Herpes Simplex Virus Infection Alters the Immunological Properties of Adipose-Tissue-Derived Mesenchymal-Stem Cells
Abstract
1. Introduction
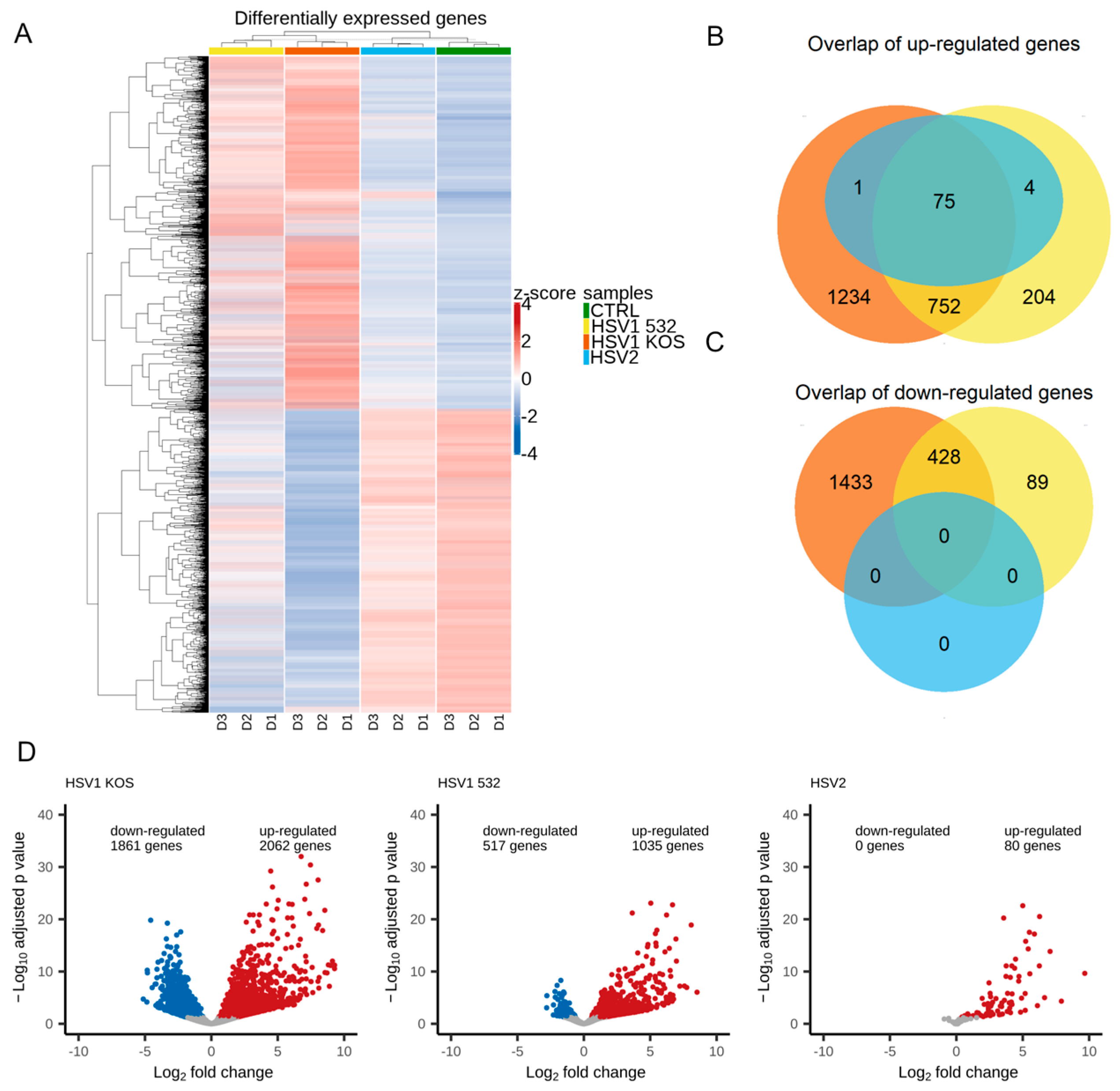
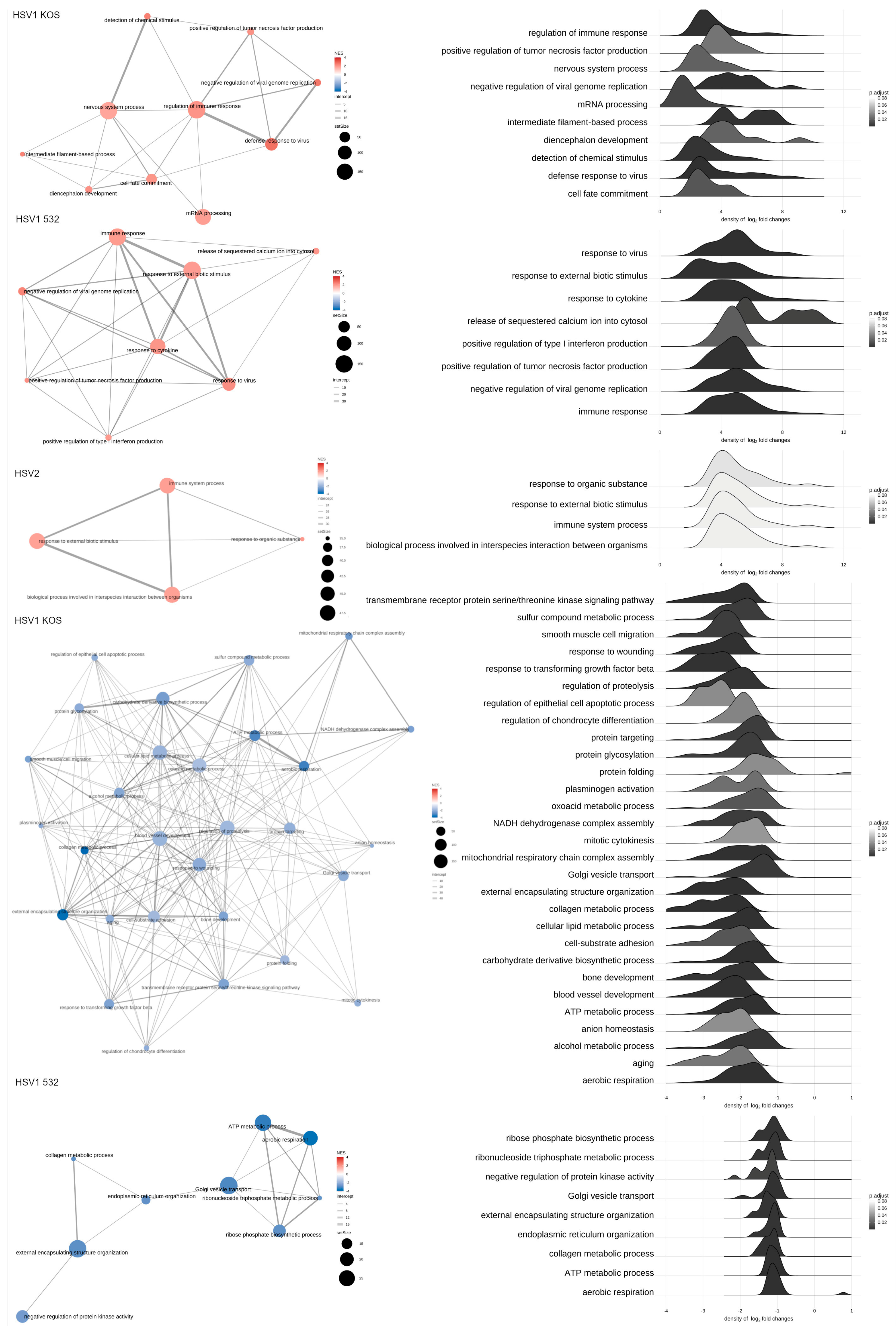
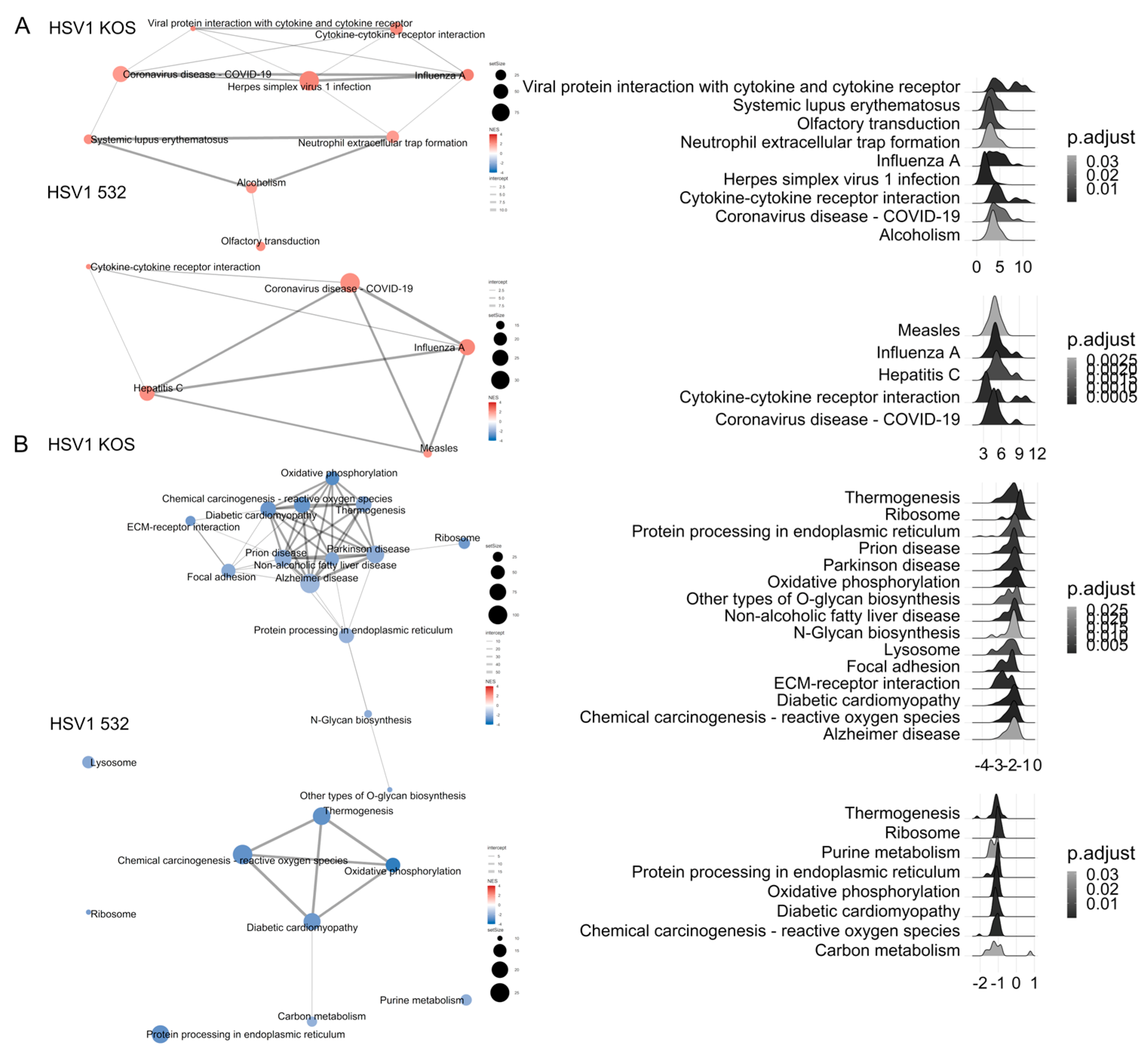
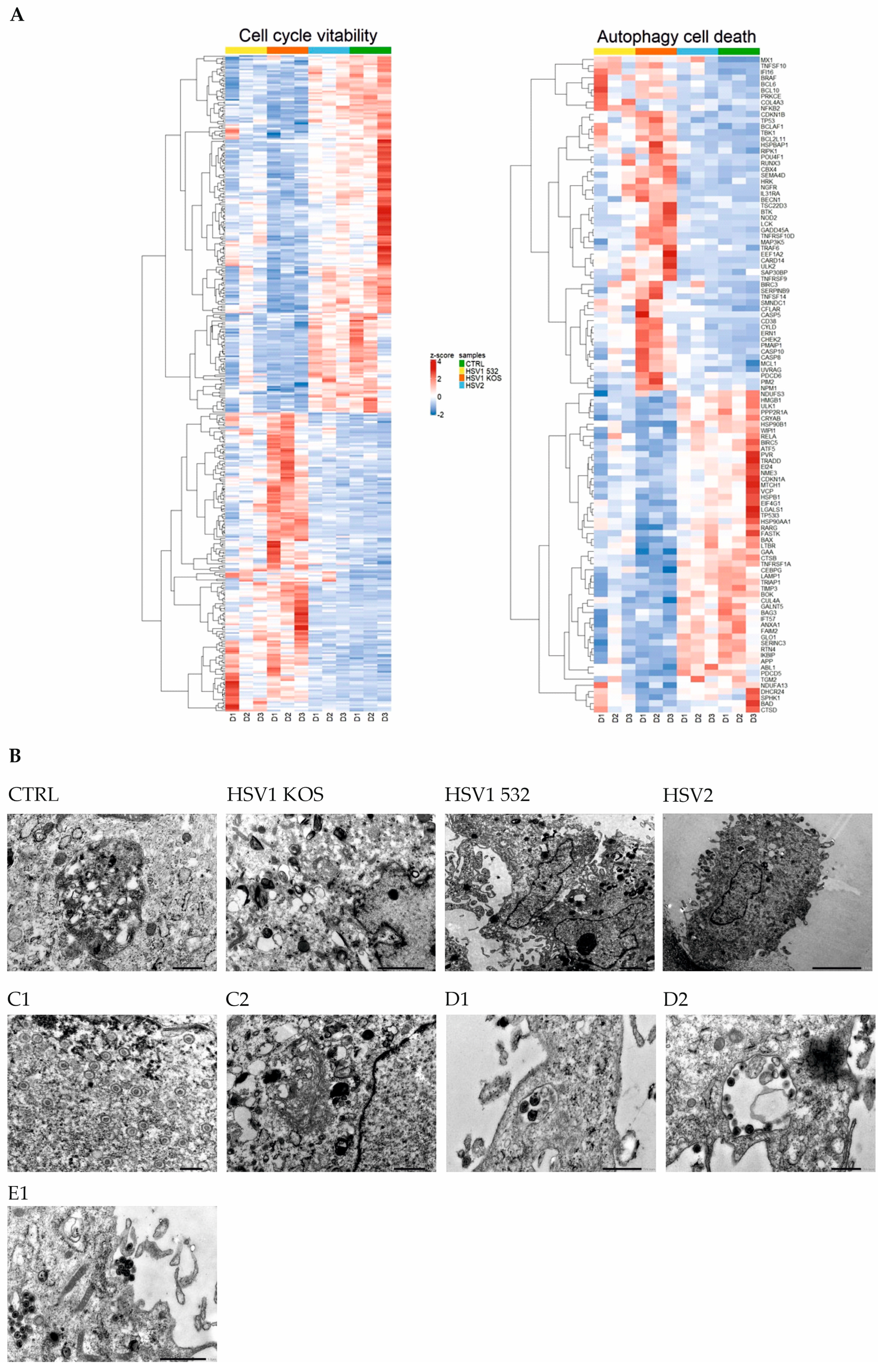
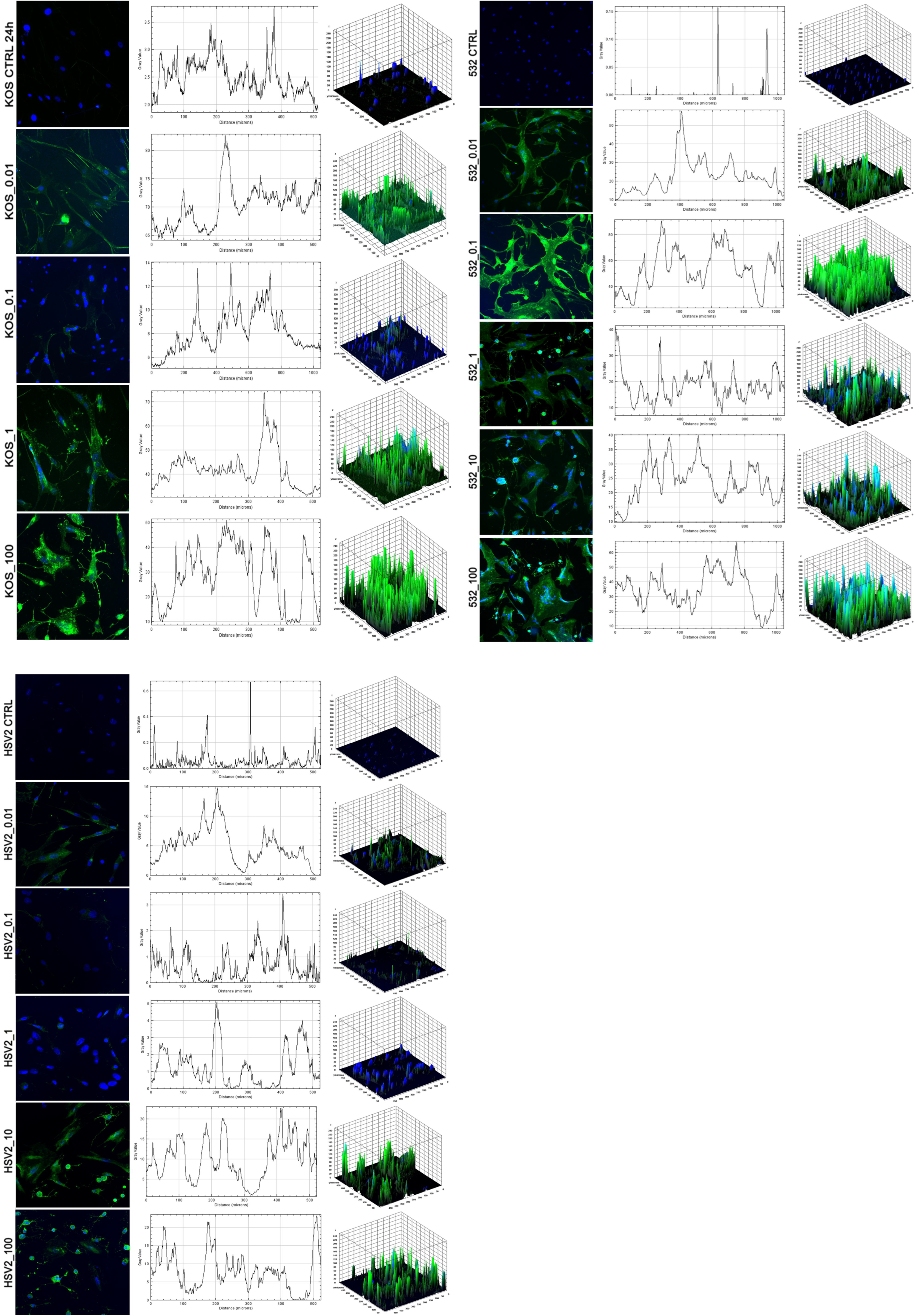
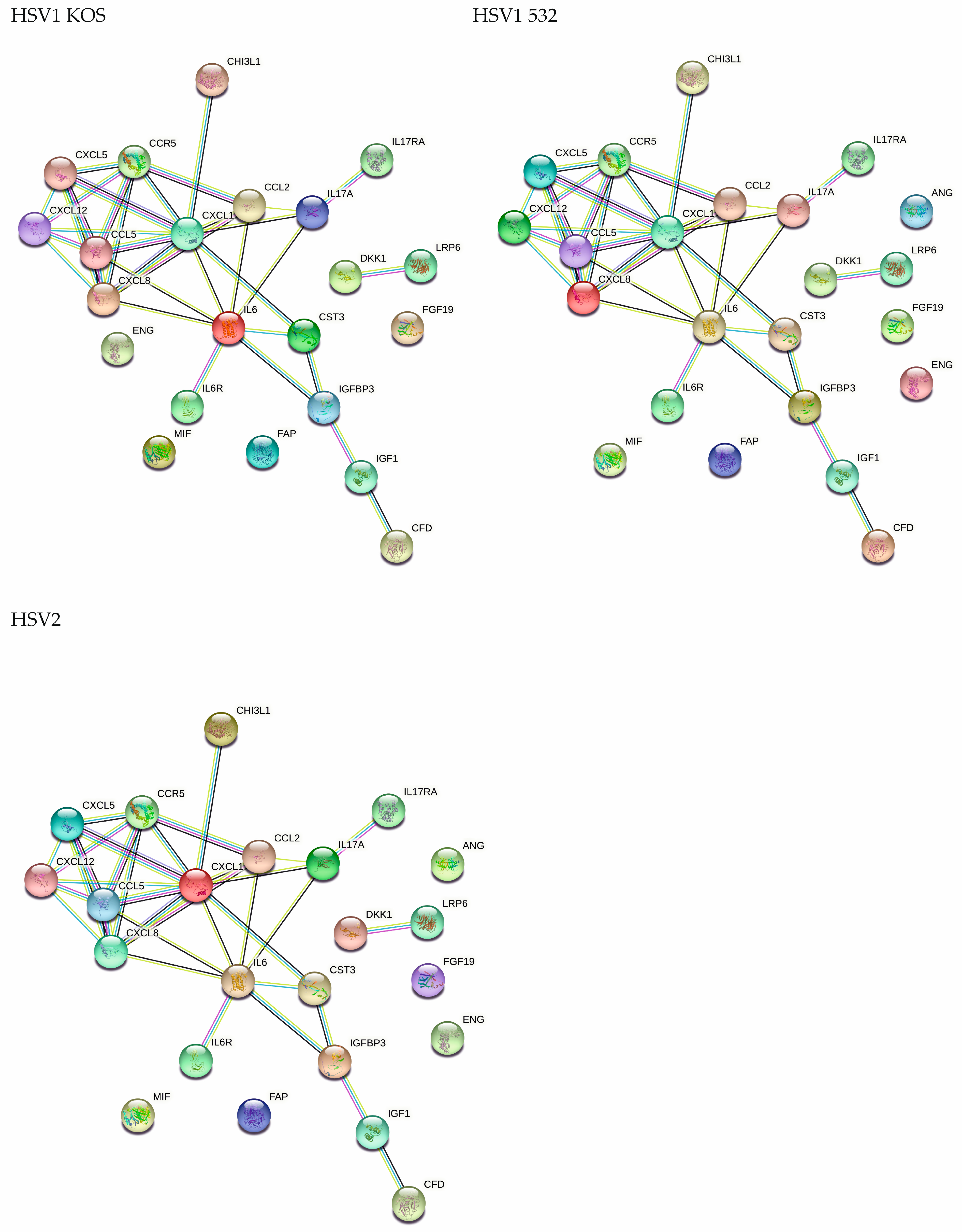
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. HSV Infection of Adipose-Derived Mesenchymal Stem Cellls
4.3. RNA-Seq Methods and Data Analysis
4.4. Fluorescent Microscopy
4.5. Quantification of Virus-Mediated Morphological Changes
4.6. Flow Cytometry
4.7. TEM Analysis
4.8. Detection of Autophagic Flux
4.9. Western Blot Analysis of LC3
4.10. Protein Array for the Detection of Cytokines
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, X.; Jiang, J.; Gu, Z.; Zhang, J.; Chen, Y.; Liu, X. Mesenchymal stromal cell therapies: Immunomodulatory properties and clinical progress. Stem Cell Res. Ther. 2020, 11, 345. [Google Scholar] [CrossRef] [PubMed]
- Shiley, K.; Blumberg, E. Herpes viruses in transplant recipients: HSV, VZV, human herpes viruses, and EBV. Hematol./Oncol. Clin. 2011, 25, 171–191. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Dummer, J.S.; Kusne, S.; Breinig, M.K.; Armstrong, J.A.; Makowka, L.; Starzl, T.E.; Ho, M. Infections with cytomegalovirus and other herpesviruses in 121 liver transplant recipients: Transmission by donated organ and the effect of OKT3 antibodies. J. Infect. Dis. 1988, 158, 124–131. [Google Scholar] [CrossRef]
- Lohr, J.M. Gastric herpes simplex virus type 1 infection is associated with functional gastrointestinal disorders in the presence and absence of comorbid fibromyalgia. Infection 2022, 50, 1043. [Google Scholar] [CrossRef] [PubMed]
- Jacob, H.S.; Visser, M.; Key, N.S.; Goodman, J.L.; Moldow, C.F.; Vercellotti, G.M. Herpes virus infection of endothelium: New insights into atherosclerosis. Trans. Am. Clin. Climatol. Assoc. 1992, 103, 95–104. [Google Scholar]
- Jung, S.H.; Lee, K.T. Atherosclerosis by Virus Infection-A Short Review. Biomedicines 2022, 10, 2634. [Google Scholar] [CrossRef] [PubMed]
- Dummer, J.S.; Armstrong, J.; Somers, J.; Kusne, S.; Carpenter, B.J.; Rosenthal, J.T.; Ho, M. Transmission of infection with herpes simplex virus by renal transplantation. J. Infect. Dis. 1987, 155, 202–206. [Google Scholar] [CrossRef]
- Koneru, B.; Tzakis, A.G.; DePuydt, L.E.; Demetris, A.J.; Armstrong, J.A.; Dummer, J.S.; Starzl, T.E. Transmission of fatal herpes simplex infection through renal transplantation. Transplantation 1988, 45, 653–656. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Pillay, D.; Ratcliffe, D.; Cane, P.A.; Collingham, K.E.; Milligan, D.W. Resistance to antiviral drugs in herpes simplex virus infections among allogeneic stem cell transplant recipients: Risk factors and prognostic significance. J. Infect. Dis. 2000, 181, 2055–2058. [Google Scholar] [CrossRef][Green Version]
- Balfour, H.H., Jr.; Chace, B.A.; Stapleton, J.T.; Simmons, R.L.; Fryd, D.S. A randomized, placebo-controlled trial of oral acyclovir for the prevention of cytomegalovirus disease in recipients of renal allografts. N. Engl. J. Med. 1989, 320, 1381–1387. [Google Scholar] [CrossRef]
