Modeling of the Peptide Release during Proteolysis of β-Lactoglobulin by Trypsin with Consideration of Peptide Bond Demasking
Abstract
1. Introduction
2. Results
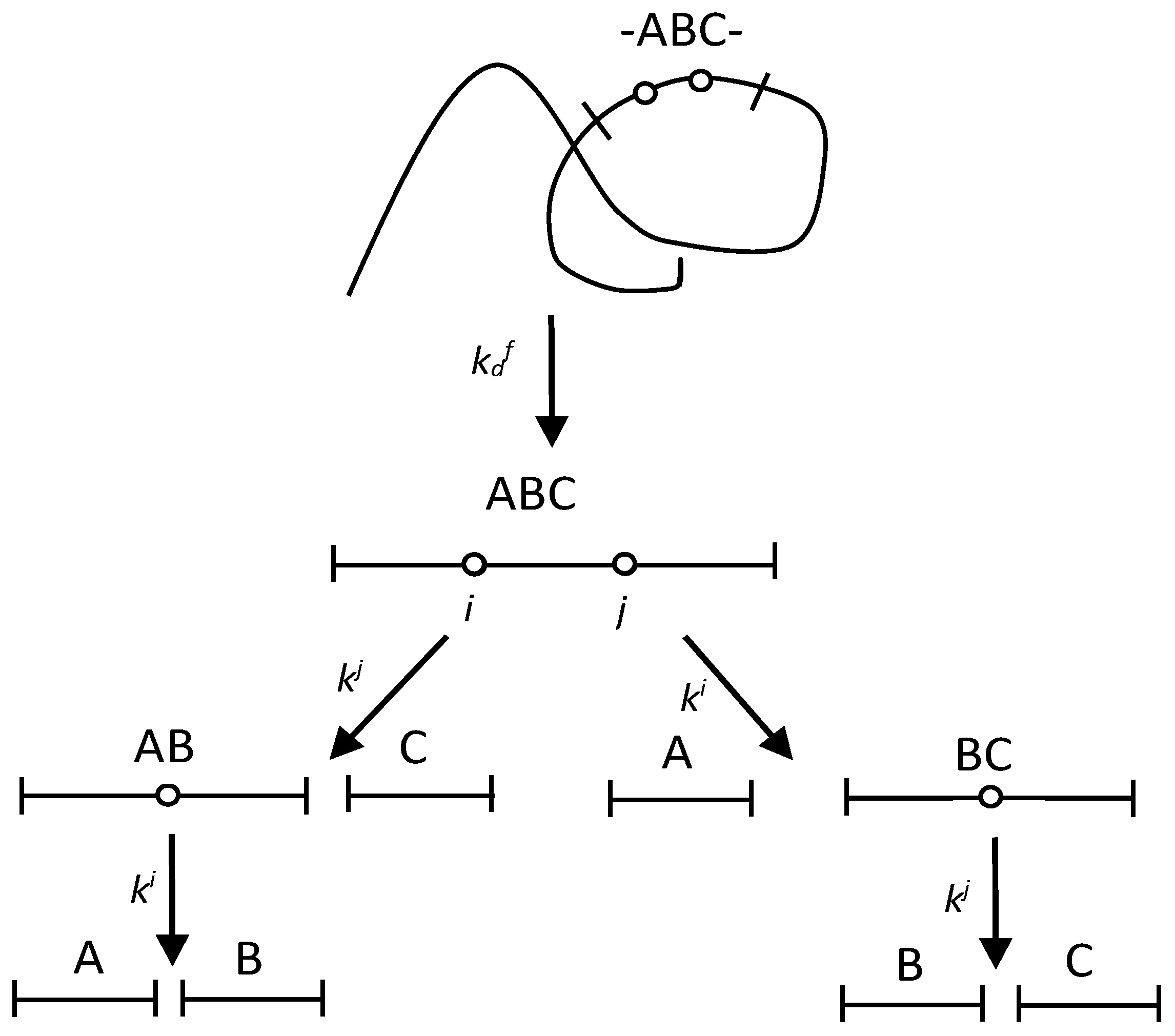
2.1. The Fragmentation with One Demasking Step
2.2. The Fragmentation with Two Demasking Steps
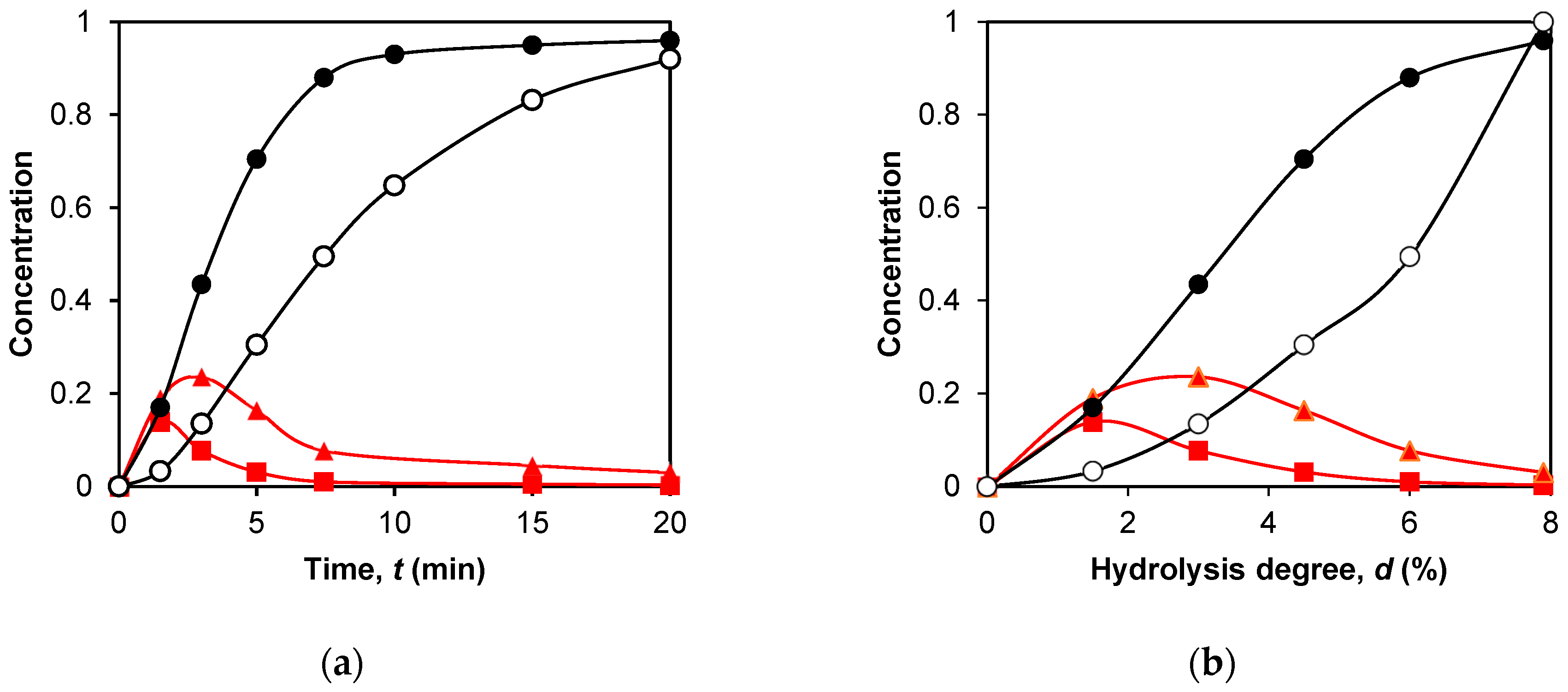
2.3. Application of Peptide Release Schemes to β-LG Proteolysis by Trypsin
2.4. Simulation of Peptide Release during β-LG Proteolysis by Trypsin
3. Discussion
4. Materials and Methods
4.1. Quantitative Modelling of Proteolysis with One-Stage Demasking
4.2. Quantitative Modelling of Proteolysis with Two-Stage Demasking
4.3. Estimation of the Parameters for Concentration Dependences
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Niemann, C. Alpha-chymotrypsin and the nature of enzyme catalysis. Science 1964, 143, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Vorob’ev, M.M.; Dalgalarrondo, M.; Chobert, J.-M.; Haertle, T. Kinetics of β-casein hydrolysis by wild-type and engineered trypsin. Biopolymers 2000, 54, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Vorob’ev, M.M. Kinetics of peptide bond demasking in enzymatic hydrolysis of casein substrates. J. Mol. Catal. B 2009, 58, 146–152. [Google Scholar] [CrossRef]
- Antonov, V.K. New data on pepsin mechanism and specificity. In Acid Proteases: Structure, Function, and Biology. Advances in Experimental Medicine and Biology; Tang, J., Ed.; Springer: New York, NY, USA, 1977; Volume 3. [Google Scholar] [CrossRef]
- Vorob’ev, M.M.; Goncharova, I.A. Computer simulation of proteolysis. Peptic hydrolysis of partially demasked β-Lactoglobulin. Nahrung-Food 1998, 42, 61–67. [Google Scholar] [CrossRef]
- Suwareh, O.; Causeur, D.; Jardin, J.; Briard-Bion, V.; Le Feunteun, S.; Pezennec, S.; Nau, F. Statistical modeling of in vitro pepsin specificity. Food Chem. 2021, 362, 130098. [Google Scholar] [CrossRef]
- Tonda, A.; Grosvenor, A.; Clerens, S.; Le Feunteun, S. In silico modeling of protein hydrolysis by endoproteases: A case study on pepsin digestion of bovine lactoferrin. Food Funct. 2017, 8, 4404–4413. [Google Scholar] [CrossRef]
