Secretory Proteomic Responses of Endometrial Epithelial Cells to Trophoblast-Derived Extracellular Vesicles
Abstract
1. Introduction
2. Results
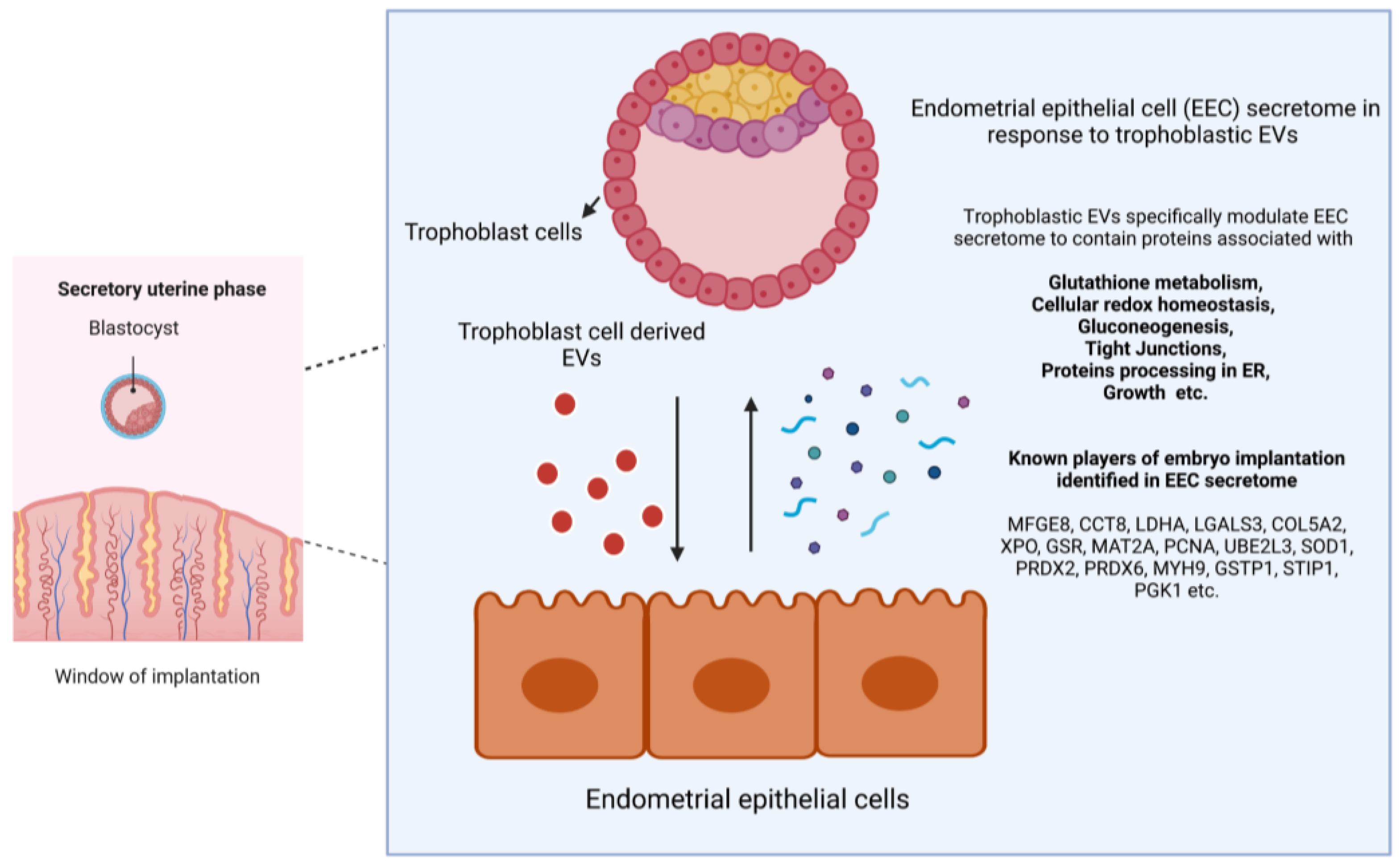
2.1. Trophoblast Cell-Derived EVs Induce Specific Secretory Protein Response in Endometrial Epithelial Cells Compared to Non-Trophoblast Cell-Derived EVs
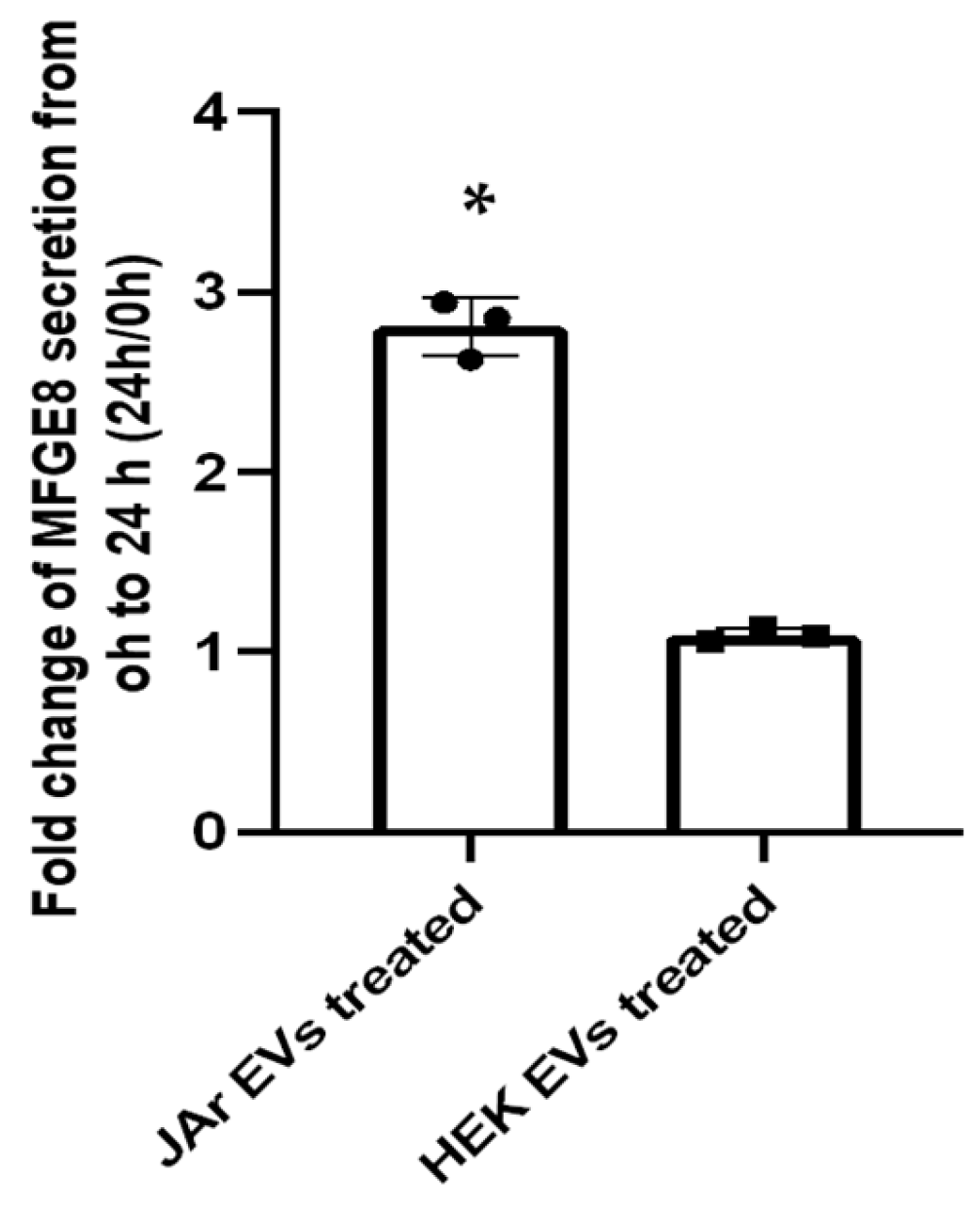
2.2. Specific Secretory Protein Changes in RL95-2 Cells in Response to JAr Cell-Derived EVs Reveal Potential Players of Embryo Implantation
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Preparation of EV-Depleted Media
4.3. EV Isolation and Characterization
4.4. Collection of Cell Culture Supernatants for Secretory Proteome Analysis and Enzyme-Linked Immunosorbent Assay
4.5. Protein Quantification and Identification with Liquid Chromatography and Tandem Mass Spectroscopy (LC-MS/MS)
4.6. Differential Protein Expression and Bioinformatics Analysis
4.7. Verification of Proteomic Data Using Enzyme-Linked Immunosorbent Assay (ELISA)
4.8. Experimental Design
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Busnelli, A.; Somigliana, E.; Cirillo, F.; Baggiani, A.; Levi-Setti, P.E. Efficacy of Therapies and Interventions for Repeated Embryo Implantation Failure: A Systematic Review and Meta-Analysis. Sci. Rep. 2021, 11, 1747. [Google Scholar] [CrossRef] [PubMed]
- Timeva, T.; Shterev, A.; Kyurkchiev, S. Recurrent Implantation Failure: The Role of the Endometrium. J. Reprod. Infertil. 2014, 15, 173–183. [Google Scholar] [PubMed]
- Kim, S.-M.; Kim, J.-S. A Review of Mechanisms of Implantation. Dev. Reprod. 2017, 21, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Fritz, R.; Jain, C.; Randall Armant, D. Cell Signaling in Trophoblast-Uterine Communication. Int. J. Dev. Biol. 2014, 58, 261–271. [Google Scholar] [CrossRef]
- Singh, M.; Chaudhry, P.; Asselin, E. Bridging Endometrial Receptivity and Implantation: Network of Hormones, Cytokines, and Growth Factors. J. Endocrinol. 2011, 210, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lin, H.; Kong, S.; Wang, S.; Wang, H.; Wang, H.; Armant, D.R. Physiological and Molecular Determinants of Embryo Implantation. Mol. Asp. Med. 2014, 34, 939–980. [Google Scholar] [CrossRef]
