Whole-Exome and Transcriptome Sequencing Expands the Genotype of Majewski Osteodysplastic Primordial Dwarfism Type II
Abstract
1. Introduction
2. Results
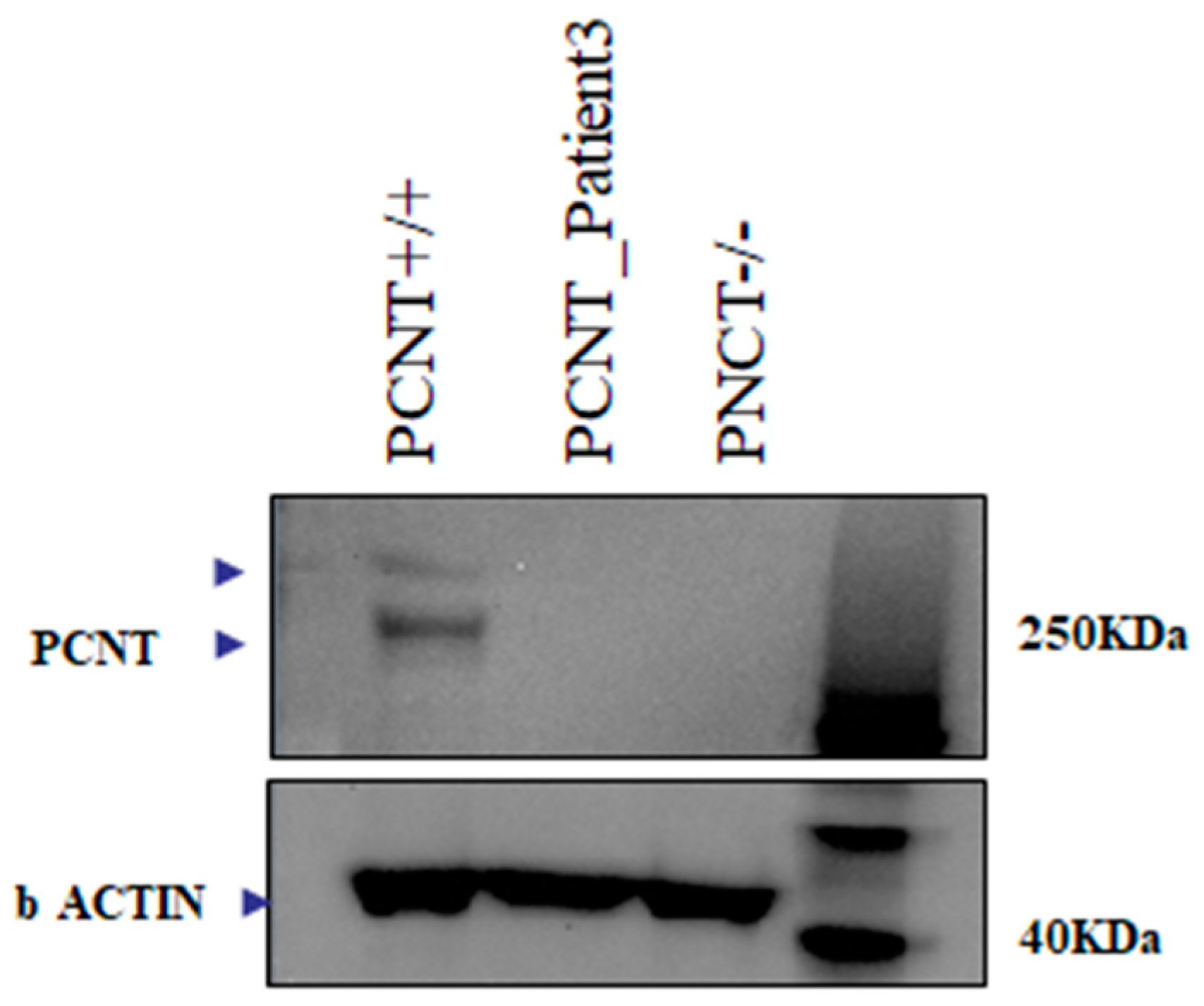
2.1. Array CGH and Exome Sequencing
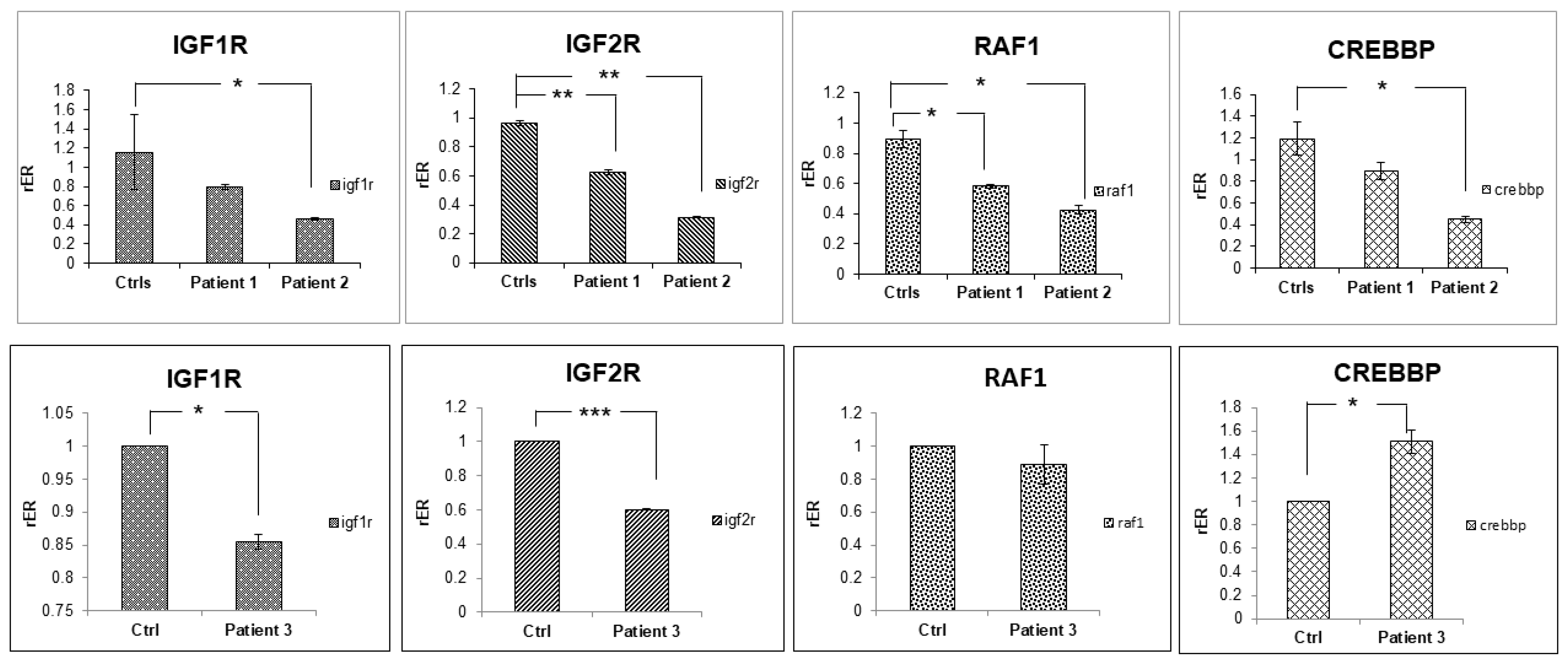
2.2. Transcriptome Sequencing
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Comparative Genomic Hybridization Array
4.3. DNA and RNA Extraction
4.4. Exome and Transcriptome Sequencing
4.5. Reverse Transcription and Real Time PCR Analysis
4.6. Bioinformatics and Statistical Analyses
4.6.1. Exome Analyses
4.6.2. Post-Processing of Exome Data and Variant Prioritization
4.6.3. RNA-Seq Analyses
4.6.4. Western Blot Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hall, J.G.; Flora, C.; Scott, C.I., Jr.; Pauli, R.M.; Tanaka, K.I. Majewski osteodysplastic primordial dwarfism type II (MOPD II), natural history and clinical findings. Am. J. Med. Genet. A 2004, 130, 55–72. [Google Scholar] [CrossRef]
- Huang-Doran, I.; Bicknell, L.S.; Finucane, F.M.; Rocha, N.; Porter, K.M.; Tung, Y.L.; Szekeres, F.; Krook, A.; Nolan, J.J.; Majewski Osteodysplastic Primordial Dwarfism Study Group; et al. Genetic defects in human pericentrin are associated with severe insulin resistance and diabetes. Diabetes 2011, 60, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Rauch, A. The shortest of the short, pericentrin mutations and beyond. Best Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Piane, M.; Della Monica, M.; Piatelli, G.; Lulli, P.; Lonardo, F.; Chessa, L.; Scarano, G. Majewski osteodysplastic primordial dwarfism type II (MOPD II) syndrome previously diagnosed as Seckel syndrome, report of a novel mutation of the PCNT gene. Am. J. Med. Genet. A 2009, 149, 2452–2456. [Google Scholar] [CrossRef]
- Willems, M.; Genevieve, D.; Borck, G.; Baumann, C.; Baujat, G.; Bieth, E.; Edery, P.; Farra, C.; Gerard, M.; Heron, D.; et al. Molecular analysis of pericentrin gene (PCNT) in a series of 24 Seckel/microcephalic osteodysplastic primordial dwarfism type II (MOPD II) families. J. Med. Genet. 2010, 47, 797–802. [Google Scholar] [CrossRef]
- Zimmerman, W.C.; Sillibourne, J.; Rosa, J.; Doxsey, S.J. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 2004, 15, 3642–3657. [Google Scholar] [CrossRef]
- Faienza, M.F.; Acquafredda, A.; D’Aniello, M.; Soldano, L.; Marzano, F.; Ventura, A.; Cavallo, L. Effect of recombinant insulin-like growth factor-1 treatment on short-term linear growth in a child with Majewski osteodysplastic primordial dwarfism type II and hepatic insufficiency. JPEM 2013, 26, 771–774. [Google Scholar] [CrossRef]