- Ariza-Heredia, E.J.; Chemaly, R.F.; Shahani, L.R.; Jang, Y.; Champlin, R.E.; Mulanovich, V.E. Delay of alternative antiviral therapy and poor outcomes of acyclovir-resistant herpes simplex virus infections in recipients of allogeneic stem cell transplant—A retrospective study. Transpl. Int. 2018, 31, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Malkin, J.E. Epidemiology of genital herpes simplex virus infection in developed countries. Herpes J. IHMF 2004, 11 (Suppl. S1), 2A–23A. [Google Scholar]
- Malkin, J.E.; Morand, P.; Malvy, D.; Ly, T.D.; Chanzy, B.; de Labareyre, C.; El Hasnaoui, A.; Hercberg, S. Seroprevalence of HSV-1 and HSV-2 infection in the general French population. Sex. Transm. Infect. 2002, 78, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Vyse, A.J.; Gay, N.J.; Slomka, M.J.; Gopal, R.; Gibbs, T.; Morgan-Capner, P.; Brown, D.W. The burden of infection with HSV-1 and HSV-2 in England and Wales: Implications for the changing epidemiology of genital herpes. Sex. Transm. Infect. 2000, 76, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Sauerbrei, A.; Schmitt, S.; Scheper, T.; Brandstadt, A.; Saschenbrecker, S.; Motz, M.; Soutschek, E.; Wutzler, P. Seroprevalence of herpes simplex virus type 1 and type 2 in Thuringia, Germany, 1999 to 2006. Eurosurveillance 2011, 16, 20005. [Google Scholar] [CrossRef]
- Alareeki, A.; Osman, A.M.M.; Khandakji, M.N.; Looker, K.J.; Harfouche, M.; Abu-Raddad, L.J. Epidemiology of herpes simplex virus type 2 in Europe: Systematic review, meta-analyses, and meta-regressions. Lancet Reg. Health-Eur. 2023, 25, 100558. [Google Scholar] [CrossRef]
- Haik, J.; Weissman, O.; Stavrou, D.; Ben-noon, H.I.; Liran, A.; Tessone, A.; Zmora, N.; Zilinsky, I.; Winkler, E.; Gur, E.; et al. Is prophylactic acyclovir treatment warranted for prevention of herpes simplex virus infections in facial burns? A review of the literature. J. Burn. Care Res. 2011, 32, 358–362. [Google Scholar] [CrossRef]
- Saunsbury, T.; Harte, M.; Ion, D. Unusual oral presentation of acyclovir-resistant herpes simplex in an allogeneic haematopoietic stem cell transplant recipient. BMJ Case Rep. 2021, 14, e247109. [Google Scholar] [CrossRef]
- Czerwiec, K.; Zawrzykraj, M.; Deptula, M.; Skoniecka, A.; Tyminska, A.; Zielinski, J.; Kosinski, A.; Pikula, M. Adipose-Derived Mesenchymal Stromal Cells in Basic Research and Clinical Applications. Int. J. Mol. Sci. 2023, 24, 3888. [Google Scholar] [CrossRef]
- Bajek, A.; Gurtowska, N.; Olkowska, J.; Kazmierski, L.; Maj, M.; Drewa, T. Adipose-Derived Stem Cells as a Tool in Cell-Based Therapies. Arch. Immunol. Ther. Exp. 2016, 64, 443–454. [Google Scholar] [CrossRef]
- Fernandez-Francos, S.; Eiro, N.; Costa, L.A.; Escudero-Cernuda, S.; Fernandez-Sanchez, M.L.; Vizoso, F.J. Mesenchymal Stem Cells as a Cornerstone in a Galaxy of Intercellular Signals: Basis for a New Era of Medicine. Int. J. Mol. Sci. 2021, 22, 3576. [Google Scholar] [CrossRef] [PubMed]
- Li, T.H.; Li, Z.M.; Qin, X.H.; Yu, N.Z.; Huang, J.Z.; Long, X. Global Analyses and Latest Research Hot Spots of Adipose-Derived Stem Cells in Fat Grafting: A Bibliometric and Visualized Review. Aesthetic Plast. Surg. 2023, 47, 1192–1204. [Google Scholar] [CrossRef] [PubMed]