- Dey, S.K.; Lim, H.; Das, S.K.; Reese, J.; Paria, B.C.; Daikoku, T.; Wang, H. Molecular Cues to Implantation. Endocr. Rev. 2004, 25, 341–373. [Google Scholar] [CrossRef] [PubMed]
- Giacomini, E.; Vago, R.; Sanchez, A.M.; Podini, P.; Zarovni, N.; Murdica, V.; Rizzo, R.; Bortolotti, D.; Candiani, M.; Viganò, P. Secretome of in Vitro Cultured Human Embryos Contains Extracellular Vesicles That Are Uptaken by the Maternal Side. Sci. Rep. 2017, 7, 5210. [Google Scholar] [CrossRef]
- Dissanayake, K.; Nõmm, M.; Lättekivi, F.; Ressaissi, Y.; Godakumara, K.; Lavrits, A.; Midekessa, G.; Viil, J.; Bæk, R.; Jørgensen, M.M.; et al. Individually Cultured Bovine Embryos Produce Extracellular Vesicles That Have the Potential to Be Used as Non-Invasive Embryo Quality Markers. Theriogenology 2020, 149, 104–116. [Google Scholar] [CrossRef]
- Saadeldin, I.M.; Kim, S.J.; Choi, Y.B.; Lee, B.C. Improvement of Cloned Embryos Development by Co-Culturing with Parthenotes: A Possible Role of Exosomes/Microvesicles for Embryos Paracrine Communication. Cell. Reprogram. 2014, 16, 223–234. [Google Scholar] [CrossRef]
- Pavani, K.C.; Hendrix, A.; Van Den Broeck, W.; Couck, L.; Szymanska, K.; Lin, X.; De Koster, J.; Van Soom, A.; Leemans, B. Isolation and Characterization of Functionally Active Extracellular Vesicles from Culture Medium Conditioned by Bovine Embryos in Vitro. Int. J. Mol. Sci. 2019, 20, 38. [Google Scholar] [CrossRef]
- Es-Haghi, M.; Godakumara, K.; Häling, A.; Lättekivi, F.; Lavrits, A.; Viil, J.; Andronowska, A.; Nafee, T.; James, V.; Jaakma, Ü.; et al. Specific Trophoblast Transcripts Transferred by Extracellular Vesicles Affect Gene Expression in Endometrial Epithelial Cells and May Have a Role in Embryo-Maternal Crosstalk. Cell Commun. Signal. 2019, 17, 146. [Google Scholar] [CrossRef]
- Godakumara, K.; Ord, J.; Lättekivi, F.; Dissanayake, K.; Viil, J.; Boggavarapu, N.R.; Faridani, O.R.; Jääger, K.; Meikas, A.V.; Jaakma, Ü. Trophoblast Derived Extracellular Vesicles Specifically Alter the Transcriptome of Endometrial Cells and May Constitute a Critical Component of Embryo-Maternal Communication. Reprod. Biol. Endocrinol. 2021, 19, 115. [Google Scholar] [CrossRef]