- Warr, A.; Robert, C.; Hume, D.; Archibald, A.; Deeb, N.; Watson, M. Exome Sequencing, Current and Future Perspectives. G3 2015, 5, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, A.; Anyane-Yeboa, K.; Wynn, J.; Wilson, A.; Cho, M.T.; Guzman, E.; Sisson, R.; Egan, C.; Chung, W.K. The usefulness of whole-exome sequencing in routine clinical practice. Genet. Med. 2014, 16, 922–931. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, B.; Mahdieh, N.; Hosomichi, K.; Nakaoka, H.; Inoue, I. Next-generation sequencing, impact of exome sequencing in characterizing Mendelian disorders. J. Hum. Genet. 2012, 57, 621–632. [Google Scholar] [CrossRef]
- Casamassimi, A.; Federico, A.; Rienzo, M.; Esposito, S.; Ciccodicola, A. Transcriptome Profiling in Human Diseases, New Advances and Perspectives. Int. J. Mol. Sci. 2017, 18, 1652. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.W.; Lee, I.H.; Liu, X.; Hirschhorn, J.N.; Mandl, K.D. Measuring coverage and accuracy of whole-exome sequencing in clinical context. Genet. Med. 2018, 20, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Samuels, D.C.; Han, L.; Li, J.; Quanghu, S.; Clark, T.A.; Shyr, Y.; Guo, Y. Finding the lost treasures in exome sequencing data. Trends Genet. 2013, 29, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, T.N.; Krepischi, A.C.; da Costa, S.S.; da Silva, I.T.; Vianna-Morgante, A.M.; Valieris, R.; Ezquina, S.A.; Bertola, D.R.; A Otto, P.; Rosenberg, C. Utility of trio-based exome sequencing in the elucidation of the genetic basis of isolated syndromic intellectual disability, illustrative cases. Appl. Clin. Genet. 2018, 11, 93–98. [Google Scholar] [CrossRef]
- Faienza, M.F.; Marzano, F.; Ventura, A.M.; Wasniewska, M.; Valenzise, M.; Valletti, A. Regulation of IGFBP3 gene expression in short children born small for gestational age. Growth Horm IGF Res. 2011, 21, 349–355. [Google Scholar] [CrossRef]
- Marzano, F.; Ventura, A.; Francesco Caratozzolo, M.; Aiello, I.; Mastropasqua, F.; Brunetti, G.; Cavallo, L.; Sbisà, E.; Faienza, M.F.; Tullo, A. The p53 family member p73 modulates the pro-proliferative role of IGFBP3 in short children born SGA. Mol. Biol. Cell 2015, 26, 2733–2741. [Google Scholar] [CrossRef]
- Boles, R.G.; Teebi, A.S.; Schwartz, D.; Harper, J.F. Further delineation of the ear, patella, short stature syndrome (Meier-Gorlin syndrome). Clin. Dysmorphol. 1994, 3, 207–214. [Google Scholar] [CrossRef]
- Taybi, H. Microcephalic osteodysplastic primordial dwarfism and cephalo-skeletal dysplasia (Taybi-Linder syndrome). Am. J. Med. Genet. 1992, 43, 628–629. [Google Scholar] [CrossRef]
- Majewski, F.; Ranke, M.; Schinzel, A. Studies of microcephalic primordial dwarfism II, the osteodysplastic type II of primordial dwarfism. Am. J. Med. Genet. 1982, 12, 23–35. [Google Scholar] [CrossRef]
- Bober, M.B.; Jackson, A.P. Microcephalic Osteodysplastic Primordial Dwarfism, Type II, a Clinical Review. Curr. Osteoporos. Rep. 2017, 15, 61–69. [Google Scholar] [CrossRef]