- Lebeau, G.; Ah-Pine, F.; Daniel, M.; Bedoui, Y.; Vagner, D.; Frumence, E.; Gasque, P. Perivascular Mesenchymal Stem/Stromal Cells, an Immune Privileged Niche for Viruses? Int. J. Mol. Sci. 2022, 23, 8038. [Google Scholar] [CrossRef] [PubMed]
- Taechangam, N.; Kol, A.; Arzi, B.; Borjesson, D.L. Multipotent Stromal Cells and Viral Interaction: Current Implications for Therapy. Stem Cell Rev. Rep. 2022, 18, 214–227. [Google Scholar] [CrossRef]
- Rollin, R.; Alvarez-Lafuente, R.; Marco, F.; Jover, J.A.; Hernandez-Garcia, C.; Rodriguez-Navas, C.; Lopez-Duran, L.; Fernandez-Gutierrez, B. Human parvovirus B19, varicella zoster virus, and human herpesvirus-6 in mesenchymal stem cells of patients with osteoarthritis: Analysis with quantitative real-time polymerase chain reaction. Osteoarthr. Cartil. 2007, 15, 475–478. [Google Scholar] [CrossRef]
- Costanzo, D.; Romeo, A.; Marena, F. Autologous Fat Grafting in Plastic and Reconstructive Surgery: An Historical Perspective. Eplasty 2022, 22, e4. [Google Scholar]
- Bora, P.; Majumdar, A.S. Adipose tissue-derived stromal vascular fraction in regenerative medicine: A brief review on biology and translation. Stem Cell Res. Ther. 2017, 8, 145. [Google Scholar] [CrossRef]
- Vargel, I.; Tuncel, A.; Baysal, N.; Hartuc-Cevik, I.; Korkusuz, F. Autologous Adipose-Derived Tissue Stromal Vascular Fraction (AD-tSVF) for Knee Osteoarthritis. Int. J. Mol. Sci. 2022, 23, 13517. [Google Scholar] [CrossRef]
- Bellei, B.; Migliano, E.; Picardo, M. Therapeutic potential of adipose tissue-derivatives in modern dermatology. Exp. Dermatol. 2022, 31, 1837–1852. [Google Scholar] [CrossRef]
- Coulange Zavarro, A.; Velier, M.; Arcani, R.; Abellan Lopez, M.; Simoncini, S.; Benyamine, A.; Gomes De Pinho, Q.; Coatmeur, R.; Wang, J.; Xia, J.; et al. Adipose Tissue and Adipose-Tissue-Derived Cell Therapies for the Treatment of the Face and Hands of Patients Suffering from Systemic Sclerosis. Biomedicines 2023, 11, 348. [Google Scholar] [CrossRef]
- Ouchi, N.; Parker, J.L.; Lugus, J.J.; Walsh, K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 2011, 11, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Clemente-Suarez, V.J.; Redondo-Florez, L.; Beltran-Velasco, A.I.; Martin-Rodriguez, A.; Martinez-Guardado, I.; Navarro-Jimenez, E.; Laborde-Cardenas, C.C.; Tornero-Aguilera, J.F. The Role of Adipokines in Health and Disease. Biomedicines 2023, 11, 1290. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.; Speretta, G.F.; Teixeira, A.M.; Lira, F.S. Editorial: Adipose tissue and skeletal muscle as endocrine organs: Role of cytokines in health and disease. Front. Physiol. 2022, 13, 1069431. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Lewis, D.E. HIV Persistence in Adipose Tissue Reservoirs. Curr. HIV AIDS Rep. 2018, 15, 60–71. [Google Scholar] [CrossRef]
- Davison, A.J.; Eberle, R.; Ehlers, B.; Hayward, G.S.; McGeoch, D.J.; Minson, A.C.; Pellett, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The order Herpesvirales. Arch. Virol. 2009, 154, 171–177. [Google Scholar] [CrossRef]
- Kamakura, M.; Nawa, A.; Ushijima, Y.; Goshima, F.; Kawaguchi, Y.; Kikkawa, F.; Nishiyama, Y. Microarray analysis of transcriptional responses to infection by herpes simplex virus types 1 and 2 and their US3-deficient mutants. Microbes Infect. 2008, 10, 405–413. [Google Scholar] [CrossRef]