- Poh, Q.H.; Rai, A.; Carmichael, I.I.; Salamonsen, L.A.; Greening, D.W. Proteome Reprogramming of Endometrial Epithelial Cells by Human Trophectodermal Small Extracellular Vesicles Reveals Key Insights into Embryo Implantation. Proteomics 2021, 21, 13–14. [Google Scholar] [CrossRef]
- Ouyang, Y.; Bayer, A.; Chu, T.; Tyurin, V.A.; Kagan, V.E.; Morelli, A.E.; Coyne, C.B.; Sadovsky, Y. Isolation of Human Trophoblastic Extracellular Vesicles and Characterization of Their Cargo and Antiviral Activity. Physiol. Behav. 2017, 176, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Segura-Benítez, M.; Bas-Rivas, A.; Juárez-Barber, E.; Carbajo-Garcia, M.C.; Faus, A.; Pellicer, A.; Ferrero, H. Human Blastocysts Uptake Extracellular Vesicles Secreted By Primary Endometrial Epithelial Cells Containing miRNAs Related To Implantation and Early Embryo Development. Fertil. Steril. 2022, 118, e73–e74. [Google Scholar] [CrossRef]
- Hua, R.; Liu, Q.; Lian, W.; Kang, T.; Gao, D.; Huang, C.; Wang, Y.; Lei, M. Extracellular Vesicles Derived from Endometrial Epithelial Cells Deliver Exogenous miR-92b-3p to Affect the Function of Embryonic Trophoblast Cells via Targeting TSC1 and DKK3. Reprod. Biol. Endocrinol. 2022, 20, 152. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Shi, S.; Liang, J.; Cao, D.; Wang, S.; Wang, Z. Endometrial Cell-Derived Small Extracellular Vesicle miR-100-5p Promotes Functions of Trophoblast during Embryo Implantation. Mol. Ther. Nucleic Acids. 2021, 23, 217–231. [Google Scholar] [CrossRef]
- Evans, J.; Rai, A.; Nguyen, H.P.T.; Poh, Q.H.; Elglass, K.; Simpson, R.J.; Salamonsen, L.A.; Greening, D.W. Human Endometrial Extracellular Vesicles Functionally Prepare Human Trophectoderm Model for Implantation: Understanding Bidirectional Maternal-Embryo Communication. Proteomics 2019, 19, 1800423. [Google Scholar] [CrossRef]
- Salamonsen, L.A.; Evans, J.; Nguyen, H.P.T.; Edgell, T.A. The Microenvironment of Human Implantation: Determinant of Reproductive Success. Am. J. Reprod. Immunol. 2016, 75, 218–225. [Google Scholar] [CrossRef]