- Doxsey, S.J.; Stein, P.; Evans, L.; Calarco, P.D.; Kirschner, M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 1994, 76, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Masahiko, K.; Atsushi, H.; Tomoyasu, K.; Shuang-Lin, X.; Keiko, M.; Yasunari, H.; Osamu, M.; Hiroko, S.; Kenichi, Y. Critical role for chicken Rad17 and Rad9 in the cellular response to DNA damage and stalled DNA replication. Genes Cells 2004, 9, 291–303. [Google Scholar]
- Iffland, P.H.; E Everett, M.; Cobb-Pitstick, K.M.; E Bowser, L.; E Barnes, A.; Babus, J.K.; Romanowski, A.J.; Baybis, M.; Elziny, S.; Puffenberger, E.G.; et al. NPRL3 loss alters neuronal morphology, mTOR localization, cortical lamination and seizure threshold. Brain 2022, 145, 3872–3885. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Viltrop, T.; Tiirats, A.; Rajashekar, B.; Reimann, E.; Kõks, S.; Rull, K.; Milani, L.; Acharya, G.; Basnet, P.; et al. Using RNA sequencing for identifying gene imprinting and random monoallelic expression in human placenta. Epigenetics 2014, 9, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Tsai, S.; James, A.H.; Thames, B.H.; Shytle, S.; Piedrahita, J.A. Lack of genomic imprinting of DNA primase, polypeptide 2 (PRIM2) in human term placenta and white blood cells. Epigenetics 2012, 7, 429–431. [Google Scholar] [CrossRef]
- Walenkamp, M.J.; Losekoot, M.; Wit, J.M. Molecular IGF-1 and IGF-1 receptor defects, from genetics to clinical management. Endocr. Dev. 2013, 24, 128–137. [Google Scholar]
- Finken, M.J.J.; van der Steen, M.; Smeets, C.C.J.; Walenkamp, M.J.E.; de Bruin, C.; Hokken-Koelega, A.C.S.; Wit, J.M. Children Born Small for Gestational Age, Differential Diagnosis, Molecular Genetic Evaluation, and Implications. Endocr. Rev. 2018, 39, 851–894. [Google Scholar] [CrossRef]
- Renes, J.S.; van Doorn, J.; Hokken-Koelegaa, A.C.S. Current Insights into the Role of the Growth Hormone-Insulin-Like Growth Factor System in Short Children Born Small for Gestational Age. Horm. Res. Paediatr. 2019, 92, 15–27. [Google Scholar] [CrossRef]
- Razzaque, M.A.; Nishizawa, T.; Komoike, Y.; Yagi, H.; Furutani, M.; Amo, R.; Kamisago, M.; Momma, K.; Katayama, H.; Nakagawa, M.; et al. Germline Gain-Of-Function Mutations in RAF1 Cause Noonan Syndrome. Nat. Genet. 2007, 39, 1013–1017. [Google Scholar] [CrossRef]
- Seltzer, L.E.; Paciorkowski, A.R. Genetic Disorders Associated with Postnatal Microcephaly. Am. J. Med. Genet. C Semin. Med. Genet. 2014, 166, 140–155. [Google Scholar] [CrossRef]
- Calabrese, C.; Mangiulli, M.; Manzari, C.; Paluscio, A.M.; Caratozzolo, M.F.; Marzano, F.; Kurelac, I.; D’erchia, A.M.; D’elia, D.; Licciulli, F.; et al. A platform independent RNA-Seq protocol for the detection of transcriptome complexity. BMC Genom. 2013, 14, 855. [Google Scholar] [CrossRef] [PubMed]
