The Evolution of Life Is a Road Paved with the DNA Quadruplet Symmetry and the Supersymmetry Genetic Code
Abstract
1. Introduction
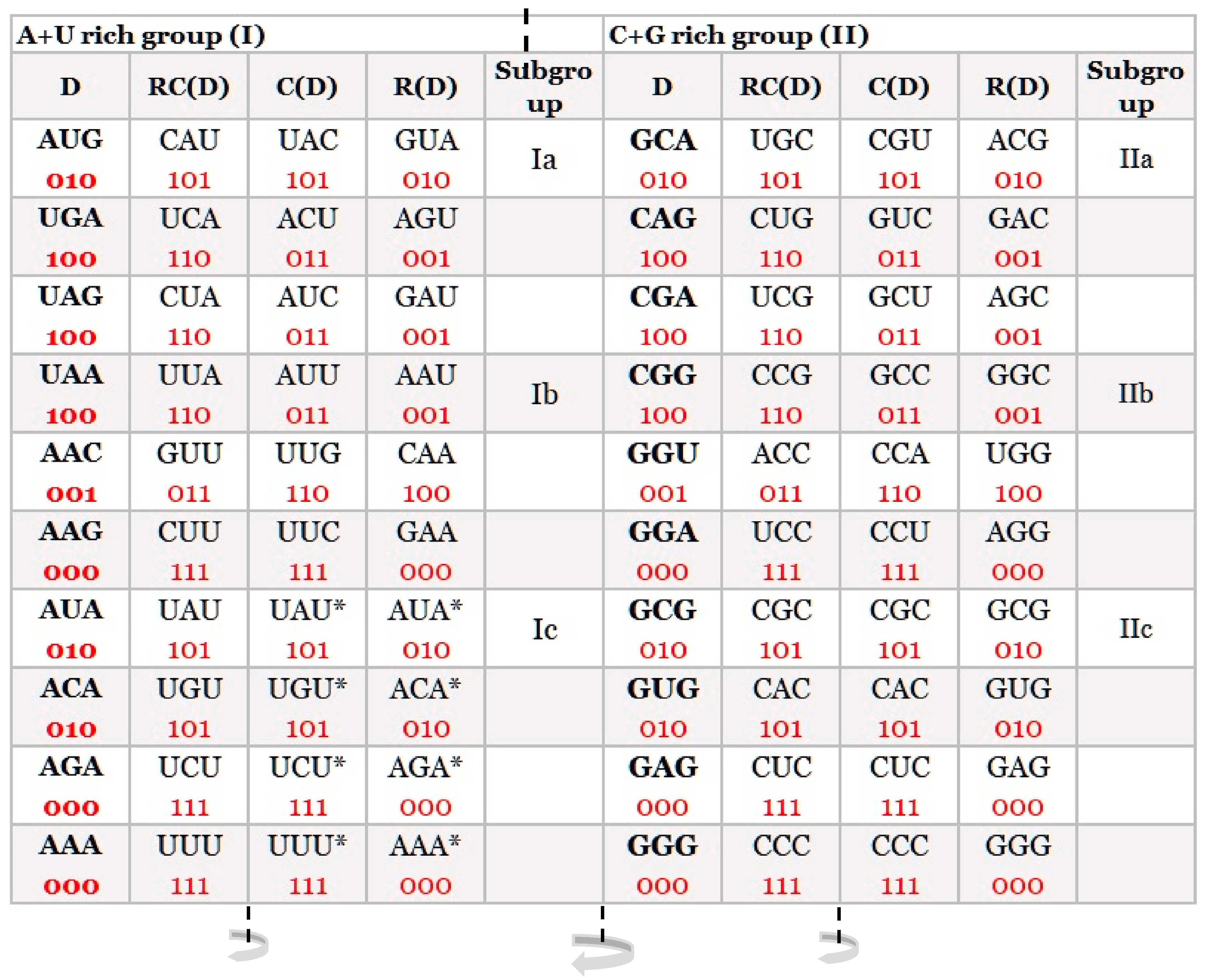
2. Classification of Trinucleotides/Codons
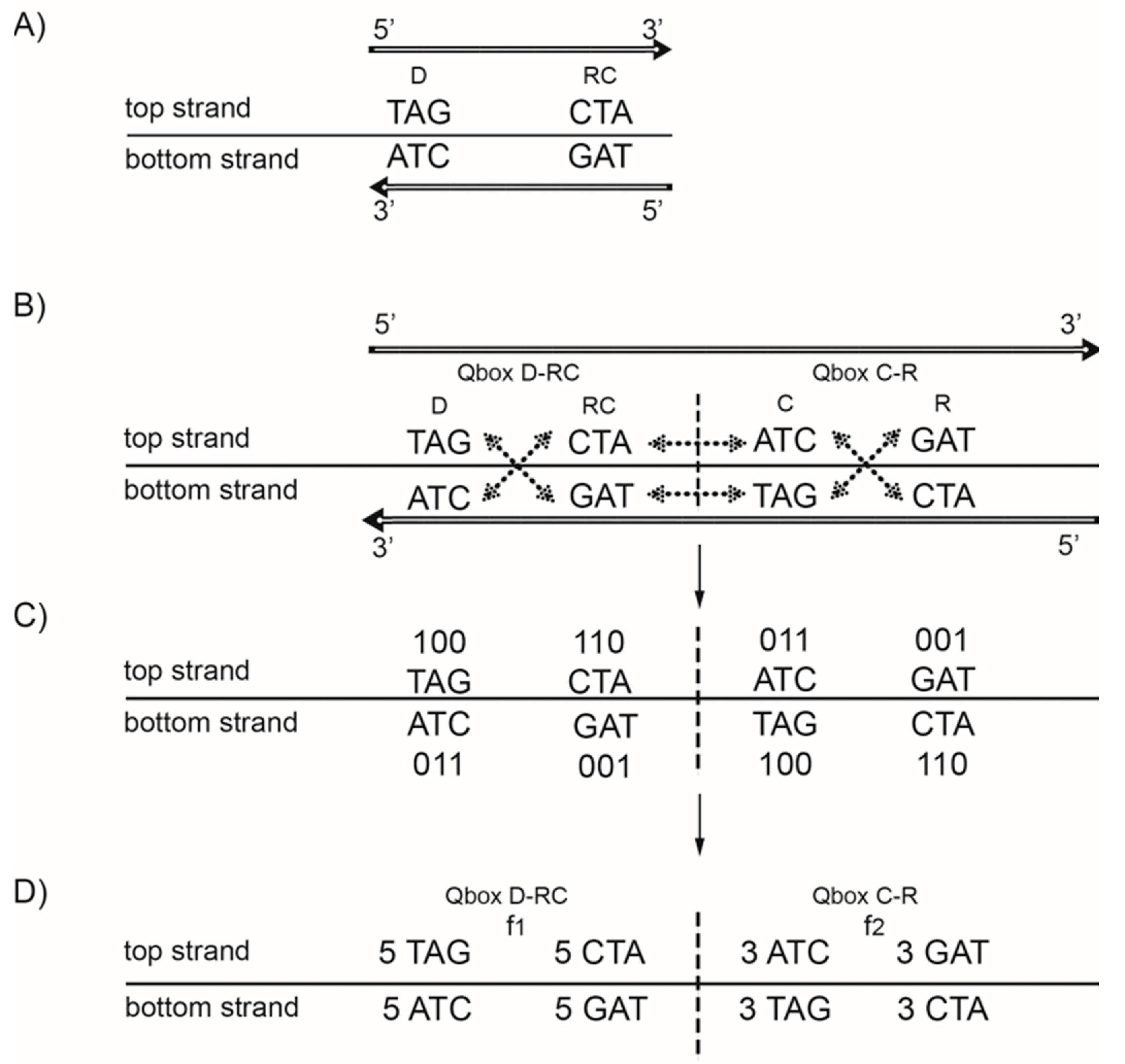
3. Symmetries of DNA Molecule—Chargaff’s First and Second Parity Rules
4. DNA Strand Symmetry/Chargaff’s Second Parity Rule (CSPR)
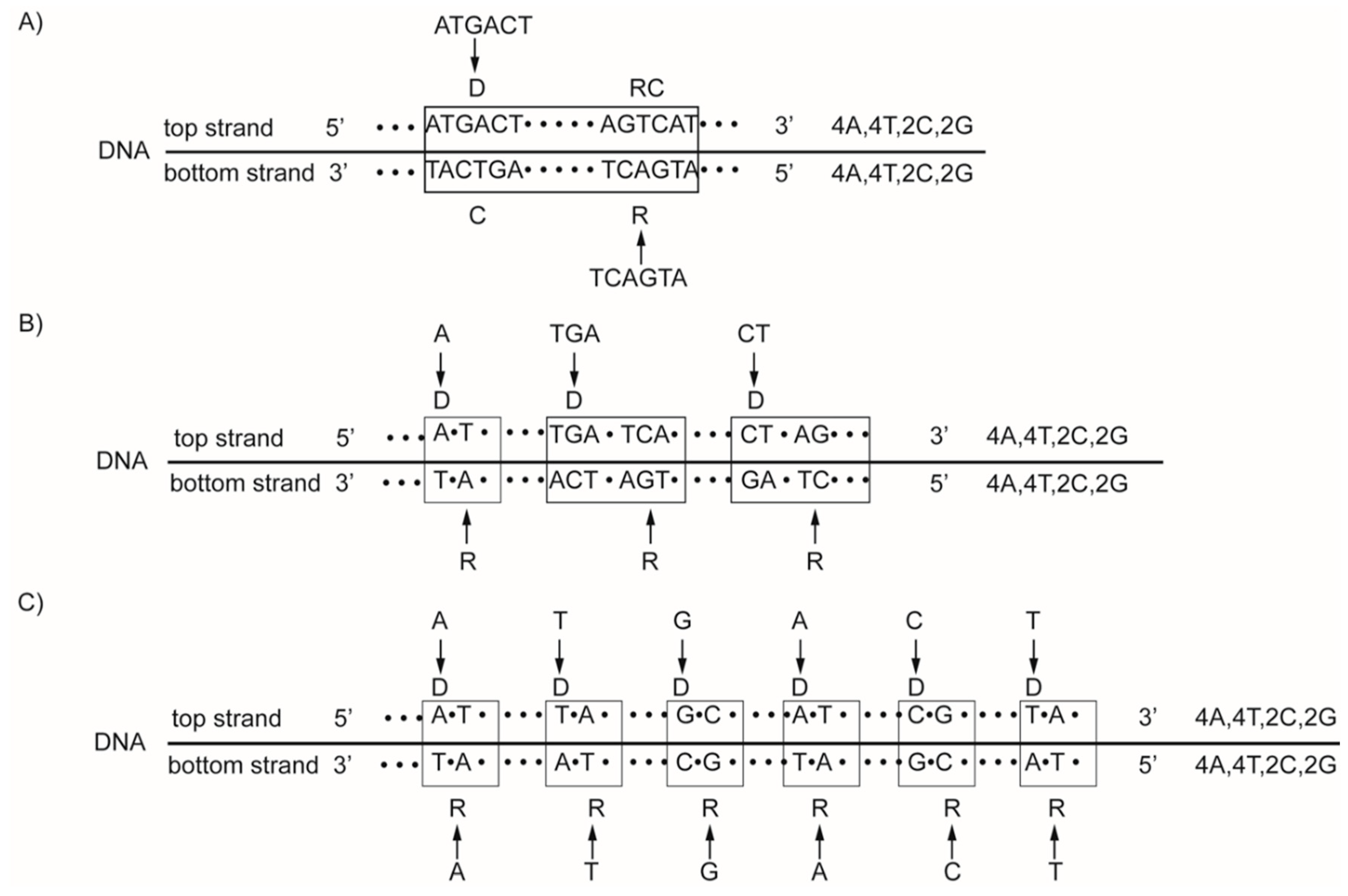
5. DNA Quadruplet Symmetry
6. The Natural Law of DNA Creation and Conservation
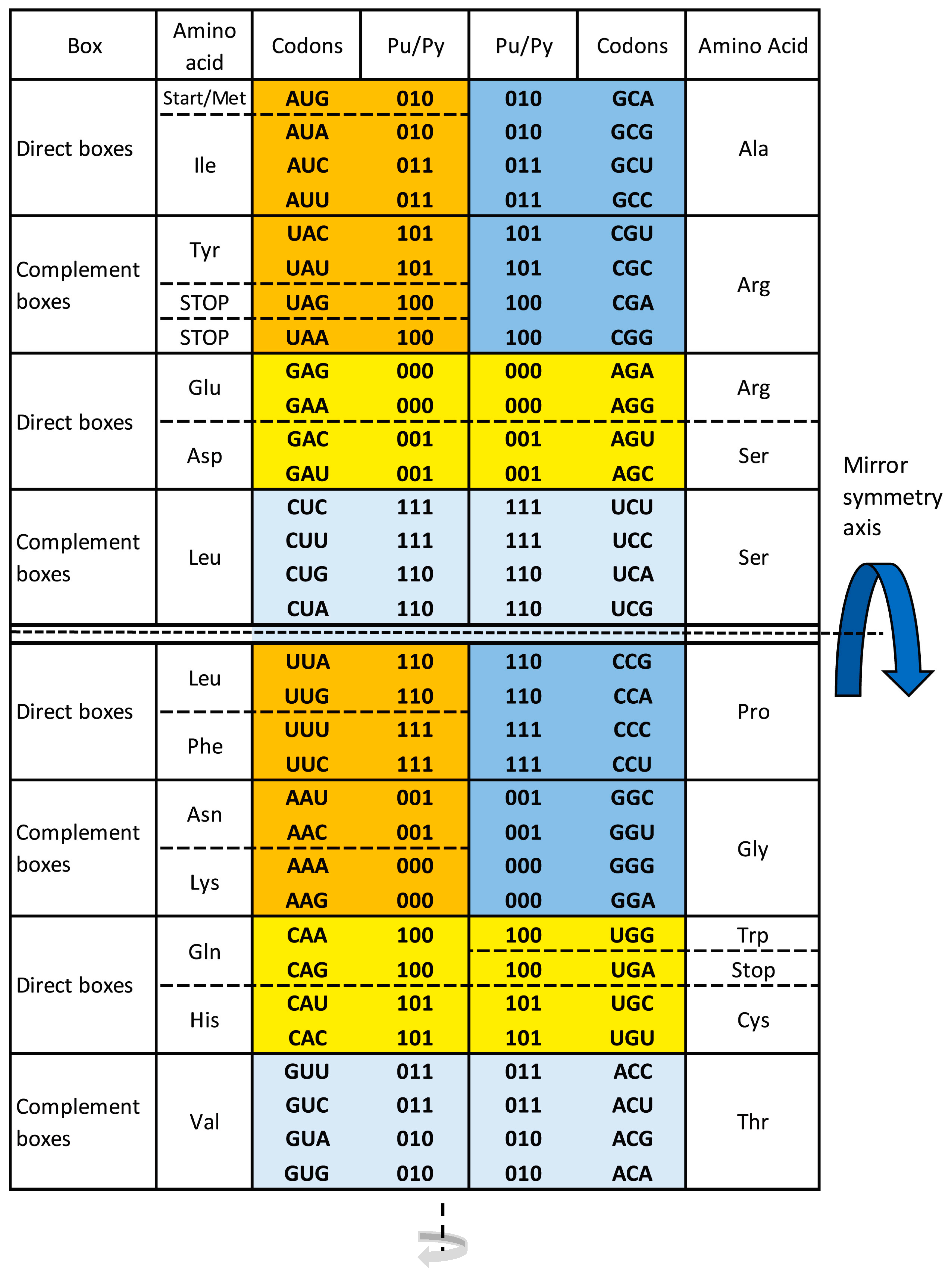
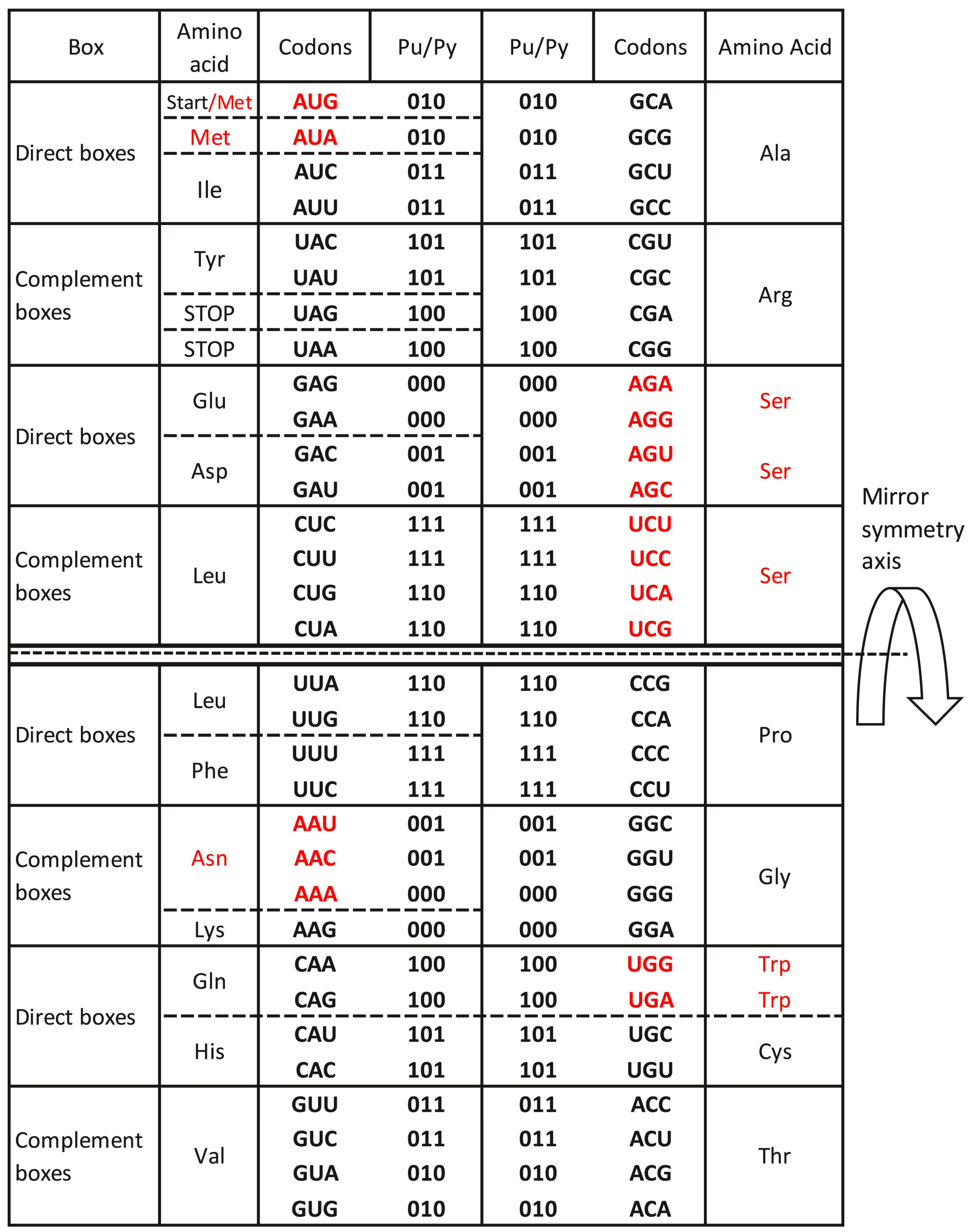
7. The Supersymmetry Genetic Code Table