- Trusek-Holownia, A.; Noworyta, A. A model of kinetics of the enzymatic hydrolysis of biopolymers—A concept for determination of hydrolysate composition. Chem. Eng. Process. Process Intensif. 2015, 89, 54–61. [Google Scholar] [CrossRef]
- Deng, Y.; van der Veer, F.; Sforza, S.; Gruppen, H.; Wierenga, P.A. Towards predicting protein hydrolysis by bovine trypsin. Process Biochem. 2018, 65, 81–92. [Google Scholar] [CrossRef]
- Butré, C.I. Introducing Enzyme Selectivity as a Quantitative Parameter to Describe the Effects of Substrate Concentration on Protein Hydrolysis. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2014. [Google Scholar]
- Muñoz-Tamayo, R.; De Groot, J.; Wierenga, P.A.; Gruppen, H.; Zwietering, M.H.; Sijtsma, L. Modeling peptide formation during the hydrolysis of β-casein by Lactococcus lactis. Process Biochem. 2012, 47, 83–93. [Google Scholar] [CrossRef]
- Polgár, L. The catalytic triad of serine peptidases. Cell. Mol. Life Sci. 2005, 62, 2161–2172. [Google Scholar] [CrossRef]
- Olsen, J.V.; Ong, S.-E.; Mann, M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteom. 2004, 3, 608–614. [Google Scholar] [CrossRef]
- Stryer, L. Biochemistry, 3rd ed.; W.H. Freeman and Company: New York, NY, USA, 1988; ISBN 0-7167-1843-x. [Google Scholar]
- Vorob’ev, M.M.; Vogel, V.; Güler, G.; Mäntele, W. Monitoring of demasking of peptide bonds during proteolysis by analysis of the apparent spectral shift of intrinsic protein fluorescence. Food Biophys. 2011, 6, 519–526. [Google Scholar] [CrossRef]
- Vorob’ev, M.M. Proteolysis of β-lactoglobulin by trypsin: Simulation by two-step model and experimental verification by intrinsic tryptophan fluorescence. Symmetry 2019, 11, 153. [Google Scholar] [CrossRef]
- Vorob’ev, M.M. Modeling of proteolysis of β-lactoglobulin and β-casein by trypsin with consideration of secondary masking of intermediate polypeptides. Int. J. Mol. Sci. 2022, 23, 8089. [Google Scholar] [CrossRef]
- Vorob’ev, M.M. Tryptophan fluorescence and time-lag hydrolysis of peptide bonds during degradation of β-lactoglobulin by trypsin. Catalysts 2020, 10, 1368. [Google Scholar] [CrossRef]