- van der Weijden, V.A.; Bick, J.T.; Bauersachs, S.; Arnold, G.J.; Fröhlich, T.; Drews, B.; Ulbrich, S.E. Uterine Fluid Proteome Changes during Diapause and Resumption of Embryo Development in Roe Deer (Capreolus capreolus). Reproduction 2019, 158, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Hart, A.R.; Liaqat, N.; Khan, A.; Dissanayake, K.; Godakumara, K.; Andronowska, A.; Eapen, S.; Heath, P.R.; Fazeli, A. The Extracellular Vesicles Proteome of Endometrial Cells Simulating the Receptive Menstrual Phase Differs from That of Endometrial Cells Simulating the Non-Receptive Menstrual Phase. Biomolecules 2023, 13, 279. [Google Scholar] [CrossRef]
- Salleh, N.; Baines, D.L.; Naftalin, R.J.; Milligan, S.R. The Hormonal Control of Uterine Luminal Fluid Secretion and Absorption. J. Membr. Biol. 2005, 206, 17–28. [Google Scholar] [CrossRef]
- Chattopadhyay, R.; Juneau, C.R.; Landis, J.; Morin, S.J.; Neal, S.A.; Scott, R.T. Persistent Fluid in the Endometrial Cavity That Resolves after Progesterone Administration Prior to Transfer Does Impact Live Birth Rate. Fertil. Steril. 2017, 107, e11. [Google Scholar] [CrossRef]
- Gurung, S.; Greening, D.W.; Catt, S.; Salamonsen, L.; Evans, J. Exosomes and Soluble Secretome from Hormone-Treated Endometrial Epithelial Cells Direct Embryo Implantation. Mol. Hum. Reprod. 2020, 26, 510–520. [Google Scholar] [CrossRef]
- Ruiz-González, I.; Xu, J.; Wang, X.; Burghardt, R.C.; Dunlap, K.A.; Bazer, F.W. Exosomes, Endogenous Retroviruses and Toll-like Receptors: Pregnancy Recognition in Ewes. Reproduction 2015, 149, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Tian, F.; Zeng, W.; Liu, X.; Fan, J.; Lin, Y.; Zhang, Y. Role of Peroxiredoxin 2 Downregulation in Recurrent Miscarriage through Regulation of Trophoblast Proliferation and Apoptosis. Cell Death Dis. 2017, 8, e2908. [Google Scholar] [CrossRef] [PubMed]
- Arcuri, F.; Ricci, C.; Ietta, F.; Cintorino, M.; Tripodi, S.A.; Cetin, I.; Garzia, E.; Schatz, F.; Klemi, P.; Santopietro, R.; et al. Macrophage Migration Inhibitory Factor in the Human Endometrium: Expression and Localization during the Menstrual Cycle and Early Pregnancy. Biol. Reprod. 2001, 64, 1200–1205. [Google Scholar] [CrossRef]