- Ahmed, M.; Huh, J.R. Cutting edge: Interleukin-17a prompts HIF1alpha for wound healing. Trends Immunol. 2022, 43, 861–863. [Google Scholar] [CrossRef]
- Aubert, M.; Blaho, J.A. Modulation of apoptosis during herpes simplex virus infection in human cells. Microbes Infect. 2001, 3, 859–866. [Google Scholar] [CrossRef]
- Whitley, R.J.; Kimberlin, D.W.; Roizman, B. Herpes simplex viruses. Clin. Infect. Dis. 1998, 26, 541–553; quiz 554–555. [Google Scholar] [CrossRef]
- Boldogkoi, Z.; Szucs, A.; Balazs, Z.; Sharon, D.; Snyder, M.; Tombacz, D. Transcriptomic study of Herpes simplex virus type-1 using full-length sequencing techniques. Sci. Data 2018, 5, 180266. [Google Scholar] [CrossRef] [PubMed]
- Megyeri, K. Modulation of Apoptotic Pathways by Herpes Simplex Viruses. In Latency Strategies of Herpesviruses; Minarovits, J., Gonczol, E., Valyi-Nagy, T., Eds.; Springer: Boston, MA, USA, 2007; pp. 37–54. [Google Scholar]
- Petrovski, G.; Pasztor, K.; Orosz, L.; Albert, R.; Mencel, E.; Moe, M.C.; Kaarniranta, K.; Facsko, A.; Megyeri, K. Herpes simplex virus types 1 and 2 modulate autophagy in SIRC corneal cells. J. Biosci. 2014, 39, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Kouroupis, D.; Sanjurjo-Rodriguez, C.; Jones, E.; Correa, D. Mesenchymal Stem Cell Functionalization for Enhanced Therapeutic Applications. Tissue Eng. Part. B Rev. 2019, 25, 55–77. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Dai, H.; Li, J. Immunomodulatory properties of mesenchymal stromal/stem cells: The link with metabolism. J. Adv. Res. 2023, 45, 15–29. [Google Scholar] [CrossRef]
- Yagi, H.; Soto-Gutierrez, A.; Parekkadan, B.; Kitagawa, Y.; Tompkins, R.G.; Kobayashi, N.; Yarmush, M.L. Mesenchymal stem cells: Mechanisms of immunomodulation and homing. Cell Transplant. 2010, 19, 667–679. [Google Scholar] [CrossRef]
- Wang, G.; Cao, K.; Liu, K.; Xue, Y.; Roberts, A.I.; Li, F.; Han, Y.; Rabson, A.B.; Wang, Y.; Shi, Y. Kynurenic acid, an IDO metabolite, controls TSG-6-mediated immunosuppression of human mesenchymal stem cells. Cell Death Differ. 2018, 25, 1209–1223. [Google Scholar] [CrossRef]
- Song, N.; Scholtemeijer, M.; Shah, K. Mesenchymal Stem Cell Immunomodulation: Mechanisms and Therapeutic Potential. Trends Pharmacol. Sci. 2020, 41, 653–664. [Google Scholar] [CrossRef]
- Eissner, G. Mesenchymal Stromal Cells “Think” Globally, but Act Locally. Cells 2023, 12, 509. [Google Scholar] [CrossRef]
- Shirjang, S.; Mansoori, B.; Solali, S.; Hagh, M.F.; Shamsasenjan, K. Toll-like receptors as a key regulator of mesenchymal stem cell function: An up-to-date review. Cell. Immunol. 2017, 315, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, Q.; Shi, J.; Xu, X.; Xu, J. The role of TNF-alpha in the fate regulation and functional reprogramming of mesenchymal stem cells in an inflammatory microenvironment. Front. Immunol. 2023, 14, 1074863. [Google Scholar] [CrossRef]
- Thanunchai, M.; Hongeng, S.; Thitithanyanont, A. Mesenchymal Stromal Cells and Viral Infection. Stem Cells Int. 2015, 2015, 860950. [Google Scholar] [CrossRef]