- Butre, C.I.; Sforza, S.; Gruppen, H.; Wierenga, P.A. Introducing enzyme selectivity: A quantitative parameter to describe enzymatic protein hydrolysis. Anal. Bioanal. Chem. 2014, 406, 5827–5841. [Google Scholar] [CrossRef]
- Vorob’ev, M.M.; Butré, C.I.; Sforza, S.; Wierenga, P.A.; Gruppen, H. Demasking kinetics of peptide bond cleavage for whey protein isolate hydrolysed by Bacillus licheniformis protease. J. Mol. Catal. B. 2016, 133, 426–431. [Google Scholar] [CrossRef]
- Caessens, P.; Visser, S.; Gruppen, H.; Voragen, A. β-Lactoglobulin hydrolysis. 1. Peptide composition and functional properties of hydrolysates obtained by the action of plasmin, trypsin, and Staphylococcus aureus V8 protease. J. Agric. Food Chem. 1999, 47, 2973–2979. [Google Scholar] [CrossRef]
- Cheison, S.; Schmitt, M.; Leeb, E.; Letzel, T.; Kulozik, U. Influence of temperature and degree of hydrolysis on the peptide composition of trypsin hydrolysates of β-lactoglobulin: Analysis by LC-ESI-TOF/MS. Food Chem. 2010, 121, 457–467. [Google Scholar] [CrossRef]
- Fernandez, A.; Riera, F. β-Lactoglobulin tryptic digestion: A model approach for peptide release. Biochem. Eng. J. 2013, 70, 88–96. [Google Scholar] [CrossRef]
- Mao, Y.; Krischke, M.; Kulozik, U. β-lactoglobulin hydrolysis by immobilized trypsin in ethanol/aqueous solvents. Process Biochem. 2019, 82, 84–93. [Google Scholar] [CrossRef]
- Cheison, S.; Lai, M.; Leeb, E.; Kulozik, U. Hydrolysis of β-lactoglobulin by trypsin under acidic pH and analysis of the hydrolysates with MALDI–TOF–MS/MS. Food Chem. 2011, 125, 1241–1248. [Google Scholar] [CrossRef]
- Emsley, A.M.; Heywood, R.J. Computer modeling of the degradation of linear polymers. Polym. Degrad. Stab. 1995, 49, 145–149. [Google Scholar] [CrossRef]
- Leeb, E.; Stefan, T.; Letzel, T.; Hinrichs, J.; Kulozik, U. Tryptic hydrolysis of β-lactoglobulin: A generic approach to describe the hydrolysis kinetic and release of peptides. Int. Dairy J. 2020, 105, 104666. [Google Scholar] [CrossRef]
- Cheison, S.C.; Leeb, E.; Letzel, T.; Kulozik, U. Influence of buffer type and concentration on the peptide composition of trypsin hydrolysates of β-lactoglobulin. Food Chem. 2011, 125, 121–127. [Google Scholar] [CrossRef]