8. During Sixty Years, What Has Hindered the Discovery of Complete Physicochemical Symmetries of the Genetic Code?
9. Discussion
- The SSyGC table starts with an AUG start signal.
- All 61 codons and three stop signals are arranged in 2 × 8 boxes, which alternate in each row on the principle of A + T rich and C + G rich codons but with the same purine and pyrimidine ordering.
- The ordering of purines and pyrimidines in both columns is identical.
- Vertically, between direct and complement boxes, purines, and pyrimidines as well as codons are regularly ordered on the principle of Watson–Crick pairing.
- Horizontally in the same row, purine in the first column transforms in the purine of the second column, and analogously pyrimidine transforms in pyrimidine, creating alternate A + T rich and C + G rich codons.
- In the SSyGC table, the codons of all amino acids are not scattered, including three sextets for Serine, Arginine, and Leucine.
- The SSyGC table has a central vertical and horizontal double mirror symmetry according to the mirror symmetry axis as well as a double mirror symmetry of DNA quadruplets and a classification of trinucleotides/codons.
- In this way, the physicochemical unique symmetry net of the whole SSyGC table is structured, creating full symmetries between bases, codons, and amino acids.
- The symmetry net is unique and common for all RNA and DNA living species on Earth.
- The symmetry net is also common for more than 30 nuclear and mitochondrial genetic codes, which differ from the Standard Genetic Code table.
- Due to the symmetry net, codons of the SSyGC table directly transform in the DNA molecule with Watson–Crick pairing (32 codons and 32 anticodons, direct and their complement, respectively).
- The unique symmetry net has remained unchanged during evolution and has the power of the natural law for the origin of life.
