A Sphingolipidomic Profiling Approach for Comparing X-ray-Exposed and Unexposed HepG2 Cells
Abstract
1. Introduction
2. Results and Discussion
2.1. Optimization of the Best Extraction Procedure of Sphingolipids from Cellular Pellets
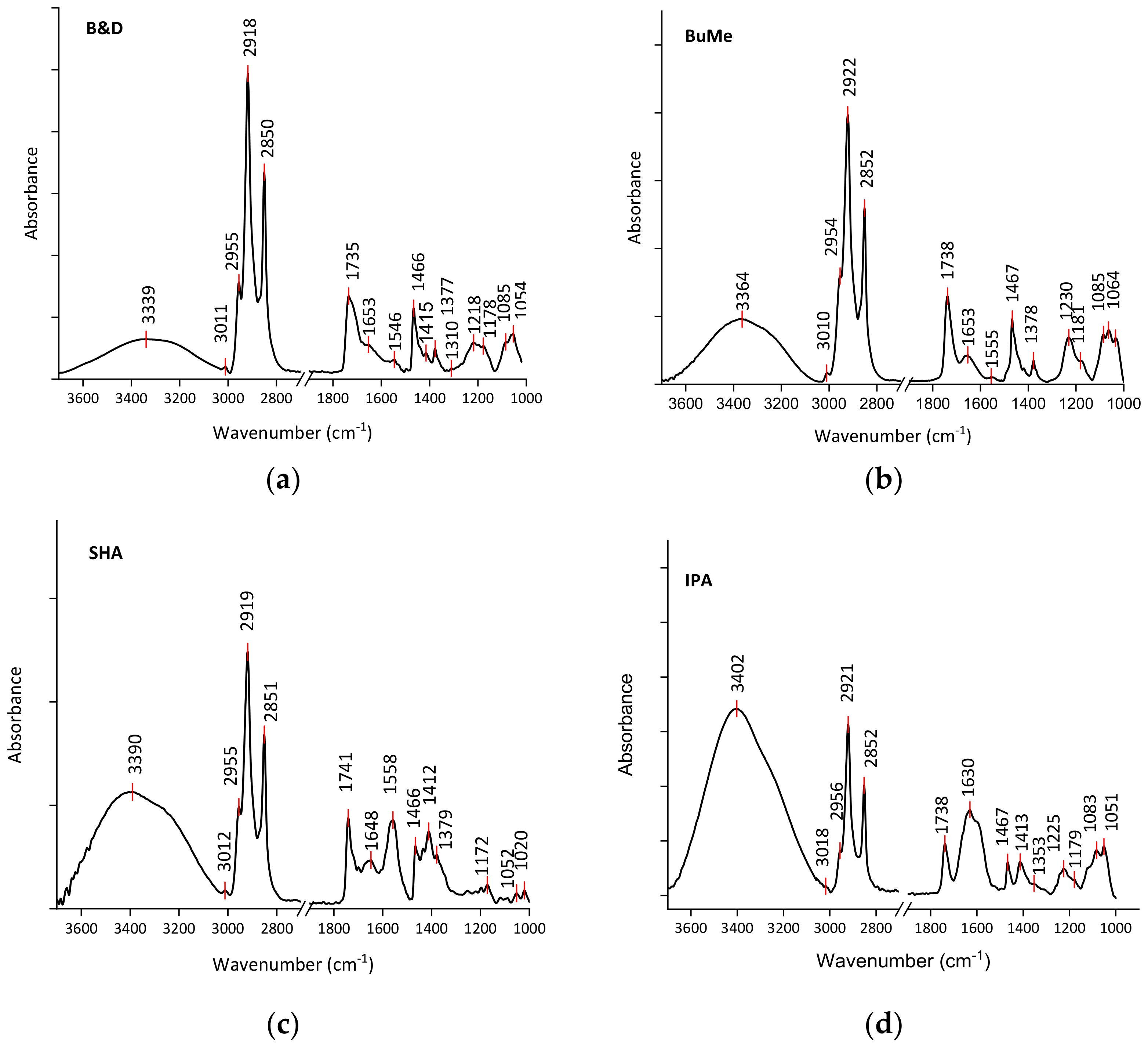
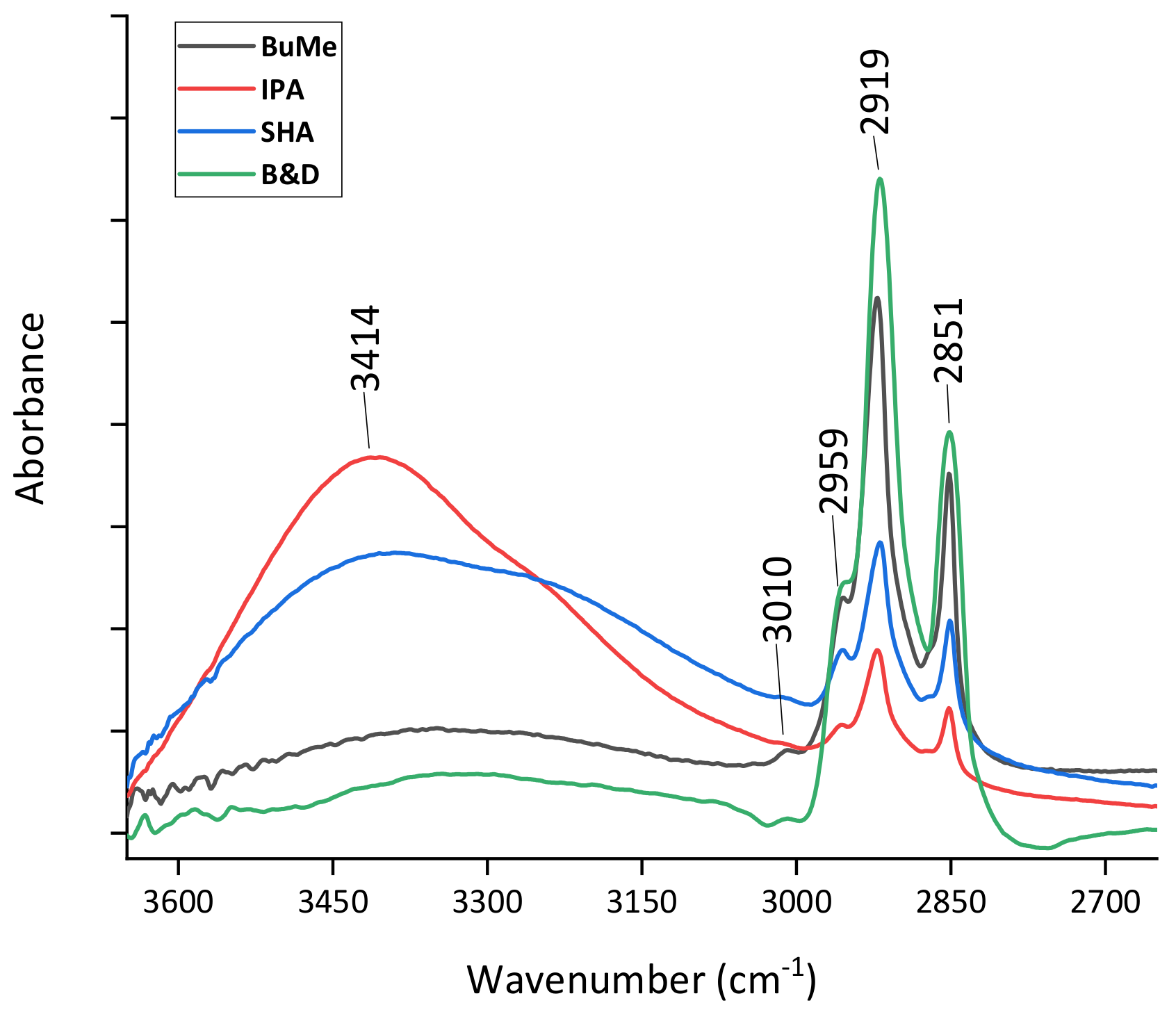
2.1.1. Evaluation of Lipid Extraction Efficiency by FT-IR Spectroscopy
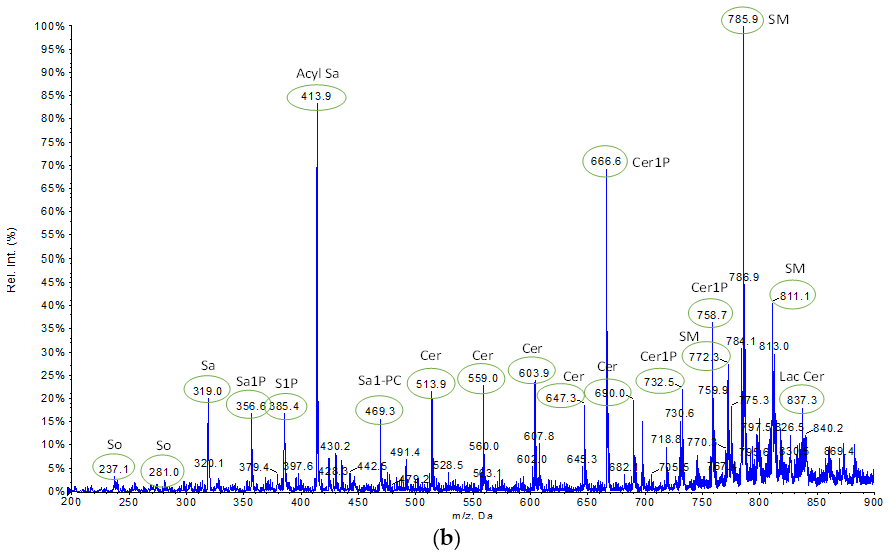
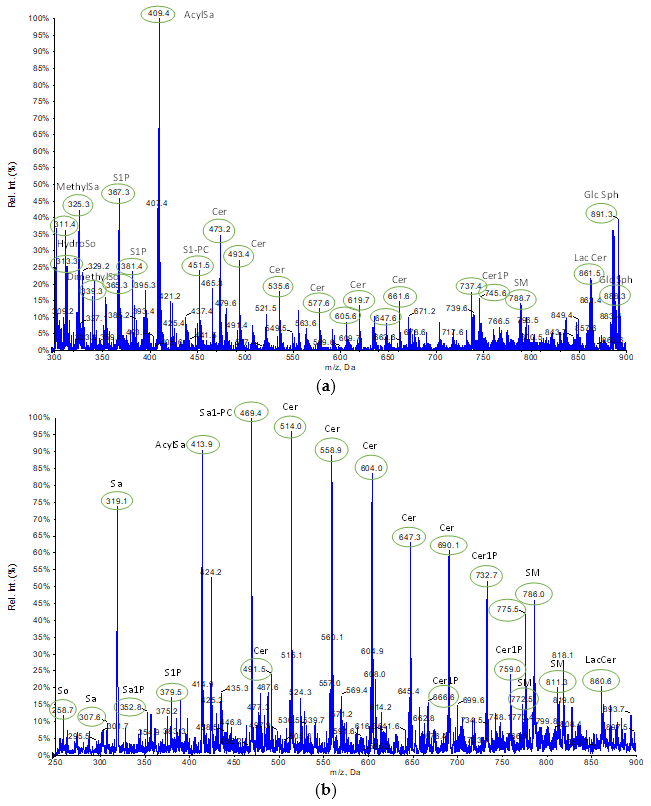
2.1.2. Characterization of Lipid Extracts from HepG2 Pellets Employing Different Protocols Using Tandem Mass Spectrometry-Shotgun
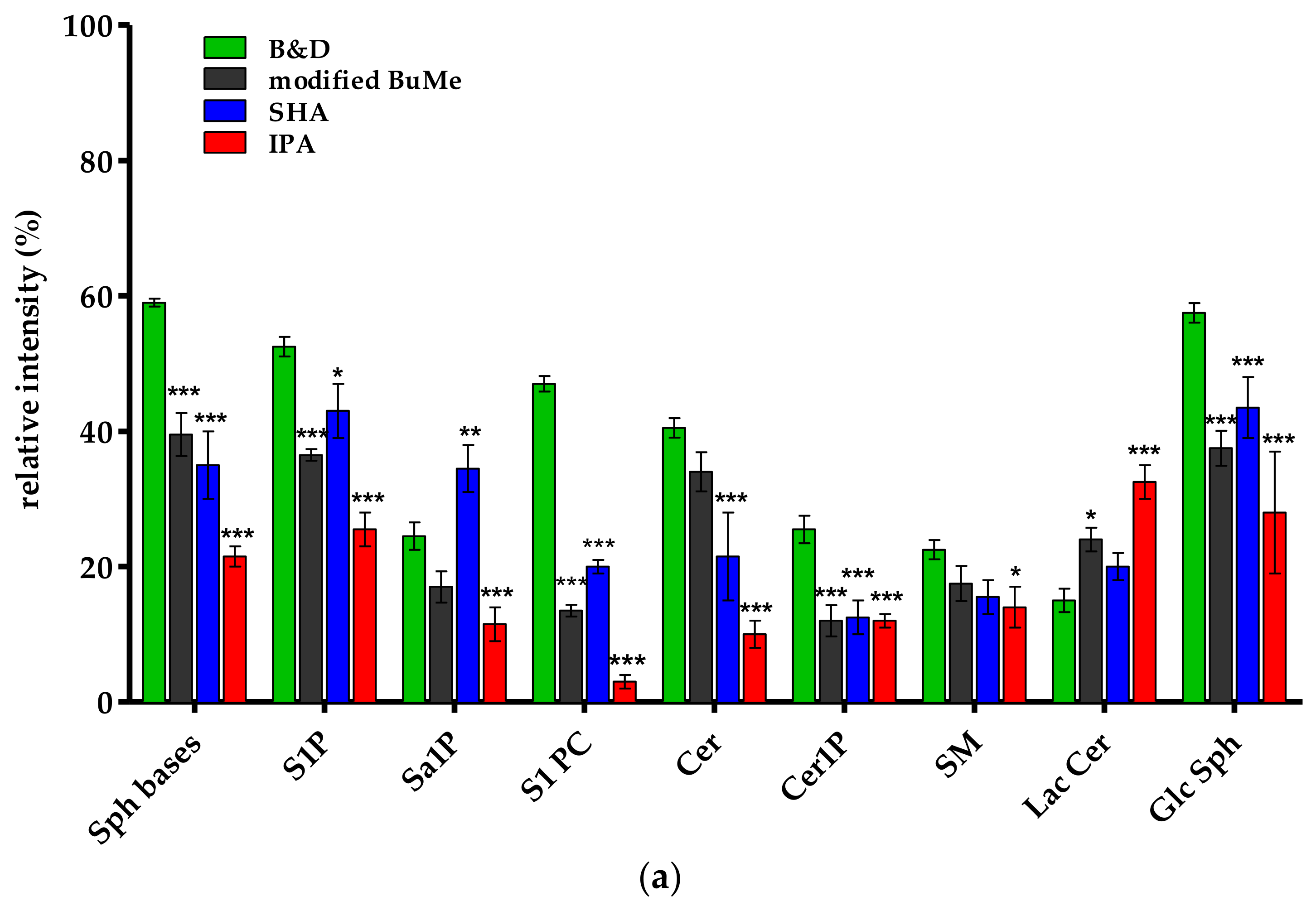
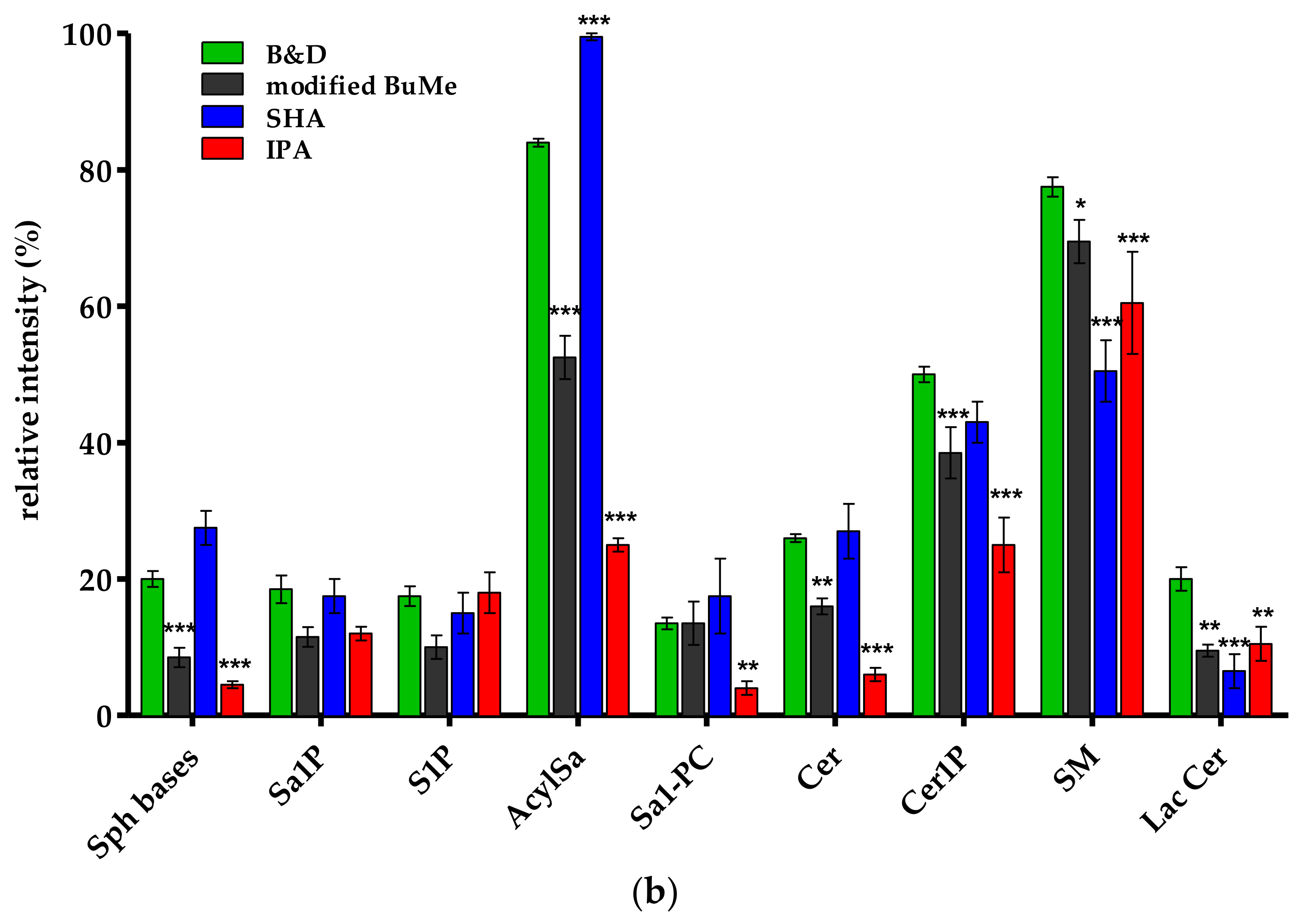
2.1.3. Comparison of Extraction Procedures from HepG2 Pellets Using Tandem Mass Spectrometry-Shotgun
Efficiency and Specificity
Recovery
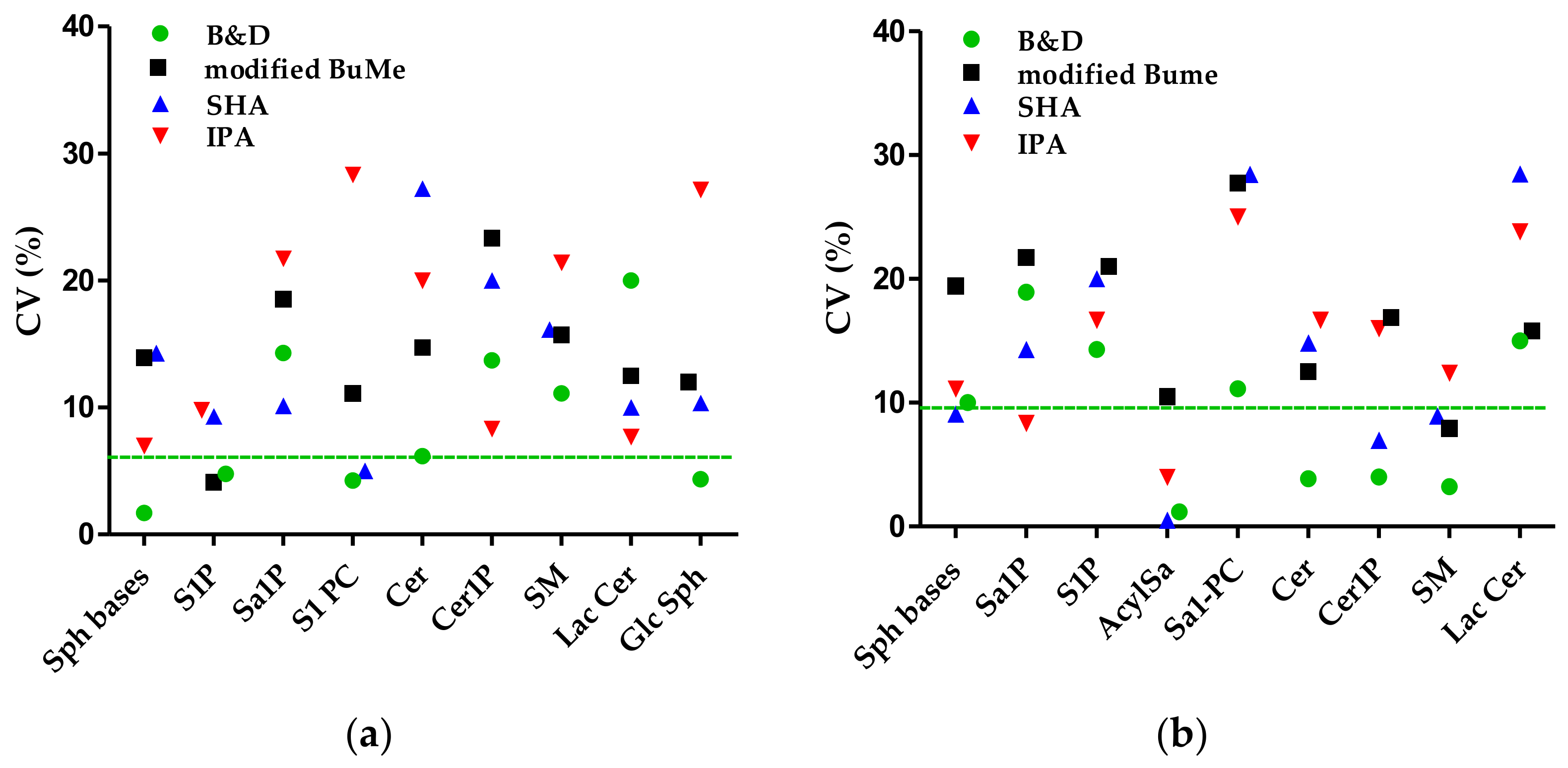
Precision and Reproducibility
2.2. Analyses of Lipidomic Profiles of Irradiated HepG2 Cells
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Cell Culture Preparation
3.2.2. X-ray Irradiation Treatment
3.2.3. Lipid Extraction Protocols
B&D Method
BuMe Modified Method
SHA Method
IPA Method
3.2.4. FT-IR Spectral Analysis
3.2.5. Mass Spectrometry Analysis
3.2.6. Performance Evaluation of Extraction Procedures
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Filippo, V.; Song, M.A.; Zhang, K.; Vinters, H.V.; Tung, S.; Kirsch, W.M.; Yang, J.; Duerksen-Hughes, P.J. Increased ceramide in brains with Alzheimer’s and other neurodegenerative diseases. J. Alzheimers Dis. 2012, 29, 537–547. [Google Scholar] [CrossRef]
- Custodia, A.; Romaus-Sanjurjo, D.; Aramburu-Núñez, M.; Álvarez-Rafael, D.; Vázquez-Vázquez, L.; Camino-Castiñeiras, J.; Leira, Y.; Pías-Peleteiro, J.M.; Aldrey, J.M.; Sobrino, T.; et al. Ceramide/Sphingosine 1-Phosphate Axis as a Key Target for Diagnosis and Treatment in Alzheimer’s Disease and Other Neurodegenerative Diseases. Int. J. Mol. Sci. 2022, 23, 8082. [Google Scholar] [CrossRef]
- Piccoli, M.; Cirillo, F.; Ghiroldi, A.; Rota, P.; Coviello, S.; Tarantino, A.; La Rocca, P.; Lavota, I.; Creo, P.; Signorelli, P.; et al. Sphingolipids and Atherosclerosis: The Dual Role of Ceramide and Sphingosine-1-Phosphate. Antioxidants 2023, 12, 143. [Google Scholar] [CrossRef] [PubMed]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2017, 18, 33–50. [Google Scholar] [CrossRef]