- Vorob’ev, M.M.; Paskonova, E.A.; Vitt, S.V.; Belikov, V.M. Kinetic description of proteolysis. Part 2. Substrate regulation of peptide bond demasking and hydrolysis. Liquid chromatography of hydrolyzates. Nahrung-Food 1986, 30, 995–1001. [Google Scholar] [CrossRef]
- Chabanon, G.; Chevalot, I.; Framboisier, X.; Chenu, S.; Marc, I. Hydrolysis of rapeseed protein isolates: Kinetics, characterization and functional properties of hydrolysates. Process Biochem. 2007, 42, 1419–1428. [Google Scholar] [CrossRef]
- Vorob’ev, M.M. Quantification of two-step proteolysis model with consecutive demasking and hydrolysis of peptide bonds using casein hydrolysis by chymotrypsin. Biochem. Eng. J. 2013, 74, 60–68. [Google Scholar] [CrossRef]
- Rivera-Burgos, D.; Regnier, F.E. Disparities between immobilized enzyme and solution based digestion of transferrin with trypsin. J. Sep. Sci. 2013, 36, 454–460. [Google Scholar] [CrossRef]
- Melikishvili, S.; Dizon, M.; Hianik, T. Application of high-resolution ultrasonic spectroscopy for real-time monitoring of trypsin activity in β-casein solution. Food Chem. 2021, 337, 127759. [Google Scholar] [CrossRef]
- Buckin, V.; Altas, M.C. Ultrasonic monitoring of biocatalysis in solutions and complex dispersions. Catalysts 2017, 7, 336. [Google Scholar] [CrossRef]
- Vreeke, G.J.C.; Vincken, J.-P.; Wierenga, P.A. The path of proteolysis by bovine chymotrypsin. Food Res. Int. 2023, 165, 112485. [Google Scholar] [CrossRef]
- Vorob’ev, M.M.; Vitt, S.V.; Belikov, V.M. Kinetic description of proteolysis. Part 3. Total kinetics of peptide bonds hydrolysis in peptide mixtures. Nahrung-Food 1987, 31, 331–340. [Google Scholar] [CrossRef]
- Marquez, M.C.; Fernandez, V. Enzymic hydrolysis of vegetable proteins: Mechanism and kinetics. Process Biochem. 1993, 28, 481–490. [Google Scholar] [CrossRef]



| Bond Index j | Cleavage Site 1 | Selectivity 2 (%) | kj/k8 5,6 | Most Rapidly Hydrolyzed Bonds | Most Slowly Hydrolyzed Bonds | Peptide Fragments in Trimer Type of Demasking | |