- Liang, X.; Tang, S.; Li, D.; Song, Y.; He, M.; Duan, Y.; Du, H. Shoutai Wan Improves Embryo Survival by Regulating Aerobic Glycolysis of Trophoblast Cells in a Mouse Model of Recurrent Spontaneous Abortion. Evid.-Based Complement. Altern. Med. 2022, 2022, 8251503. [Google Scholar] [CrossRef]
- Guérin, P.; El Mouatassim, S.; Ménézo, Y. Oxidative Stress and Protection against Reactive Oxygen Species in the Pre-Implantation Embryo and Its Surroundings. Hum. Reprod. Update 2001, 7, 175–189. [Google Scholar] [CrossRef]
- Gardner, D.K. Lactate Production by the Mammalian Blastocyst: Manipulating the Microenvironment for Uterine Implantation and Invasion? BioEssays 2015, 37, 364–371. [Google Scholar] [CrossRef]
- Haouzi, D.; Dechaud, H.; Assou, S.; Monzo, C.; De Vos, J.; Hamamah, S. Transcriptome Analysis Reveals Dialogues between Human Trophectoderm and Endometrial Cells during the Implantation Period. Hum. Reprod. 2011, 26, 1440–1449. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Zhao, M.; Peng, Y.; Fu, Y.S. Identification of Gene Expression Changes in Rabbit Uterus during Embryo Implantation. Genomics 2016, 107, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Öner, H.; Öner, J.; Demir, R. Distributions of PCNA and Cas-3 in Rat Uterus during Early Pregnancy. Folia Histochem. Cytobiol. 2010, 48, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Wathlet, S.; Adriaenssens, T.; Segers, I.; Verheyen, G.; van Landuyt, L.; Coucke, W.; Devroey, P.; Smitz, J. Pregnancy Prediction in Single Embryo Transfer Cycles after ICSI Using QPCR: Validation in Oocytes from the Same Cohort. PLoS ONE 2013, 8, e54226. [Google Scholar] [CrossRef]
- Zarbakhsh, S. Effect of Antioxidants on Preimplantation Embryo Development in Vitro: A Review. Zygote 2021, 29, 179–193. [Google Scholar] [CrossRef]
- Ikeda, S.; Kawahara-Miki, R.; Iwata, H.; Sugimoto, M.; Kume, S. Role of Methionine Adenosyltransferase 2A in Bovine Preimplantation Development and Its Associated Genomic Regions. Sci. Rep. 2017, 7, 3800. [Google Scholar] [CrossRef]
- Cai, S.; Ye, Q.; Zeng, X.; Yang, G.; Ye, C.; Chen, M.; Yu, H.; Wang, Y.; Wang, G.; Huang, S.; et al. CBS and MAT2A Improve Methionine-Mediated DNA Synthesis through SAMTOR/MTORC1/S6K1/CAD Pathway during Embryo Implantation. Cell Prolif. 2021, 54, e12950. [Google Scholar] [CrossRef]