- Chiara, M.; Gioiosa, S.; Chillemi, G.; D’Antonio, M.; Flati, T.; Picardi, E.; Zambelli, F.; Horner, D.S.; Pesole, G.; Castrignanò, T. CoVaCS, a consensus variant calling system. BMC Genom. 2018, 19, 120. [Google Scholar] [CrossRef] [PubMed]
- Marco-Sola, S.; Sammeth, M.; Guigó, R.; Ribeca, P. The GEM map per, fast, accurate and versatile alignment by filtration. Nat. Methods 2012, 9, 1185–1188. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR, Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP, the NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI, current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar, public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Evans, K.; Hayden, M.; Heywood, S.; Hussain, M.; Phillips, A.D.; Cooper, D.N. The Human Gene Mutation Database, towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017, 136, 665–677. [Google Scholar] [CrossRef]
- Liu, X.; Wu, C.; Li, C.; Boerwinkle, E. dbNSFPv3.0, a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 2016, 37, 235–241. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools, a flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Consiglio, A.; Mencar, C.; Grillo, G.; Marzano, F.; Caratozzolo, M.F.; Liuni, S. A fuzzy method for RNA-Seq differential expression analysis in presence of multireads. BMC Bioinform. 2016, 17 (Suppl. 12), 345. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools, paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards suite, from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
| Case | Refseq | Gene | Genotype | Exon/ Intron | cDNA (HGVS) | Protein Change (HGVS) | Mutation Type | Gnom AD | dbSNP | ACMG | Class | Gene Function |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NM_006031.6 | PCNT | Hmz | Intron 18 | c.3608-2A>G | - | splicing | - | - | PVS1, PM2 | C4 | The protein encoded by this gene binds to calmodulin and is expressed in the centrosome. It is an integral component of the pericentriolar material (PCM). The protein interacts with the microtubule nucleation component gamma-tubulin and is likely important to normal functioning of the centrosomes, cytoskeleton, and cell-cycle progression. MutationPathogenic variants in this gene cause Seckel syndrome-4 and microcephalic osteodysplastic primordial dwarfism type II |
| 2 | NM_006031.6 | PCNT | Hmz | Exon 10 | c.1523dupA | p.(Thr510AsnfsTer4) | indel | 0.0000319 | rs1369869782 | PVS1, PM2, PP5 | C5 | |
