar] [CrossRef]
- Bromberg, J.; Darnell, J.E., Jr. The role of STATs in transcriptional control and their impact on cellular function. Oncogene 2000, 19, 2468–2473. [Google Scholar] [CrossRef]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.E., Jr. STATs and gene regulation. Science 1997, 277, 1630–1635. [Google Scholar] [CrossRef]
- van Pel, D.M.; Stirling, P.C.; Minaker, S.W.; Sipahimalani, P.; Hieter, P. Saccharomyces cerevisiae genetics predicts candidate therapeutic genetic interactions at the mammalian replication fork. G3 2013, 3, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Behan, F.M.; Iorio, F.; Picco, G.; Goncalves, E.; Beaver, C.M.; Migliardi, G.; Santos, R.; Rao, Y.; Sassi, F.; Pinnelli, M.; et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 2019, 568, 511–516. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, J.J. STAT3 plays an important role in DNA replication by turning on WDHD1. Cell Biosci. 2021, 11, 10. [Google Scholar] [CrossRef]
| Tumor | Mechanisms | Functions | Refs. |
|---|---|---|---|
| Nasopharyngeal carcinoma | Regulating ITGAV expression | Affect cell apoptosis | [78] |
| Laryngeal squamous cell carcinoma | Regulating Skp2 expression | Tumor diagnostic biomarkers and tumor formation | [79] |
| Esophageal cancer | As the downstream target of the PI3K/AKT pathway | The occurrence and survival of tumors | [80] |
| As a cell cycle regulator and a downstream molecule in the PI3K/AKT pathway | The occurrence and survival of tumors | [19] | |
| Weighted gene co-expression network analysis | As a hub gene | [81] | |
| Lung adenocarcinoma | Enhancing the ubiquitination degradation of MAPRE2 | Cisplatin resistance | [24] |
| As a cell cycle regulator and a downstream molecule in the PI3K/AKT pathway | The occurrence and survival of tumors | [19] | |
| Regulating the ATM signaling pathway | Radiosensitivity of cancer | [25] | |
| Triple-negative breast cancer | As a downstream target of PTEN-AKT signaling | Tumor cells survival | [22] |
| Hepatocellular carcinoma | Co-expressed genes analysis | As a biomarker for diagnosis and prognosis | [82] |
| Cholangiocarcinoma | Inhibited by miR-494 | Tumor formation and metastasis | [20] |
| Pancreatic adenocarcinoma | Weighted gene co-expression network analysis | As a hub gene for tumor progression | [83] |
| Cervical cancer | Weighted gene co-expression network analysis | As a hub gene for cancer lymph node metastasis | [21] |
| Ovarian cancer | Regulated by ATM- and Rad3-related pathway | Cisplatin resistance | [23] |
| Activating the FA pathway | Cisplatin resistance | [84] | |
| Acute myeloid leukemia | A large-scale loss of-function RNA interference | Tumor growth and viability | [85] |
| Upstream or Downstream | Related Factors | Function and Mechanism | Refs. |
|---|---|---|---|
| Upstream | miR-494 | miR-494 overexpression suppresses EMT, tumor formation, and LNM while promoting CCA cell apoptosis through inhibiting WDHD1 in CCA. | [20] |
| JAK-STAT pathway | WDHD1 is a novel STAT3 target gene and mediated the DNA replication function of STAT3. | [120] | |
| PI3K/AKT pathway | As a downstream molecule in the PI3K/AKT pathway, WDHD1 can be phosphorylated by AKT1. | [19,22,80] | |
| Downstream | MAPRE2 | WDHD1 induces cisplatin resistance in LUAD by promoting MAPRE2 ubiquitination. | [24] |
| ITGAV | ITGAV may act as a potential target gene for WDHD1 and affect cell cycle in nasopharyngeal carcinoma. | [78] | |
| UBA52 | WDHD1 may regulate UBA52 and contribute to the progress of HCC. | [82] | |
| SKP2 | WDHD1 may affect cell cycle in LSCC by regulating SKP2. | [79] | |
| ATM | WDHD1 activates the ATM-CHK1-CDC25C pathway and regulates the G2/M phase block. | [23] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Zhu, Q. WD Repeat and HMG Box DNA Binding Protein 1: An Oncoprotein at the Hub of Tumorigenesis and a Novel Therapeutic Target. Int. J. Mol. Sci. 2023, 24, 12494. https://doi.org/10.3390/ijms241512494
Zhang Z, Zhu Q. WD Repeat and HMG Box DNA Binding Protein 1: An Oncoprotein at the Hub of Tumorigenesis and a Novel Therapeutic Target. International Journal of Molecular Sciences. 2023; 24(15):12494. https://doi.org/10.3390/ijms241512494
Chicago/Turabian StyleZhang, Zhiwei, and Qing Zhu. 2023. "WD Repeat and HMG Box DNA Binding Protein 1: An Oncoprotein at the Hub of Tumorigenesis and a Novel Therapeutic Target" International Journal of Molecular Sciences 24, no. 15: 12494. https://doi.org/10.3390/ijms241512494
APA StyleZhang, Z., & Zhu, Q. (2023). WD Repeat and HMG Box DNA Binding Protein 1: An Oncoprotein at the Hub of Tumorigenesis and a Novel Therapeutic Target. International Journal of Molecular Sciences, 24(15), 12494. https://doi.org/10.3390/ijms241512494