- The protection of the genetic code and DNA molecule symmetries during all of evolution reveals their role in decreasing entropy (disorder) and the preservation of species integrity.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chargaff, E. Structure and function of nucleic acids as cell constituents. Fed. Proc. 1951, 10, 654–659. [Google Scholar]
- Koonin, E.V. Are there laws of genome evolution? PLoS Comput. Biol. 2011, 7, 1002173. [Google Scholar] [CrossRef]
- Wigner, E.P. Problems of symmetry in old and new physics. Bull. Amer. Math. Soc. 1969, 75, 891. [Google Scholar]
- Wigner, E.P. Physics and the Explanation of Life. In Philosophical Foundations of Science; Seeger, R.J., Cohen, R.S., Eds.; Boston Studies in the Phylosophy of Science; Springer: Dordrecht, The Netherlands, 1969; Volume 11, pp. 119–132. [Google Scholar]
- Nöther, E. Invariante Variantionsprobleme. In Nachrichten von der Gesellschaft der Wissenschaften zu Göttingen, Mathematisch-Physikalische Klasse; University Göttingen: Göttingen, Germany, 1918; pp. 235–257. [Google Scholar]
- Gross, D.J. The role of symmetry in fundamental physics. Proc. Natl. Acad. Sci. USA 1996, 93, 14256–14259. [Google Scholar] [CrossRef]
- Monod, J. On symmetry and function in biological systems. In Selected Papers in Molecular Biology by Jacques Monod; Ullman, A., Ed.; Academic Press: New York, NY, USA, 1978. [Google Scholar]
- Rosandić, M.; Paar, V.; Glunčić, M. Fundamental role of start/stop regulators in whole DNA and new trinucleotide classification. Gene 2013, 531, 184–190. [Google Scholar] [CrossRef]
- Rosandić, M.; Vlahović, I.; Glunčić, M.; Paar, V. Trinucleotide’s quadruplet symmetries and natural symmetry law of DNA creation ensuing Chargaff’s second parity rule. J. Biomol. Struct. Dyn. 2016, 34, 1383–1394. [Google Scholar] [CrossRef]
- Rosandić, M.; Vlahović, I.; Pilaš, I.; Glunčić, M.; Paar, V. An Explanation of Exceptions from Chargaff’s Second Parity Rule/Strand Symmetry of DNA Molecules. Genes 2022, 13, 1929. [Google Scholar] [CrossRef]
- Rosandić, M.; Paar, V. Standard Genetic code vs. Supersymmetry Genetic Code–Alphabetical table vs. physicochemical table. BioSystems 2022, 14, 110748. [Google Scholar]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Rudner, R.; Karkas, J.D.; Chargaff, E. Separation of B. subtilis DNA into complementary strands. III. Direct analysis. Proc. Natl. Acad. Sci. USA 1968, 60, 921–922. [Google Scholar] [CrossRef]
- Prabhu, V.V. Symmetry observations in long nucleotide sequences. Nucleic Acids Res. 1993, 21, 2797–2800. [Google Scholar] [CrossRef]
- Baldi, B.; Brunak, S. Bioinformatics: The Machine Learning Approach; Cambridge Mass: London, UK, 2018. [Google Scholar]
- Qi, D.; Cuticchia, A.J. Compositional symmetries in complete genomes. Bioinformatics 2001, 17, 557–559. [Google Scholar] [CrossRef]
- Baisnee, P.F.; Hampson, S.; Baldi, B. Why are complementary DNA strands symmetric? Bioinformatics 2002, 18, 1021–1033. [Google Scholar] [CrossRef]
- Albrecht-Buehler, G. Inversions and inverted transpositions as the basis for an almost universal “format” of genome sequences. Genomics 2007, 90, 297–305. [Google Scholar] [CrossRef]
- Zhang, S.H.; Huang, Y.Z. Characteristics of oligonucleotide frequencies across genomes: Conservation versus variation, strand symmetry, and evolutionary implications. Nat. Proc. 2008, 125, 28. [Google Scholar] [CrossRef]
- Poudel, B.R.; Satapathy, S.S.; Kumar, A.; Jha, P.K.; Buragohain, A.K.; Borah, M.; Ray, S.K. A study in entire chromosomes of violations of the intra-strand parity of complementary nucleotides (Chargaff’s second parity rule). DNA Res. 2009, 16, 325–343. [Google Scholar] [CrossRef]
- Perez, J.C. Codon populations in single-stranded whole human genome DNA are fractal and fine-tuned by the golden ratio 1.618. Interdisc. Sci. Comput. Life Sci. 2010, 2, 228–240. [Google Scholar] [CrossRef]
- Hart, A.; Martinez, S. Statistical testing of Chargaff’s second parity rule in bacterial genome sequences. Stoch. Model. 2011, 27, 272–317. [Google Scholar] [CrossRef]
- Yamagishi, M.E.B.; Herai, R.H. Chargaff’s “grammar of biology”: New fractal-like rules. arXiv 2011, arXiv:1112.1528. [Google Scholar]
- Zhang, H.; Li, P.; Zhong, H.S.; Zhang, S.H. Conservation vs. variation of dinucleotide frequencies across bacterial and archaeal genomes: Evolutionary implications. Front. Microbiol. 2013, 4, 269. [Google Scholar]
- Maschner, M.; Schubert, I.; Scholz, U.; Friedel, S. Patterns of nucleotide asymmetries in plant and animal genomes. BioSystems 2013, 111, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, E.A.; Trifonov, E.N. Compensatory nature of Chargaff´s second parity rule. J. Biomol. Struct. Dyn. 2013, 31, 1324–1336. [Google Scholar] [CrossRef] [PubMed]
- Rosandić, M.; Vlahović, I.; Paar, V. Novel look at DNA and life–symmetry as evolutionary forcing. J. Theor. Biol. 2019, 485, 109985. [Google Scholar] [CrossRef]