- Sheridan, M.; Ogretmen, B. The Role of Ceramide Metabolism and Signaling in the Regulation of Mitophagy and Cancer Therapy. Cancers 2021, 13, 2475. [Google Scholar] [CrossRef] [PubMed]
- Meikle, P.J.; Summers, S.A. Sphingolipids and phospholipids in insulin resistance and related metabolic disorders. Nat. Rev. Endocrinol. 2017, 13, 79–91. [Google Scholar] [CrossRef]
- Hammerschmidt, P.; Brüning, J.C. Contribution of specific ceramides to obesity-associated metabolic diseases. Cell Mol. Life Sci. 2022, 79, 395. [Google Scholar] [CrossRef]
- Gault, C.R.; Obeid, L.M.; Hannun, Y.A. An Overview of Sphingolipid Metabolism: From Synthesis to Breakdown. Adv. Exp. Med. Biol. 2010, 688, 1–23. [Google Scholar] [CrossRef]
- Kitatani, K.; Idkowiak-Baldys, J.; Hannun, Y.A. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell Signal. 2008, 20, 1010–1018. [Google Scholar] [CrossRef]
- Canals, D.; Salamone, S.; Hannun, Y.A. Visualizing bioactive ceramides. Chem. Phys. Lipids 2018, 216, 142–151. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Many ceramides. J. Biol. Chem. 2011, 286, 27855–27862. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, S.; Futerman, A.H. The metabolism and function of sphingolipids and glycosphingolipids. Cell Mol. Life Sci. 2007, 64, 2270–2284. [Google Scholar] [CrossRef] [PubMed]
- Companioni, O.; Mir, C.; Garcia-Mayea, Y.; Lleonart, M.E. Targeting Sphingolipids for Cancer Therapy. Front. Oncol. 2021, 11, 745092. [Google Scholar] [CrossRef]
- Espaillat, M.P.; Shamseddine, A.A.; Adada, M.M.; Hannun, Y.A.; Obeid, L.M. Ceramide and sphingosine-1-phosphate in cancer, two faces of the sphinx. Transl. Cancer Res. 2015, 4, 484–499. [Google Scholar]
- Grammatikos, G.; Schoell, N.; Ferreiros, N.; Bon, D.; Herrmann, E.; Farnik, H.; Koberle, V.; Piiper, A.; Zeuzem, S.; Kronenberger, B.; et al. Serum sphingolipidomic analyses reveal an upregulation of C16-ceramide and sphingo-sine-1-phosphate in hepatocellular carcinoma. Oncotarget 2016, 7, 18095–18105. [Google Scholar] [CrossRef] [PubMed]
- Separovic, D.; Shields, A.F.; Philip, P.A.; Bielawski, J.; Bielawska, A.; Pierce, J.S.; Tarca, A.L. Altered levels of serum ceramide, sphingosine and sphingomyelin are associated with colorectal cancer: A retrospective pilot study. Anticancer. Res. 2017, 37, 1213–1218. [Google Scholar]