|---|---|---|---|---|---|---|---|
| 8 | MK-GL | 13.7 | 0 | >>1 | + | 9–14, 15–40, 41–69/70 One-stage demasking | |
| 14 | QK-DL | 7.4 | 0.33 | 1.2 | |||
| 20 | WY-SL | 2.08 | 0.02 | + | |||
| 40 | LR-VY | 9.9 | 0.15 | 3.1 | |||
| 60 | QK-WE | 0.2 | 2.00 | 0.01 | + | ||
| 69, 70 | 10.1 7 | 21 | + | ||||
| 75 | EK-TK | 9.1 | 0.32 | 0.8 | + | 76–83, 84–91, 92–100/101 Two-stage demasking | |
| 83 | FK-ID | 2.9 | 1.21 | 0.4 | |||
| 91 | NK-VL | 3.8 | 0.89 | 0.5 | |||
| 100, 101 | 3.6 8 | 0.85 | 1.1 | + | |||
| 124 | VR-TP | 5.0 | 1.36 | 0.5 | 101/102–124, 125–135, 136–138 Two-stage demasking | ||
| 135 | EK-FD | 1.6 | 1.94 | 0.05 | |||
| 138 | DK-AL | 5.3 | 0.27 | 1.4 | + | ||
| 141 | LK-AL | 9.4 | 0.18 | 2.3 | + | ||
| 148 | IR-LS | 11.0 | 0.14 | 3.5 | + |
| Intermediate Peptide | Type of Demasking | Hydrolysis Rate Constants (min−1) | Calculated Values of dr (%) 1 | Experimental Estimation of dr 1 (%) |
|---|---|---|---|---|
| f(9–69/70), ABC | One-stage 2 | k14= 0.53, k40 = 1.41 | 2.6 | 1.5 3 |
| f(9–40), AB | One-stage | 3.5 | 3.6 | |
| f(15–69/70), BC | One-stage | 2.8 | - 4 | |
| f(76–100/101) ABC | Two-stage 5 | k83= 1, k91 = 1 | 3.9 | 3.4 |
| f(76–91), AB | Two-stage | 4.3 | 4.4 | |
| f(84–100/101), BC | Two-stage | 4.3 | 4.7 | |
| f(101/102–138), ABC | Two-stage | k124= 2, k138 = 0.2 | 3.8 | 3.4 |
| f(101/102–135), AB | Two-stage | 4.1 | - 4 | |
| f(125–138), BC | Two-stage | 4.7 | 6.1 |
| Final Peptide | Type of Demasking | Hydrolysis Rate Constants (min−1) | Calculated Values of n 1 | Experimental Estimation of n 1 |
|---|---|---|---|---|
| f(9–14), A | One-stage 2 | k14= 0.53, k40 = 1.41 | 0.85 | 0.68 |
| f(15–40), B f(41–69/70), C | One-stage One-stage | 0.92 0.44 | 0.86 0.59 | |
| f(76–83), A | Two-stage 3 | k83= 1, k91 = 1 | 2.41 | 1.46 |
| f(84–91), B | Two-stage | 2.86 | 2.37 | |
| f(92–100/101), C | Two-stage | 2.41 | 1.40 | |
| f(101/102–124), A | Two-stage | k124= 2, k135 = 0.2 | 2.21 | 1.13 |
| f(125–135), B | Two-stage | 4.84 | 5.76 | |
| f(136–138), C | Two-stage | 4.77 | 5.10 |
| Modeling Principles | Advantages | Disadvantages |
|---|---|---|