- Glunčić, M.; Vlahović, I.; Rosandić, M.; Paar, V. Tandemly repeated NBPF HOR copies (Olduvai triplets): Possible impact on human brain evolution. Life Sci. Alliance 2022, e202101306. [Google Scholar] [CrossRef]
- Sueoka, N. Intra-strand parity rules of DNA base composition and usage biases of synonymous codons. J. Mol. Evol. 1995, 40, 318–325. [Google Scholar] [CrossRef]
- Lobry, J.R. Properties of a general model of DNA evolution under no-strand bias conditions. J. Mol. Evol. 1995, 40, 326–330. [Google Scholar] [CrossRef]
- Forsdyke, D.R.; Bell, S.J. Purine loading, stem-loops and Chargaff’s second parity rule: A discussion of the application of elementary principles to easy chemical observations. Appl. Bioinform. 2004, 3, 3–8. [Google Scholar] [CrossRef]
- Nirenberg, M.W.; Matthaei, J.H. The dependence of cell-free protein synthesis in E. coli upon naturally polyribonucleotides. Proceed. Nat. Acad. Sci. USA 1961, 47, 101588. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H. Evolution of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Rosandić, M.; Paar, V. The Novel Ideal Symmetry Genetic Code table–common purine-pyrimidine symmetry net for all RNA and DNA species. J. Theor. Biol. 2021, 524, 110748. [Google Scholar] [CrossRef]
- Koonin, E.V.; Novozhilov, A.S. Origin and evolution of the genetic code: The universal enigma. NIH Public Access 2009, 61, 99–111. [Google Scholar] [CrossRef]
- Freeland, S.J.; Hurst, I.D. The genetic code is one in a million. J. Mol. Evol. 1998, 47, 238–248. [Google Scholar] [CrossRef]
- Bashford, J.D.; Tsohantjis, I.; Jarvis, P.D. A supersymmetric model for the evolution on the genetic code. Proc. Natl. Acad. Sci. USA 1998, 95, 987–992. [Google Scholar] [CrossRef]
- Ahmed, A.; Frey, G.; Michel, C.J. Essential molecular functions associated with circular code evolution. J. Theor. Biol. 2010, 264, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Michael, C.J. The maximal C3 self-complementary trinucleotide circular code X in genes of bacteria, archaea, eukaryotes, plasmids and viruses. Life 2017, 7, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Castro-Chavez, F.A. A tetrahedral representation of the genetic code emphasizing aspects of symmetry. BIOcomplexity 2012, 2, 1–6. [Google Scholar]
- Nemzer, A. A binary representation of the genetic code. BioSystems 2017, 155, 10–19. [Google Scholar] [CrossRef]
- Shu, J.-J. A new integrated symmetrical table for genetic codes. BioSystems 2017, 151, 21–26. [Google Scholar] [CrossRef][Green Version]
- Saier, M.H. Understanding the genetic code. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed]
- Štambuk, N.; Konjevoda, N. Determining amino acid scores of the genetic code table: Complementarity, structure function and evolution. BioSystems 2020, 187, 104026. [Google Scholar] [CrossRef]
- Di Giulio, M. The theory of the genetic code and origin of ideas. J. Theor. Biol. 2021, 516, 110615. [Google Scholar] [CrossRef]
- Barbieri, M. Introduction to code biology. Biosemiotics 2014, 7, 167–179. [Google Scholar] [CrossRef][Green Version]
- Findley, G.L.; Findley, A.M.; McGlynn, S.P. Symmetry characteristics of the genetic code. Proc. Natl. Acad. Sci. USA 1982, 79, 7061–7065. [Google Scholar] [CrossRef]
- Hornos, J.E.M.; Hornos, Y.M.M. Algebraic model for the evolution of the genetic code. Phys. Rev. Lett. 1993, 71, 4401. [Google Scholar] [CrossRef]
- Hornos, J.E.M.; Hornos, Y.M.M.; Forger, M. Symmetry and symmetry breaking: An algebraic approach to the genetic code. Int. J. Mod. Phys. 1999, 13, 2795–2885. [Google Scholar] [CrossRef]
- Meddox, J. The genetic code by numbers. Nature 1994, 367, 111. [Google Scholar] [CrossRef] [PubMed]
- Forger, M.; Hornos YM, M.; Hornos JE, M. Global aspects in the algebraic approach to the genetic code. Rev. E Phys. 1997, 56, 7078–7082. [Google Scholar] [CrossRef]
- Kent, R.D.; Schlesinger, M.; Wybourne, B.G. On algebraic approaches to the genetic code. Canad. J. Phys. 1998, 76, 445–452. [Google Scholar] [CrossRef]
- Antoneli, F.; Forger, N. Symmetry breaking in the genetic code: Finite groups. Math. Comput. Model. 2011, 53, 1469–1488. [Google Scholar] [CrossRef]
- Lenstra, R. Evolution of the genetic code through progressive symmetry breaking. J. Theor. Biol. 2014, 347, 95–108. [Google Scholar] [CrossRef]
- Yang, Y. Algebraic principle of natural bases from the standard genetic code degeneracy. bioRxiv 2023. [Google Scholar] [CrossRef]
- Gonzalez, G.L.; Giannerini, S.; Rosa, R. On the origin of degeneracy in the genetic code. Interface Focus 2019, 9, 20190038. [Google Scholar] [CrossRef] [PubMed]
- Rosandić, M.; Paar, V. Codon sextets with leading role of serine create “ideal” symmetry classification scheme of the genetic code. Gene 2014, 543, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Novozhilov, A.S. Origin and Evolution of the Universal Genetic Code. Annu. Rev. Genet. 2017, 51, 45–62. [Google Scholar] [CrossRef]