- Jin, L.; Liu, W.-R.; Tian, M.-X.; Fan, J.; Shi, Y.-H. The SphKs/S1P/S1PR1 axis in immunity and cancer: More ore to be mined. World J. Surg. Oncol. 2016, 14, 131. [Google Scholar] [CrossRef]
- Alshaker, H.; Sauer, L.; Monteil, D.; Ottaviani, S.; Srivats, S.; Böhler, T.; Pchejetski, D. Therapeutic Potential of Targeting SK1 in Human Cancers. Adv. Cancer Res. 2013, 117, 143–200. [Google Scholar] [CrossRef]
- Cuvillier, O.; Pirianov, G.; Kleuser, B.; Vanek, P.G.; Coso, O.A.; Gutkind, S.; Spiegel, S. Suppression of ceramide-mediated pro-grammed cell death by sphingosine-1-phosphate. Nature 1996, 381, 800–803. [Google Scholar] [CrossRef]
- Chong, J.R.; Xiang, P.; Wang, W.; Hind, T.; Chew, W.S.; Ong, W.-Y.; Lai, M.K.; Herr, D.R. Sphingolipidomics analysis of large clinical cohorts. Part 2: Potential impact and applications. Biochem. Biophys. Res. Commun. 2018, 504, 602–607. [Google Scholar] [CrossRef]
- Han, X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 2016, 12, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Laaksonen, R.; Ekroos, K.; Sysi-Aho, M.; Hilvo, M.; Vihervaara, T.; Kauhanen, D.; Suoniemi, M.; Hurme, R.; März, W.; Scharnagl, H.; et al. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur. Heart J. 2016, 37, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Saddoughi, S.A.; Garrett-Mayer, E.; Chaudhary, U.; O’Brien, P.E.; Afrin, L.B.; Day, T.A.; Gillespie, M.B.; Sharma, A.K.; Wilhoit, C.S.; Bostick, R.; et al. Results of a Phase II Trial of Gemcitabine Plus Doxorubicin in Patients with Recurrent Head and Neck Cancers: Serum C18-Ceramide as a Novel Biomarker for Monitoring Response. Clin. Cancer Res. 2011, 17, 6097–6105. [Google Scholar] [CrossRef] [PubMed]
- Saini, R.K.; Prasad, P.; Shang, X.; Keum, Y.S. Advances in Lipid Extraction Methods—A Review. Int. J. Mol. Sci. 2021, 22, 13643. [Google Scholar] [CrossRef]
- González-Fernández, M.J.; Manzano-Agugliaro, F.; Zapata-Sierra, A.; Belarbi, E.H.; Guil-Guerrero, J.L. Green argan oil extraction from roasted and unroasted seeds by using various polarity solvents allowed by the EU legislation. J. Clean. Prod. 2020, 276, 123081. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Löfgren, L.; Ståhlman, M.; Forsberg, G.-B.; Saarinen, S.; Nilsson, R.; Hansson, G.I. The BUME method: A novel automated chloroform-free 96-well total lipid extraction method for blood plasma. J. Lipid Res. 2012, 53, 1690–1700. [Google Scholar] [CrossRef]