| NM_031419.4 | NFKBIZ | Hmz | Exon 11 | c.1635+2-ACTTTTAGAA | - | splicing | 0.0000489 | - | PP3, PM2 | C3 | This gene is a member of the ankyrin-repeat family and is induced by lipopolysaccharide (LPS). The C-terminal portion of the encoded product which contains the ankyrin repeats, shares high sequence similarity with the I kappa B family of proteins. The latter are known to play a role in inflammatory responses to LPS by their interaction with NF-B proteins through ankyrin-repeat domains. Studies in mouse indicate that this gene product is one of the nuclear I kappa B proteins and an activator of IL-6 production | |
| 3 | NM_014739.3 | BCLAF1 | Hmz | Intron 10 | c.2397+1G>C | - | splicing | - | - | PVS1, PM2 | C4 | This gene encodes a transcriptional repressor that interacts with several members of the BCL2 family of proteins. Overexpression of this protein induces apoptosis. The protein localizes to dot-like structures throughout the nucleus, and redistributes to a zone near the nuclear envelope. Diseases associated with BCLAF1 include Emery-Dreifuss Muscular Dystrophy and Uterine Adnexa Cancer. Among its related pathways are Interactome of polycomb repressive complex 2 (PRC2). |
| NM_002723 | PRB4 | Hmz | Exon 3 | c.363_364ins GACGACCC… | - | frameshift insertion | - | - | PVS1_Moderate + PM1 + PM2 + PP3 | - | This gene encodes a member of the heterogeneous family of basic, proline-rich, human salivary glycoproteins. The encoded preproprotein undergoes proteolytic processing to generate one or more mature peptides before secretion from the parotid glands. | |
| NM_000552 | VWF | Hmz | Exon 28 | c.4165_4166ins ACCAGCGAGGTC… | - | stopgain | - | - | PVS1 | - | This gene encodes a member of the heterogeneous family of basic, proline-rich, human salivary glycoproteins. The encoded preproprotein undergoes proteolytic processing to generate one or more mature peptides before secretion from the parotid glands. | |
| 2 * | NM_001077624.3 | ZNF846 | Het | c.885_886insGA | p.Tyr296AspfsTer63 | Frameshift | 0 | - | PM2 | 3 | This gene encodes a predicted protein to enable DNA-binding transcription repressor activity, RNA polymerase II-specific and RNA polymerase II transcription regulatory region sequence-specific DNA binding activity. Predicted to be involved in negative regulation of transcription by RNA polymerase II. | |
| 2 * | NM_001077624.3 | ZNF846 | Het | c.884C>A | Ser295Leu | Missense | 0.000008 | rs765545468 | PM2 | 3 | ||
| 3 * | M_001374736.1 | DST | Het | c.9207A>T | Arg3069Ser | Missense | 0 | rs1364606135 | PM2, BP1 | 3 | This gene encodes a member of the plakin protein family of adhesion junction plaque proteins. Multiple alternatively spliced transcript variants encoding distinct isoforms have been found for this gene, but the full-length nature of some variants has not been defined. It has been reported that some isoforms are expressed in neural and muscle tissue, anchoring neural intermediate filaments to the actin cytoskeleton, and some isoforms are expressed in epithelial tissue, anchoring keratin-containing intermediate filaments to hemidesmosomes. Consistent with the expression, mice defective for this gene show skin blistering and neurodegeneration. | |