- Wang, F.; Liu, Y. Identification of Key Genes, Regulatory Factors, and Drug Target Genes of Recurrent Implantation Failure (RIF). Gynecol. Endocrinol. 2020, 36, 448–455. [Google Scholar] [CrossRef]
- Nangia-Makker, P.; Honjo, Y.; Sarvis, R.; Akahani, S.; Hogan, V.; Pienta, K.J.; Raz, A. Galectin-3 Induces Endothelial Cell Morphogenesis and Angiogenesis. Am. J. Pathol. 2000, 156, 899–909. [Google Scholar] [CrossRef]
- Blidner, A.G.; Rabinovich, G.A. “Sweetening” Pregnancy: Galectins at the Fetomaternal Interface. Am. J. Reprod. Immunol. 2013, 69, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lei, C.; Zhang, W. Expression of Galectin-3 in Mouse Endometrium and Its Effect during Embryo Implantation. Reprod. Biomed. Online 2012, 24, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.X.; Zhang, W.; Zhou, J.P.; Liu, Y.K. Interactions between Galectin-3 and Integrinβ3 in Regulating Endometrial Cell Proliferation and Adhesion. Hum. Reprod. 2009, 24, 2879–2889. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Grzmil, P.; Altmann, M.E.; Adham, I.M.; Engel, U.; Jarry, H.; Schweyer, S.; Wolf, S.; Mänz, J.; Engel, W. Embryo Implantation Failure and Other Reproductive Defects in Ube2q1-Deficient Female Mice. Reproduction 2013, 145, 45–56. [Google Scholar] [CrossRef]
- Altmäe, S.; Reimand, J.R.; Hovatta, O.; Zhang, P.; Kere, J.; Laisk, T.; Saare, M.; Peters, M.; Vilo, J.; Stavreus-Evers, A.; et al. Implantation: Identification of Gene Expression Pathways, Regulation, and Integrated Regulatory Networks. Mol. Endocrinol. 2012, 26, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Prašnikar, E.; Kunej, T.; Knez, J.; Repnik, K.; Potočnik, U.; Kovačič, B. Determining the Molecular Background of Endometrial Receptivity in Adenomyosis. Biomolecules 2020, 10, 1311. [Google Scholar] [CrossRef]
- Franchi, A.; Bocca, S.; Anderson, S.; Riggs, R.; Oehninger, S. Expression of Milk Fat Globule EGFfactor 8 (MFG-E8) mRNA and Protein in the Human Endometrium and Its Regulation by Prolactin. Mol. Hum. Reprod. 2011, 17, 360–371. [Google Scholar] [CrossRef][Green Version]