- Yang, K.; Wang, J.; Xiang, A.P.; Zhan, X.; Wang, Y.; Wu, M.; Huang, X. Functional RIG-I-like receptors control the survival of mesenchymal stem cells. Cell Death Dis. 2013, 4, e967. [Google Scholar] [CrossRef]
- Soland, M.A.; Keyes, L.R.; Bayne, R.; Moon, J.; Porada, C.D.; St. Jeor, S.; Almeida-Porada, G. Perivascular stromal cells as a potential reservoir of human cytomegalovirus. Am. J. Transplant. 2014, 14, 820–830. [Google Scholar] [CrossRef]
- Khatri, M.; Saif, Y.M. Influenza virus infects bone marrow mesenchymal stromal cells in vitro: Implications for bone marrow transplantation. Cell Transplant. 2013, 22, 461–468. [Google Scholar] [CrossRef]
- Kallmeyer, K.; Ryder, M.A.; Pepper, M.S. Mesenchymal Stromal Cells: A Possible Reservoir for HIV-1? Stem Cell Rev. Rep. 2022, 18, 1253–1280. [Google Scholar] [CrossRef]
- Pessina, A.; Bonomi, A.; Cocce, V.; Bernardo, M.E.; Cometa, A.M.; Ferrari, M.; Sisto, F.; Cavicchini, L.; Locatelli, F. Assessment of human herpesvirus-6 infection in mesenchymal stromal cells ex vivo expanded for clinical use. Transpl. Infect. Dis. 2009, 11, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.; Marquez, M.; Alencastro, F.; Spors, F.; Zhao, Y.; Tiwari, V. Herpes simplex virus type-1 (HSV-1) entry into human mesenchymal stem cells is heavily dependent on heparan sulfate. J. Biomed. Biotechnol. 2011, 2011, 264350. [Google Scholar] [CrossRef]
- Meisel, R.; Heseler, K.; Nau, J.; Schmidt, S.K.; Leineweber, M.; Pudelko, S.; Wenning, J.; Zimmermann, A.; Hengel, H.; Sinzger, C.; et al. Cytomegalovirus infection impairs immunosuppressive and antimicrobial effector functions of human multipotent mesenchymal stromal cells. Mediat. Inflamm. 2014, 2014, 898630. [Google Scholar] [CrossRef] [PubMed]
- Avanzi, S.; Leoni, V.; Rotola, A.; Alviano, F.; Solimando, L.; Lanzoni, G.; Bonsi, L.; Di Luca, D.; Marchionni, C.; Alvisi, G.; et al. Susceptibility of human placenta derived mesenchymal stromal/stem cells to human herpesviruses infection. PLoS ONE 2013, 8, e71412. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, K.; Cho, G.W. Autophagy: An evolutionarily conserved process in the maintenance of stem cells and aging. Cell Biochem. Funct. 2019, 37, 452–458. [Google Scholar] [CrossRef]
- Revuelta, M.; Matheu, A. Autophagy in stem cell aging. Aging Cell 2017, 16, 912–915. [Google Scholar] [CrossRef]
- Chandra, P.K.; Gerlach, S.L.; Wu, C.; Khurana, N.; Swientoniewski, L.T.; Abdel-Mageed, A.B.; Li, J.; Braun, S.E.; Mondal, D. Mesenchymal stem cells are attracted to latent HIV-1-infected cells and enable virus reactivation via a non-canonical PI3K-NFkappaB signaling pathway. Sci. Rep. 2018, 8, 14702. [Google Scholar] [CrossRef] [PubMed]
- Capasso, S.; Alessio, N.; Squillaro, T.; Di Bernardo, G.; Melone, M.A.; Cipollaro, M.; Peluso, G.; Galderisi, U. Changes in autophagy, proteasome activity and metabolism to determine a specific signature for acute and chronic senescent mesenchymal stromal cells. Oncotarget 2015, 6, 39457–39468. [Google Scholar] [CrossRef] [PubMed]
- Alt, E.U.; Senst, C.; Murthy, S.N.; Slakey, D.P.; Dupin, C.L.; Chaffin, A.E.; Kadowitz, P.J.; Izadpanah, R. Aging alters tissue resident mesenchymal stem cell properties. Stem Cell Res. 2012, 8, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Rubtsov, Y.; Goryunov, K.; Romanov, A.; Suzdaltseva, Y.; Sharonov, G.; Tkachuk, V. Molecular Mechanisms of Immunomodulation Properties of Mesenchymal Stromal Cells: A New Insight into the Role of ICAM-1. Stem Cells Int. 2017, 2017, 6516854. [Google Scholar] [CrossRef]