| 3 * | M_001374736.1 | DST | Het | c.6371C>A | p.Ala2124Glu | Missense | PM2, BP1 | 3 |
| Case | Gene | Function |
|---|---|---|
| 1-2-3 | RAD17 | Cell cycle checkpoint protein. Essential for sustained cell growth, maintenance of chromosomal stability, and ATR-dependent checkpoint activation upon DNA damage. Has a weak ATPase activity required for binding to chromatin. May also serve as a sensor of DNA replication progression, and may be involved in homologous recombination. |
| 2-3 | NPRL3 | As a component of the GATOR1 complex functions as an inhibitor of the amino acid-sensing branch of the TORC1 pathway. Important role in cortical development. |
| 2-3 | BCLAF1 | Death-promoting transcriptional repressor. Bclaf1 Promotes Maintenance and Self-Renewal of Fetal Hematopoietic Stem Cells. |
| 3-1 | PRIM2 | Regulatory subunit of the DNA primase complex and component of the DNA polymerase alpha complex (also known as the alpha DNA polymerase-primase complex) which play an essential role in the initiation of DNA synthesis. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marzano, F.; Chiara, M.; Consiglio, A.; D’Amato, G.; Gentile, M.; Mirabelli, V.; Piane, M.; Savio, C.; Fabiani, M.; D’Elia, D.; et al. Whole-Exome and Transcriptome Sequencing Expands the Genotype of Majewski Osteodysplastic Primordial Dwarfism Type II. Int. J. Mol. Sci. 2023, 24, 12291. https://doi.org/10.3390/ijms241512291
Marzano F, Chiara M, Consiglio A, D’Amato G, Gentile M, Mirabelli V, Piane M, Savio C, Fabiani M, D’Elia D, et al. Whole-Exome and Transcriptome Sequencing Expands the Genotype of Majewski Osteodysplastic Primordial Dwarfism Type II. International Journal of Molecular Sciences. 2023; 24(15):12291. https://doi.org/10.3390/ijms241512291
Chicago/Turabian StyleMarzano, Flaviana, Matteo Chiara, Arianna Consiglio, Gabriele D’Amato, Mattia Gentile, Valentina Mirabelli, Maria Piane, Camilla Savio, Marco Fabiani, Domenica D’Elia, and et al. 2023. "Whole-Exome and Transcriptome Sequencing Expands the Genotype of Majewski Osteodysplastic Primordial Dwarfism Type II" International Journal of Molecular Sciences 24, no. 15: 12291. https://doi.org/10.3390/ijms241512291
APA StyleMarzano, F., Chiara, M., Consiglio, A., D’Amato, G., Gentile, M., Mirabelli, V., Piane, M., Savio, C., Fabiani, M., D’Elia, D., Sbisà, E., Scarano, G., Lonardo, F., Tullo, A., Pesole, G., & Faienza, M. F. (2023). Whole-Exome and Transcriptome Sequencing Expands the Genotype of Majewski Osteodysplastic Primordial Dwarfism Type II. International Journal of Molecular Sciences, 24(15), 12291. https://doi.org/10.3390/ijms241512291