| The technique for determining the concentrations of peptide bonds during proteolysis is well established | In the transition from the concentrations of peptides to the concentrations of peptide bonds, a part of the information is lost |
| Quantification of rate constants for hydrolysis of peptide bonds is well tested | The determination of the hydrolysis rate constants is limited by accuracy of determining experimental curves |
| Accounting for demasking improves the accuracy of proteolysis description | Precise determination of the demasking rate constants requires the use of new analytical methods |
| The equations describing the release of peptides are relatively simple | The model does not describe the release of four-dimensional or longer peptide blocks. |
| The volume of calculations is reduced | The release of minor peptide fragments is not predicted |
| Peptide Fragment | Constant Term C0 | |||
|---|---|---|---|---|
| A | 1 | 0 | ||
| B | 1 | |||
| C | 1 | 0 | ||
| AB | 0 | |||
| BC | 0 | |||
| ABC | 0 |
| Peptide Fragment | ||
|---|---|---|
| A | 0 | |
| B | 0 | 0 |
| C | 0 | |
| AB | 0 | |
| BC | 0 | |
| ABC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vorob’ev, M.M. Modeling of the Peptide Release during Proteolysis of β-Lactoglobulin by Trypsin with Consideration of Peptide Bond Demasking. Int. J. Mol. Sci. 2023, 24, 11929. https://doi.org/10.3390/ijms241511929
Vorob’ev MM. Modeling of the Peptide Release during Proteolysis of β-Lactoglobulin by Trypsin with Consideration of Peptide Bond Demasking. International Journal of Molecular Sciences. 2023; 24(15):11929. https://doi.org/10.3390/ijms241511929
Chicago/Turabian StyleVorob’ev, Mikhail M. 2023. "Modeling of the Peptide Release during Proteolysis of β-Lactoglobulin by Trypsin with Consideration of Peptide Bond Demasking" International Journal of Molecular Sciences 24, no. 15: 11929. https://doi.org/10.3390/ijms241511929
APA StyleVorob’ev, M. M. (2023). Modeling of the Peptide Release during Proteolysis of β-Lactoglobulin by Trypsin with Consideration of Peptide Bond Demasking. International Journal of Molecular Sciences, 24(15), 11929. https://doi.org/10.3390/ijms241511929