- Barbieri, M. What is code biology? BioSystems 2017, 164, 1–10. [Google Scholar] [CrossRef]
- Konjevoda, P.; Štambuk, N. Relational model of the standard genetic code. Biosystems 2021, 210, 104529. [Google Scholar] [CrossRef]
- Lei, L.; Burton, Z.F. Evolution of Life on Earth: tRNA, Aminoacyl-tRNA Synthetases and the Genetic Code. Life 2020, 10, 21. [Google Scholar] [CrossRef]
- Ikehara, K. Towards Revealing the Origin of Life–Presenting the GADV Hypothesis; Springer Nature: Cham, Switzerland, 2021. [Google Scholar]
- Ikehara, K. Why Were [GADV]-amino Acids and GNC Codons Selected and How Was GNC Primeval Genetic Code Established? Genes 2023, 14, 375. [Google Scholar] [CrossRef]
- Wong, J.T.; Ng, S.K.; Mat, W.K.; Hu, T.; Xue, H. Coevolution theory of the genetic code at age forty: Pathway to translation and the synthetic life. Life 2016, 6, 12. [Google Scholar] [CrossRef]
- Fredens, J.; Wang, K.; de la Torre, D.; Funke, L.F.H.; Robertson, W.E.; Christova, Y.; Chia, T.; Schmied, W.H.; Dunkelmann, D.L.; Beranek, V.; et al. Total synthesis of Escherichia coli with a recoded genome. Nature 2019, 569, 514–518. [Google Scholar] [CrossRef]
- Bito, T.; Okumura, E.; Fujishima, M.; Watanabe, F. Potential of Chlorella as a Dietary Supplement to Promote Human Health. Nutrients 2020, 12, 2524. [Google Scholar] [CrossRef]
- José, M.V.; Zamudio, G.S. Symmetrical Properties of Graph Representations of Genetic Codes: From Genotype to Phenotype. Symmetry 2018, 10, 388. [Google Scholar] [CrossRef]
- Błażej, P.; Kowalski, D.R.; Mackiewicz, D.; Wnetrzak, M.; Aloqalaa, D.A.; Mackiewicz, P. The structure of the genetic code as an optimal graph clustering problem. J. Math. Biol. 2022, 85, 9. [Google Scholar] [CrossRef]
- Fimmel, E.; Strüngmann, L. Mathematical fundamentals for the noise immunity of the genetic code. Biosystems 2018, 164, 186–198. [Google Scholar] [CrossRef]
- Seligmann, H. Bijective codon transformations show genetic code symmetries centred on cytosine’s coding properties. Theory Biosci. 2017, 137, 17–31. [Google Scholar] [CrossRef]
- Tripathi, S.; Deem, M.W. The standard genetic code facilities exploration of the space of functional nucleotide sequences. J. Mol. Evol. 2018, 86. [Google Scholar] [CrossRef] [PubMed]
- Hornos, Y.E.M.; Braggion, L.; Magini, M.; Forger, M. Summary presentation in the evolution of the genetic code. IUBMB Life 2004, 56, 125–130. [Google Scholar] [CrossRef]
- Aloqalaa, D.A.; Kowalski, D.R.; Błażej, P.; Wnȩtrzak, M.; Mackiewicz, D.; Mackiewicz, P. The Properties of the Standard Genetic Code and Its Selected Alternatives in Terms of the Optimal Graph Partition. Biomed. Eng. Syst. Technol. 2020, 1211, 170–191. [Google Scholar] [CrossRef]
- Gonzalez, D.L.; Giannerini, S.; Rosa, R. Rumer’s transformation: A symmetry puzzle standing for half a century. Biosystems 2019, 187, 104036. [Google Scholar] [CrossRef] [PubMed]
- Fimmel, E.; Strüngmann, L. Linear codes and the mitochondrial genetic code. Biosystems 2019, 184, 103990. [Google Scholar] [CrossRef] [PubMed]
- Błażej, P.; Wnętrzak, M.; Mackiewicz, D.; Gagat, P.; Mackiewicz, P. Many alternative and theoretical genetic codes are more robust to amino acid replacements than the standard genetic code. J. Theor. Biol. 2019, 464, 21–32. [Google Scholar] [CrossRef]
- Radavičius, M.; Rekašius, T.; Židanavičiūtė, J. Local Symmetry of Non-Coding Genetic Sequences. Informatica 2019, 30, 553–571. [Google Scholar] [CrossRef]
- Sergienko, I.V.; Biletskyy, B.A.; Gupal, A.M.; Gupal, M.A. Optimal Noise-Immune Genetic Codes. Cybern. Syst. Anal. 2019, 55, 34–39. [Google Scholar] [CrossRef]
- Planat, M.; Aschheim, R.; Amaral, M.M.; Fang, F.; Irwin, K. Complete Quantum Information in the DNA Genetic Code. Symmetry 2020, 12, 1993. [Google Scholar] [CrossRef]
- Wang, B. The Pattern of Occurrence of Cytosine in the Genetic Code Minimizes Deleterious Mutations and Favors Proper Function of the Translational Machinery. Open J. Genet. 2020, 10, 8–15. [Google Scholar] [CrossRef]
- Petoukhov, S.V.; Verevkin, V.V. Bisymmetric matrices and symmetrologic researches in genetic biomechanics. Symmetry Cult. Sci. 2020, 31, 105–115. [Google Scholar] [CrossRef]
- Shulgina, Y.; Eddy, S.R. A computational screen for alternative genetic codes in over 250,000 genomes. Elife 2021, 10, e71402. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Nagao, A. Genetic Code and Its Variations. Encycl. Life Sci. 2021, 2, 147–157. [Google Scholar] [CrossRef]
- Yarus, M. Evolution of the Standard Genetic Code. J. Mol. Evol. 2021, 89, 19–44. [Google Scholar] [CrossRef] [PubMed]
- Jarus, M. A crescendo of competent coding (c3) contains the Standard Genetic Code. RNA 2022, 28, 1337–1347. [Google Scholar] [CrossRef]