- Cruz, M.; Wang, M.; Frisch-Daiello, J.; Han, X. Improved Butanol-Methanol (BUME) Method by Replacing Acetic Acid for Lipid Extraction of Biological Samples. Lipids 2016, 51, 887–896. [Google Scholar] [CrossRef]
- Breil, C.; Abert Vian, M.; Zemb, T.; Kunz, W.; Chemat, F. “Bligh and Dyer” and Folch Methods for Solid-Liquid-Liquid Extraction of Lipids from Microorganisms. Comprehension of Solvatation Mechanisms and towards Substitution with Alternative Solvents. Int. J. Mol. Sci. 2017, 18, 708. [Google Scholar] [CrossRef] [PubMed]
- Hammad, S.M.; Pierce, J.S.; Soodavar, F.; Smith, K.J.; Al Gadban, M.M.; Rembiesa, B.; Klein, R.L.; Hannun, Y.A.; Bielawski, J.; Bielawska, A. Blood sphingolipidomics in healthy humans: Impact of sample collection methodology. J. Lipid Res. 2010, 51, 3074–3087. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Brennan, M.-L.; Shen, Z.; MacPherson, J.C.; Schmitt, D.; Molenda, C.E.; Hazen, S.L. Myeloperoxidase Functions as a Major Enzymatic Catalyst for Initiation of Lipid Peroxidation at Sites of Inflammation. J. Biol. Chem. 2002, 277, 46116–46122. [Google Scholar] [CrossRef] [PubMed]
- Shaner, R.L.; Allegood, J.C.; Park, H.; Wang, E.; Kelly, S.; Haynes, C.A.; Sullards, M.C.; Merrill, A.H., Jr. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 2009, 50, 1692–1707. [Google Scholar] [CrossRef]
- Forfang, K.; Zimmermann, B.; Kosa, G.; Kohler, A.; Shapaval, V. FTIR Spectroscopy for Evaluation and Monitoring of Lipid Extraction Efficiency for Oleaginous Fungi. PLoS ONE 2017, 12, e0170611. [Google Scholar] [CrossRef]
- Strug, I.; Utzat, C.; Cappione, A.; Gutierrez, S.; Amara, R.; Lento, J.; Capito, F.; Skudas, R.; Chernokalskaya, E.; Nadler, T. Development of a Univariate Membrane-Based Mid-Infrared Method for Protein Quantitation and Total Lipid Content Analysis of Biological Samples. J. Anal. Methods Chem. 2014, 2014, 657079. [Google Scholar] [CrossRef]
- Shapaval, V.; Brandenburg, J.; Blomqvist, J.; Tafintseva, V.; Passoth, V.; Sandgren, M.; Kohler, A. Biochemical profiling, prediction of total lipid content and fatty acid profile in oleaginous yeasts by FTIR spectroscopy. Biotechnol. Biofuels 2019, 12, 140. [Google Scholar] [CrossRef]
- Cajka, T.; Fiehn, O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. TrAC Trends Anal. Chem. 2014, 61, 192–206. [Google Scholar] [CrossRef]
- Wu, Z.; Shon, J.C.; Liu, K.-H. Mass Spectrometry-based Lipidomics and Its Application to Biomedical Research. J. Lifestyle Med. 2014, 4, 17–33. [Google Scholar] [CrossRef]