- Ozcan, S.; Alessio, N.; Acar, M.B.; Mert, E.; Omerli, F.; Peluso, G.; Galderisi, U. Unbiased analysis of senescence associated secretory phenotype (SASP) to identify common components following different genotoxic stresses. Aging 2016, 8, 1316–1329. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, F.; Zhao, H.; Zhang, X.; Chen, H.; Zhang, K. Human adipose-derived mesenchymal stem cells are resistant to HBV infection during differentiation into hepatocytes in vitro. Int. J. Mol. Sci. 2014, 15, 6096–6110. [Google Scholar] [CrossRef]
- Lopez-Garcia, L.; Castro-Manrreza, M.E. TNF-alpha and IFN-gamma Participate in Improving the Immunoregulatory Capacity of Mesenchymal Stem/Stromal Cells: Importance of Cell—Cell Contact and Extracellular Vesicles. Int. J. Mol. Sci. 2021, 22, 9531. [Google Scholar] [CrossRef]
- Aguiar Koga, B.A.; Fernandes, L.A.; Fratini, P.; Sogayar, M.C.; Carreira, A.C.O. Role of MSC-derived small extracellular vesicles in tissue repair and regeneration. Front. Cell Dev. Biol. 2022, 10, 1047094. [Google Scholar] [CrossRef]
- Lanzoni, G.; Linetsky, E.; Correa, D.; Messinger Cayetano, S.; Alvarez, R.A.; Kouroupis, D.; Alvarez Gil, A.; Poggioli, R.; Ruiz, P.; Marttos, A.C.; et al. Umbilical cord mesenchymal stem cells for COVID-19 acute respiratory distress syndrome: A double-blind, phase 1/2a, randomized controlled trial. Stem Cells Transl. Med. 2021, 10, 660–673. [Google Scholar] [CrossRef]
- Leng, Z.; Zhu, R.; Hou, W.; Feng, Y.; Yang, Y.; Han, Q.; Shan, G.; Meng, F.; Du, D.; Wang, S.; et al. Transplantation of ACE2(-) Mesenchymal Stem Cells Improves the Outcome of Patients with COVID-19 Pneumonia. Aging Dis. 2020, 11, 216–228. [Google Scholar] [CrossRef]
- Megyeri, K.; Orosz, L.; Kormos, B.; Pasztor, K.; Seprenyi, G.; Ocsovszki, I.; Mandi, Y.; Bata-Csorgo, Z.; Kemeny, L. The herpes simplex virus-induced demise of keratinocytes is associated with a dysregulated pattern of p63 expression. Microbes Infect. 2009, 11, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Orosz, L.; Gallyas, E.; Kemeny, L.; Mandi, Y.; Facsko, A.; Megyeri, K. Involvement of p63 in the herpes simplex virus-1-induced demise of corneal cells. J. Biomed. Sci. 2010, 17, 47. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Gene Ontology Semantic Similarity Analysis Using GOSemSim. Methods Mol. Biol. 2020, 2117, 207–215. [Google Scholar]
- Yu, G.; Li, F.; Qin, Y.; Bo, X.; Wu, Y.; Wang, S. GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 2010, 26, 976–978. [Google Scholar] [CrossRef]
- Yu, G.; Gao, C.-H. Enrichplot: Visualization of Functional Enrichment Result, R package version 1161; R Foundation for Statistical Computing: Vienna, Austria, 2022.
- Sayols, S. Rrvgo: A Bioconductor package for interpreting lists of Gene Ontology terms. microPubl. Biol. 2023, 2023. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Turkes, E.; Ostendorf, B.; Grioni, A.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling, R package version 1180; R Foundation for Statistical Computing: Vienna, Austria, 2022.