- Wnetrzak, M.; Błażej, P.; Mackiewicz, P. Properties of the Standard Genetic Code and Its Alternatives Measured by Codon Usage from Corresponding Genomes. Bioinformatics 2020, 3, 44–51. [Google Scholar] [CrossRef]
- Karasev, V.A. The Canonical Table of the Genetic Code as a periodic system of triplets. Biosystems 2022, 214, 104636. [Google Scholar] [CrossRef]
- He, M. Symmetry and asymmetry in bioinformatics – from genetic code to life. Symmetry Cult. Sci. 2021, 32, 292. [Google Scholar] [CrossRef]
- Lei, L.; Burton, Z.F. Evolution of the genetic code. Transcription 2021, 12, 28–53. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, J. Is the Genetic Code Optimized for Resource Conservation? Mol. Biol. Evol. 2021, 38, 5122–5126. [Google Scholar] [CrossRef]
- Parvathy, S.T.; Udayasuriyan, V.; Bhadana, V. Codon usage bias. Mol. Biol. Rep. 2021, 49, 539–565. [Google Scholar] [CrossRef]
- Taylor, N.C.; Nikić-Spiegel, I. Film-like organelles equip cells with multiple genetic codes. Trends Biochem. Sci. 2021, 47, 369–371. [Google Scholar] [CrossRef]
- Qin, X.; Liu, T. Recent Advances in Genetic Code Expansion Techniques for Protein Phosphorylation Studies. J. Mol. Biol. 2022, 434, 167406. [Google Scholar] [CrossRef]
- Guo, J.; Niu, W. Genetic Code Expansion Through Quadruplet Codon Decoding. J. Mol. Biol. 2022, 434, 167346. [Google Scholar] [CrossRef]
- Michel, C.J.; Sereni, J.-S. Trinucleotide k-circular codes II: Biology. Biosystems 2022, 217, 104668. [Google Scholar] [CrossRef]
- Kimoto, M.; Hirao, I. Genetic Code Engineering by Natural and Unnatural Base Pair Systems for the Site-Specific Incorporation of Non-Standard Amino Acids into Proteins. Front. Mol. Biosci. 2022, 9, 851646. [Google Scholar] [CrossRef]
- Minton, K. Locking in a synthetic genetic code. Nat. Rev. Genet. 2022, 24, 1. [Google Scholar] [CrossRef] [PubMed]
- Mishra, D.B.; Acharya, B.; Rath, D.; Gerogiannis, V.C.; Kanavos, A. A Novel Real Coded Genetic Algorithm for Software Mutation Testing. Symmetry 2022, 14, 1525. [Google Scholar] [CrossRef]
- Di Giulio, M. The error minimization of the genetic code would have been determined by natural selection and not by a neutral evolution. Biosystems 2023, 224, 104838. [Google Scholar] [CrossRef]
- Seki, M. On the origin of the genetic code. Genes Genet. Syst. 2023, 98, 9–24. [Google Scholar] [CrossRef]
- Sakamoto, K. Genetic Code Expansion: Another Solution to Codon Assignments. Int. J. Mol. Sci. 2022, 24, 361. [Google Scholar] [CrossRef] [PubMed]
- Joiret, M.; Leclercq, M.; Lambrechts, G.; Rapino, F.; Close, P.; Louppe, G.; Geris, L. Cracking the genetic code with neural networks. Front. Artif. Intell. 2023, 6, 1128153. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, T.; Nagashima, K.; Nakai, I.; Young, E.D.; Abe, Y.; Aleon, J.; O’D Alexander, C.M.; Amari, A.S.; Amelin, Y.; Bajo, K.i.; et al. Samples returned from the asteroid Ryugu are similar to Ivuna–type carbonaceous meteorites. Science 2022, 379, eabn7850. [Google Scholar] [CrossRef]
- Nakamura, E.; Kobayashi, K.; Tanaka, R.; Hunihiro, T.; Kitagawa, H.; Potiszil, C.; Ota, T.; Sakaguchi, C.; Yamanaka, M.; Ratnayake, D.M.; et al. On the origin and evolution of the asteroid Ryugu: A comprehensive geochemical perspective. Proc. Jpn. Acad. 2022, 98, 227–282. [Google Scholar] [CrossRef]
- Liu, Y.; Zhong, X.; Luo, Z.; Meng, X.; Li, R.; Zhong, W.; Yang, L.; Wang, H.; Wei, D. The identification of a robust leucine dehyd rogenase from a directed soil metagenome for efficient synthesis of L-2-aminobutyric acid. Biotechnol. J. 2023, e2200590. [Google Scholar] [CrossRef]
- Rosandić, M. Ryugu amino acid samples support genetic code supersymmetry. Science 2022, 379, 6334. [Google Scholar]
- José, M.V.; Zamudio, G.S.; Morgado, E.R. A unified model of the standard genetic code. R. Soc. Open Sci. 2017, 4, 160908. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosandić, M.; Paar, V. The Evolution of Life Is a Road Paved with the DNA Quadruplet Symmetry and the Supersymmetry Genetic Code. Int. J. Mol. Sci. 2023, 24, 12029. https://doi.org/10.3390/ijms241512029
Rosandić M, Paar V. The Evolution of Life Is a Road Paved with the DNA Quadruplet Symmetry and the Supersymmetry Genetic Code. International Journal of Molecular Sciences. 2023; 24(15):12029. https://doi.org/10.3390/ijms241512029
Chicago/Turabian StyleRosandić, Marija, and Vladimir Paar. 2023. "The Evolution of Life Is a Road Paved with the DNA Quadruplet Symmetry and the Supersymmetry Genetic Code" International Journal of Molecular Sciences 24, no. 15: 12029. https://doi.org/10.3390/ijms241512029
APA StyleRosandić, M., & Paar, V. (2023). The Evolution of Life Is a Road Paved with the DNA Quadruplet Symmetry and the Supersymmetry Genetic Code. International Journal of Molecular Sciences, 24(15), 12029. https://doi.org/10.3390/ijms241512029