- Han, X.; Gross, R.W. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 2005, 24, 367–412. [Google Scholar] [CrossRef]
- Han, X.; Yang, K.; Gross, R.W. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lip-idomic analyses. Mass Spectrom. Rev. 2012, 31, 134–178. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Rozen, S.; Boyle, S.; Hellegers, C.; Cheng, H.; Burke, J.R.; Welsh-Bohmer, K.A.; Doraiswamy, P.M.; Kaddurah-Daouk, R. Metabolomics in early Alzheimer’s disease: Identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS ONE 2011, 6, e21643. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, M.; Han, X. Comprehensive and Quantitative Analysis of Lysophospholipid Molecular Species Present in Obese Mouse Liver by Shotgun Lipidomics. Anal. Chem. 2015, 87, 4879–4887. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Han, X. Characterization and direct quantitation of sphingoid base-1-phosphates from lipid extracts: A shotgun lipidomics approach. J. Lipid Res. 2006, 47, 1865–1873. [Google Scholar] [CrossRef]
- Jiang, X.; Cheng, H.; Yang, K.; Gross, R.W.; Han, X. Alkaline methanolysis of lipid extracts extends shotgun lipidomics analyses to the low-abundance regime of cellular sphingolipids. Anal. Biochem. 2007, 371, 135–145. [Google Scholar] [CrossRef][Green Version]
- Holčapek, M.; Jirásko, R.; Lísa, M. Recent developments in liquid chromatography-mass spectrometry and related techniques. J. Chromatogr. A 2012, 1259, 3–15. [Google Scholar] [CrossRef]
- Corre, I.; Niaudet, C.; Paris, F. Plasma membrane signaling induced by ionizing radiation. Mutat. Res. Mol. Mech. Mutagen. 2010, 704, 61–67. [Google Scholar] [CrossRef]
- Haimovitz-Friedman, A.; Kan, C.C.; Ehleiter, D.; Persaud, R.S.; McLoughlin, M.; Fuks, Z.; Kolesnick, R.N. Ionizing radiation acts on cellular membranes to generate ceramide and initiate apoptosis. J. Exp. Med. 1994, 180, 525–535. [Google Scholar] [CrossRef]
- Vit, J.P.; Rosselli, F. Role of the ceramide-signaling pathways in ionizing radiation-induced apoptosis. Oncogene 2003, 22, 8645–8652. [Google Scholar] [CrossRef]
- Sharma, D.; Czarnota, G.J. Involvement of Ceramide Signalling in Radiation-Induced Tumour Vascular Effects and Vascular-Targeted Therapy. Int. J. Mol. Sci. 2022, 23, 6671. [Google Scholar] [CrossRef]