- Vereb, Z.; Mazlo, A.; Szabo, A.; Poliska, S.; Kiss, A.; Litauszky, K.; Koncz, G.; Boda, Z.; Rajnavolgyi, E.; Bacsi, A. Vessel Wall-Derived Mesenchymal Stromal Cells Share Similar Differentiation Potential and Immunomodulatory Properties with Bone Marrow-Derived Stromal Cells. Stem Cells Int. 2020, 2020, 8847038. [Google Scholar] [CrossRef] [PubMed]
- Vereb, Z.; Poliska, S.; Albert, R.; Olstad, O.K.; Boratko, A.; Csortos, C.; Moe, M.C.; Facsko, A.; Petrovski, G. Role of Human Corneal Stroma-Derived Mesenchymal-Like Stem Cells in Corneal Immunity and Wound Healing. Sci. Rep. 2016, 6, 26227. [Google Scholar] [CrossRef] [PubMed]
- Vereb, Z.; Lumi, X.; Andjelic, S.; Globocnik-Petrovic, M.; Urbancic, M.; Hawlina, M.; Facsko, A.; Petrovski, G. Functional and molecular characterization of ex vivo cultured epiretinal membrane cells from human proliferative diabetic retinopathy. Biomed. Res. Int. 2013, 2013, 492376. [Google Scholar] [CrossRef] [PubMed]
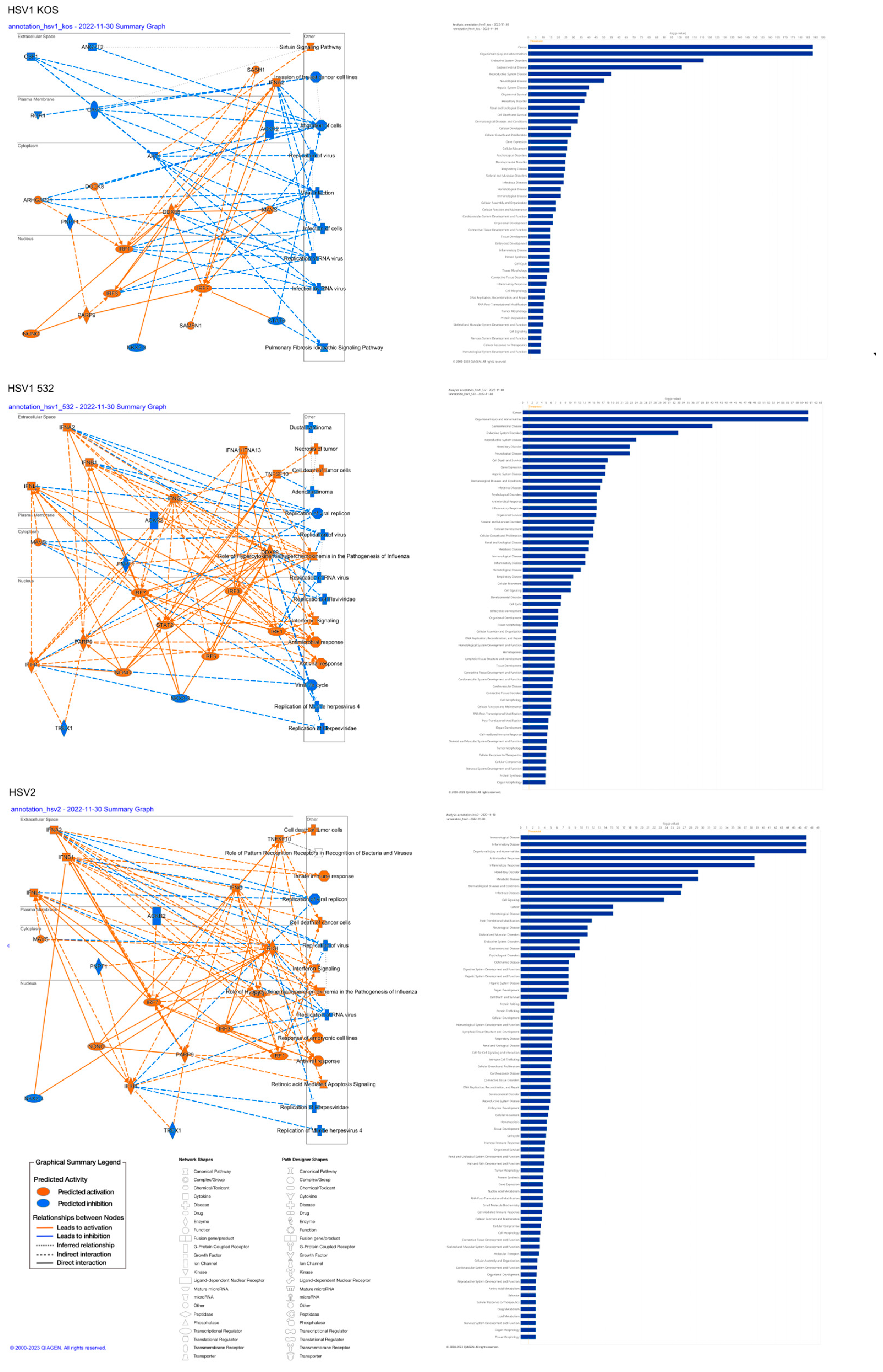





| Top Canonical Pathways | |||
|---|---|---|---|
| Name | p-value | Overlap | |
| HSV1-KOS | Mitochondrial Dysfunction | 6.00 × 10−12 | 36.3% (62/171) |
| Oxidative Phosphorylation | 2.50 × 10−10 | 39.6% (44/111) | |
| Pulmonary Fibrosis Idiopathic Signaling Pathway | 3.21 × 10−09 | 27.6% (90/326) | |
| Sirtuin Signaling Pathway | 1.83 × 10−07 | 26.6% (78/293) | |
| Huntington’s Disease Signaling | 1.24 × 10−05 | 24.7% (70/283) | |
| HSV1-532 | Interferon Signaling | 3.85 × 10−06 | 30.6% (1/36) |
| Role of Hypercytokinemia/Hyperchemokinemia in the Pathogenesis of Influenza | 7.84 × 10−06 | 19.8% (17/86) | |
| Inhibition of ARE-Mediated mRNA Degradation Pathway | 2.28 × 10−05 | 14.8% (24/162) | |
| Activation of IRF via Cytosolic Pattern Recognition Receptors | 7.92 × 10−05 | 20.0% (13/65) | |
| Sirtuin Signaling Pathway | 1.01 × 10−04 | 11.6% (34/293) | |
| HSV-2 | Role of Hypercytokinemia/hyperchemokinemia in the Pathogenesis of Influenza | 6.59 × 10−17 | 14.0% 12/86 |
| Interferon Signaling | 2.14 × 10−11 | 19.4% 7/36 | |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | 4.02 × 10−08 | 5.1% 8/156 | |
| Macrophage Classical Activation Signaling Pathway | 1.77 × 10−07 | 4.2% 8/189 | |
| Activation of IRF by Cytosolic Pattern Recognition Receptors | 2.19 × 10−06 | 7.7% 5/65 | |
| Top Upstream Regulators | |||
| Upstream Regulators | |||
| HSV1-KOS | Name | p-value | Predicted Activation |
| beta-estradiol | 2.25 × 10−33 | Inhibited | |
| MYC | 3.35 × 10−30 | ||
| TP53 | 1.97 × 10−29 | ||
| TGFB1 | 1.93 × 10−27 | Inhibited | |
| lipopolysaccharide | 3.42 × 10−26 | ||
| HSV1-532 | NONO | 2.11 × 10−38 | Activated |
| CHROMR variant 3 | 1.67 × 10−35 | Activated | |
| IFNL1 | 2.64 × 10−33 | Activated | |
| TREX1 | 4.88 × 10−33 | Inhibited | |
| pyridostatin | 2.16 × 10−32 | Activated | |
| HSV-2 | NONO | 2.61 × 10−82 | Activated |
| IFNL1 | 1.02 × 10−78 | Activated | |
| pyridostatin | 9.69 × 10−73 | Activated | |
| IFNA2 | 1.19 × 10−68 | Activated | |
| TREX1 | 5.75 × 10−67 | Inhibited | |
| Causal Network | |||
| HSV1-KOS | beta-estradiol | 1.17 × 10−48 | Inhibited |
| CCNT1 | 6.16 × 10−44 | Inhibited | |
| BMS-690514 | 7.17 × 10−44 | ||
| RPL11 | 1.58 × 10−43 | ||
| PA2G4 | 3.68 × 10−43 | ||
| HSV1-532 | NONO | 7.25 × 10−39 | Activated |
| CHROMR variant 3 | 1.67 × 10−35 | Activated | |
| IFNL1 | 1.65 × 10−34 | Activated | |
| pyridostatin | 2.16 × 10−32 | Activated | |
| STAG2 | 6.77 × 10−31 | Inhibited | |
| HSV-2 | NONO | 1.17 × 10−82 | Activated |
| IFNL1 | 8.99 × 10−80 | Activated | |
| pyridostatin | 9.69 × 10−73 | Activated | |
| IFNA2 | 8.54 × 10−69 | Activated | |
| STAG2 | 3.63 × 10−66 | Inhibited | |
| Top Diseases and Bio Functions | |||
|---|---|---|---|
| Diases and Disorders | |||
| HSV-1 KOS | Name | p-value range | # Molecules |
| Cancer | 8.05 × 10−09–5.38 × 10−189 | 3425 | |
| Organismal Injury and Abnormalities | 8.47 × 10−09–5.38 × 10−189 | 3482 | |
| Endocrine System Disorders | 4.28 × 10−09–9.25 × 10−117 | 2942 | |
| Gastrointestinal Disease | 6.99 × 10−09–2.30 × 10−102 | 3038 | |
| Reproductive System Disease | 3.98 × 10−09–8.17 × 10−56 | 2442 | |
| HSV-1 532 | Cancer | 1.92 × 10−05–4.01 × 10−61 | 1323 |