- Derenne, A.; Claessens, T.; Conus, C.; Goormaghtigh, E. Infrared spectroscopy of membrane lipids. In Encyclopedia of Biophysics; Roberts, G.C.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1074–1081. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Martinez, M.A.G.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Advances in Fourier transform infrared (FTIR) spec-troscopy of biological tissues. Appl. Spectrosc. Rev. 2017, 52, 456–506. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Fourier Transform Infrared (FTIR) Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Dreissig, I.; Machill, S.; Salzer, R.; Krafft, C. Quantification of brain lipids by FTIR spectroscopy and partial least squares regression. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2009, 71, 2069–2075. [Google Scholar] [CrossRef]
- Millipore, M. Simplified Analysis of Lipid or Detergent Content in Biological Samples Using the IR-Based Direct Detect® Spectrometer; Application Note; Merck Millipore: Burlington, MA, USA, 2013. [Google Scholar]
- Faramarzi, B.; Moggio, M.; Cardamuro, V.; Portaccio, M.; Diano, N.; Manti, L.; Lepore, M. An FTIR Spectroscopy Investigation on Different Methods of Lipid Extraction from HepG2 Cells. Eng. Proc. 2022, 27, 39. [Google Scholar] [CrossRef]
- Xie, L.-M.; Yau, L.-F.; Jiang, Z.-H.; Zhang, L.-Y.; Xia, Y.; Wang, J.-R. Sphingolipidomic study of davidiin-treated HepG2 human hepatocellular carcinoma cells using UHPLC-MS. RSC Adv. 2017, 7, 55249–55256. [Google Scholar] [CrossRef]
- Merrill, A.H., Jr.; Sullards, M.C.; Allegood, J.C.; Kelly, S.; Wang, E. Sphingolipidomics: High-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods 2005, 36, 207–224. [Google Scholar] [CrossRef]
- Alshehry, Z.H.; Mundra, P.A.; Barlow, C.K.; Mellett, N.A.; Wong, G.; McConville, M.J.; Simes, J.; Tonkin, A.M.; Sullivan, D.R.; Barnes, E.H.; et al. Plasma Lipidomic Profiles Improve on Traditional Risk Factors for the Prediction of Cardiovascular Events in Type 2 Diabetes Mellitus. Circulation 2016, 134, 1637–1650. [Google Scholar] [CrossRef]
- Wong, M.W.K.; Braidy, N.; Pickford, R.; Sachdev, P.S.; Poljak, A. Comparison of Single Phase and Biphasic Extraction Protocols for Lipidomic Studies Using Human Plasma. Front. Neurol. 2019, 10, 879. [Google Scholar] [CrossRef]
- Nava, V.E.; Cuvillier, O.; Edsall, L.C.; Kimura, K.; Milstien, S.; Gelmann, E.P.; Spiegel, S. Sphingosine enhances apoptosis of radiation-resistant prostate cancer cells. Cancer Res 2000, 60, 6671. [Google Scholar]
- Santana, P.; Peña, L.A.; Haimovitz-Friedman, A.; Martin, S.; Green, D.; McLoughlin, M.; Cordon-Cardo, C.; Schuchman, E.H.; Fuks, Z.; Kolesnick, R. Acid Sphingomyelinase-Deficient Human Lymphoblasts and Mice Are Defective in Radiation-Induced Apoptosis. Cell 1996, 86, 189–199. [Google Scholar] [CrossRef]
- Schillaci, F.; Anzalone, A.; Cirrone, G.A.P.; Carpinelli, M.; Cuttone, G.; Cutroneo, M.; De Martinis, C.; Give, D.; Korn, G.; Mag-giore, M.; et al. ELIMED, MEDical and multidisciplinary ap-plications at ELI-Beamlines. J. Phys. Conf. Ser. 2014, 508, 012010. [Google Scholar] [CrossRef]
- Delfino, I.; Perna, G.; Ricciardi, V.; Lasalvia, M.; Manti, L.; Capozzi, V.; Lepore, M. X-ray irradiation effects on nuclear and membrane regions of single SH-SY5Y human neuroblastoma cells investigated by Raman micro-spectroscopy. J. Pharm. Biomed. Anal. 2019, 164, 557–573. [Google Scholar] [CrossRef] [PubMed]
- Manti, L.; Braselmann, H.; Calabrese, M.L.; Massa, R.; Pugliese, M.; Scampoli, P.; Sicignano, G.; Grossi, G. Effects of modulated microwave radiation at cellular telephone frequency (1.95 GHz) on X-ray-induced chromosome aberrations in human lymphocytes in vitro. Radiat. Res. 2008, 169, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Lasch, P. Spectral pre-processing for biomedical vibrational spectroscopy and microspectroscopic imaging. Chemom. Intell. Lab. Syst. 2012, 117, 100–114. [Google Scholar] [CrossRef]

| Sphingolipids | m/z Molecular Ion | CE 1 | m/z Product Ions |
|---|---|---|---|
| Hydroxysphingosine | 311.3–313.3 | −35 | |
| Methylsphinganine | 323.2–325.4 | −35 | |
| Dimethylsphingosine | 339.4 | −35 | |
| C17 S1P (d17:1) | 367.5 | −30 | 367.5 → 250.3, 78.8 |
| C17 Sa1P (d17:1) | 369.3 | −25 | 369.3 → 252.2, 78.8 |
| C18 S1P (d18:1) | 381.6 | −30 | 381.6 → 266.2, 78.8 |
| AcylSa | 409.3 | −35 | 409.3 → 264.4 |
| C17 S1-PC | 451.3–459.3 | −45 | |
| Cer(d14:1/15:0) | 473.3 | −40 | 473.3 → 237.1 |
| Cer(d18:1/13:0) | 493.5 | −40 | 493.5 → 264.4 |
| Cer(d18:1/16:0) | 535.5 | −45 | 535.5 → 264.4 |
| Cer(d18:1/19:0) | 577.7 | −45 | 577.7 → 264.4 |
| Cer(d18:2/22:0) | 619.6 | −45 | 619.6 → 264.4 |
| Cer(d18:1/25:0) | 661.6 | −45 | 661.6 → 264.4 |
| GlcCer(C16:0) | 703.7 | −45 | 703.7 → 266.4 |
| DH Cer1P(d18:0/24:0) | 731.9 | −45 | 731.9 → 266.3, 78.8 |