| Organismal Injury and Abnormalities | 1.94 × 10−05–4.01 × 10−61 | 1347 | |
| Gastrointestinal Disease | 1.82 × 10−05–7.75 × 10−41 | 1181 | |
| Endocrine System Disorders | 1.08 × 10−05–1.28 × 10−33 | 1125 | |
| HSV-2 | Immunological Disease | 3.23 × 10−03–8.71 × 10−48 | 59 |
| Inflammatory Disease | 3.23 × 10−03–8.71 × 10−48 | 51 | |
| Organismal Injury and Abnormalities | 3.23 × 10−03–8.71 × 10−48 | 75 | |
| Antimicrobial Response | 3.23 × 10−03–2.90 × 10−39 | 33 | |
| Inflammatory Response | 3.23 × 10−03–2.90 × 10−39 | 53 | |
| Molecular and Cellular Functions | |||
| HSV-1 KOS | Name | p-value range | # Molecules |
| Cell Death and Survival | 3.72 × 10−09–4.41 × 10−34 | 1274 | |
| Cellular Development | 6.54 × 10−09–3.79 × 10−29 | 1045 | |
| Cellular Growth and Proliferation | 6.54 × 10−09–3.79 × 10−29 | 953 | |
| Gene Expression | 9.96 × 10−11–5.57 × 10−27 | 788 | |
| Cellular Movement | 6.39 × 10−09–9.76 × 10−27 | 939 | |
| HSV-1- 532 | Cell Death and Survival | 1.62 × 10−05–1.05 × 10−18 | 517 |
| Gene Expression | 1.29 × 10−05–3.07 × 10−18 | 356 | |
| Cellular Development | 1.79 × 10−05–1.30 × 10−15 | 448 | |
| Cellular Growth and Proliferation | 1.79 × 10−05–1.30 × 10−15 | 418 | |
| HSV-2 | Cell Signaling | 2.14 × 10−04–2.37 × 10−24 | 17 |
| Post-Translational Modification | 2.23 × 10−03–1.84 × 10-12 | 10 | |
| Cell Death and Survival | 3.23 × 10−03–1.94 × 10−08 | 36 | |
| Protein Folding | 2.68 × 10−06–2.68 × 10−06 | 3 | |
| Protein Trafficking | 3.23 × 10−03–2.68 × 10−06 | 8 | |
| Physiological System Development and Function | |||
| HSV-1 KOS | Name | p-value range | # Molecules |
| Organismal Survival | 7.18 × 10−09–2.40 × 10−39 | 1011 | |
| Cardiovascular System Development and Function | 8.04 × 10−09–5.20 × 10−17 | 410 | |
| Organismal Development | 3.02 × 10−09–5.20 × 10−17 | 1083 | |
| Connective Tissue Development and Function | 4.63 × 10−09–1.78 × 10−15 | 594 | |
| Tissue Development | 4.63 × 10−09–1.78 × 10−15 | 757 | |
| HSV-1- 532 | Organismal Survival | 1.17 × 10−05–2.74 × 10−16 | 395 |
| Embryonic Development | 1.58 × 10−05–3.05 × 10−08 | 221 | |
| Organismal Development | 1.76 × 10−05–3.05 × 10−08 | 404 | |
| Tissue Morphology | 1.82 × 10−05–3.40 × 10−08 | 289 | |
| Hematological System Development and Function | 1.58 × 10−05–1.70 × 10−07 | 212 | |
| HSV-2 | Digestive System Development and Function | 3.23 × 10−03–1.25 × 10−08 | 11 |
| Hepatic System Development and Function | 3.23 × 10−03–1.25 × 10−08 | 11 | |
| Organ Development | 3.23 × 10−03–1.25 × 10−08 | 18 | |
| Hematological System Development and Function | 3.23 × 10−03–5.14 × 10−06 | 28 | |
| Lymphoid Tissue Structure and Development | 3.23 × 10−03–5.14 × 10−06 | 24 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kun-Varga, A.; Gubán, B.; Miklós, V.; Parvaneh, S.; Guba, M.; Szűcs, D.; Monostori, T.; Varga, J.; Varga, Á.; Rázga, Z.; et al. Herpes Simplex Virus Infection Alters the Immunological Properties of Adipose-Tissue-Derived Mesenchymal-Stem Cells. Int. J. Mol. Sci. 2023, 24, 11989. https://doi.org/10.3390/ijms241511989
Kun-Varga A, Gubán B, Miklós V, Parvaneh S, Guba M, Szűcs D, Monostori T, Varga J, Varga Á, Rázga Z, et al. Herpes Simplex Virus Infection Alters the Immunological Properties of Adipose-Tissue-Derived Mesenchymal-Stem Cells. International Journal of Molecular Sciences. 2023; 24(15):11989. https://doi.org/10.3390/ijms241511989
Chicago/Turabian StyleKun-Varga, Anikó, Barbara Gubán, Vanda Miklós, Shahram Parvaneh, Melinda Guba, Diána Szűcs, Tamás Monostori, János Varga, Ákos Varga, Zsolt Rázga, and et al. 2023. "Herpes Simplex Virus Infection Alters the Immunological Properties of Adipose-Tissue-Derived Mesenchymal-Stem Cells" International Journal of Molecular Sciences 24, no. 15: 11989. https://doi.org/10.3390/ijms241511989
APA StyleKun-Varga, A., Gubán, B., Miklós, V., Parvaneh, S., Guba, M., Szűcs, D., Monostori, T., Varga, J., Varga, Á., Rázga, Z., Bata-Csörgő, Z., Kemény, L., Megyeri, K., & Veréb, Z. (2023). Herpes Simplex Virus Infection Alters the Immunological Properties of Adipose-Tissue-Derived Mesenchymal-Stem Cells. International Journal of Molecular Sciences, 24(15), 11989. https://doi.org/10.3390/ijms241511989