| Cer1P(d18:0/25:0) | 745.6 | −45 | 745.6 → 78.9 |
| SM(d18:0/22:0) | 787.8 | −40 | 787.8 → 184.4 |
| SM(d18:1/26:0) | 829.9 | −40 | 829.9 → 184.4 |
| LacCer (C16:0) | 861.6 | −45 | 861.6 → 266.2, 342.1 |
| GlcSph | 885.4 | −40 | |
| GlcSph | 891.3 | −40 |
| Sphingolipids | m/z Molecular Ion | CE 1 | m/z Product Ions |
|---|---|---|---|
| So (d14:0) | 237.1 | 20 | |
| So (d17:1) | 281.0 | 20 | |
| Sa (d19:0) | 319.0 | 45 | 319.0 → 268.3 |
| C16 Sa1P | 356.6 | 25 | 356.6 →252.2, 78.8 |
| C18 S1P (d18:1) | 385.4 | 30 | 385.4 →266.2, 78.8 |
| AcylSa | 413.9 | 30 | 413.9 → 264.4 |
| Sa1-PC | 469.3 | 30 | |
| Cer(d18:1/14:0) | 513.9 | 45 | 513.9 → 264.4 |
| Cer(d18:1/17:0) | 559.0 | 45 | 559.0 → 264.4 |
| Cer(d18:1/21:3) | 603.9 | 45 | 603.9 → 264.4 |
| Cer(d18:1/24:1) | 647.3 | 45 | 647.3 → 264.4 |
| Cer1P(d18:1/19:0) | 666.6 | 45 | 666.6 → 264.4, 78.9 |
| Cer(d18:1/26:1) | 690.0 | 45 | 690.0 → 264.4 |
| Cer1P(d18:0/24:0) | 732.5 | 45 | 732.5 → 266.3, 78.9 |
| Cer1P(d18:1/26:0) | 758.7 | 45 | 758.7 → 264.4, 78.9 |
| SM(d17:1/22:0) | 772.3 | 45 | 772.3 → 184.4 |
| SM(d18:1/22:0) | 785.9 | 45 | 785.9 → 184.4 |
| SM(d18:1/24:1) | 811.1 | 45 | 811.1 → 184.4 |
| LacCer (d16:0/16:0) | 837.3 | 45 | 837.3 → 266.2, 342.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moggio, M.; Faramarzi, B.; Portaccio, M.; Manti, L.; Lepore, M.; Diano, N. A Sphingolipidomic Profiling Approach for Comparing X-ray-Exposed and Unexposed HepG2 Cells. Int. J. Mol. Sci. 2023, 24, 12364. https://doi.org/10.3390/ijms241512364
Moggio M, Faramarzi B, Portaccio M, Manti L, Lepore M, Diano N. A Sphingolipidomic Profiling Approach for Comparing X-ray-Exposed and Unexposed HepG2 Cells. International Journal of Molecular Sciences. 2023; 24(15):12364. https://doi.org/10.3390/ijms241512364
Chicago/Turabian StyleMoggio, Martina, Bahar Faramarzi, Marianna Portaccio, Lorenzo Manti, Maria Lepore, and Nadia Diano. 2023. "A Sphingolipidomic Profiling Approach for Comparing X-ray-Exposed and Unexposed HepG2 Cells" International Journal of Molecular Sciences 24, no. 15: 12364. https://doi.org/10.3390/ijms241512364
APA StyleMoggio, M., Faramarzi, B., Portaccio, M., Manti, L., Lepore, M., & Diano, N. (2023). A Sphingolipidomic Profiling Approach for Comparing X-ray-Exposed and Unexposed HepG2 Cells. International Journal of Molecular Sciences, 24(15), 12364. https://doi.org/10.3390/ijms241512364