- Schmitz, C.; Yu, L.; Bocca, S.; Anderson, S.; Cunha-Filho, J.S.; Rhavi, B.S.; Oehninger, S. Role for the Endometrial Epithelial Protein MFG-E8 and Its Receptor Integrin Avβ3 in Human Implantation: Results of an in Vitro Trophoblast Attachment Study Using Established Human Cell Lines. Fertil. Steril. 2014, 101, 874–882. [Google Scholar] [CrossRef]
- Bocca, S.M.; Anderson, S.; Amaker, B.; Swanson, R.J.; Franchi, A.; Lattanzio, F.; Oehninger, S. Milk Fat Globule Epidermal Growth Factor 8 (MFG-E8): A Novel Protein in the Mammalian Endometrium with Putative Roles in Implantation and Placentation. Placenta 2012, 33, 795–802. [Google Scholar] [CrossRef]
- Barua, S.; Macedo, A.; Kolb, D.S.; Wynne-Edwards, K.E.; Klein, C. Milk-Fat Globule Epidermal Growth Factor 8 (MFGE8) Is Expressed at the Embryo- and Fetal-Maternal Interface in Equine Pregnancy. Reprod. Fertil. Dev. 2018, 30, 585–590. [Google Scholar] [CrossRef]
- Yu, L.; Hu, R.; Sullivan, C.; Swanson, R.J.; Oehninger, S.; Sun, Y.P.; Bocca, S. MFGE8 Regulates TGF-β-Induced Epithelial Mesenchymal Transition in Endometrial Epithelial Cells in Vitro. Reproduction 2016, 152, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Popli, P.; Shukla, V.; Kaushal, J.B.; Kumar, R.; Gupta, K.; Dwivedi, A. Peroxiredoxin 6 Plays Essential Role in Mediating Fertilization and Early Embryonic Development in Rabbit Oviduct. Reprod. Sci. 2022, 29, 1560–1576. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Yang, J.; Lv, H.; Lv, S.; Zhang, C.; Chen, Z.J. Dysfunction of Pseudogene PGK1P2 is Involved in Preeclampsia by Acting as a Competing Endogenous RNA of PGK1. Pregnancy Hypertens. 2018, 13, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Killeen, A.P.; Morris, D.G.; Kenny, D.A.; Mullen, M.P.; Diskin, M.G.; Waters, S.M. Global Gene Expression in Endometrium of High and Low Fertility Heifers during the Mid-Luteal Phase of the Estrous Cycle. BMC Genom. 2014, 15, 234. [Google Scholar] [CrossRef]
- Munch, E.M.; Sparks, A.E.; Gonzalez Bosquet, J.; Christenson, L.K.; Devor, E.J.; Van Voorhis, B.J. Differentially Expressed Genes in Preimplantation Human Embryos: Potential Candidate Genes for Blastocyst Formation and Implantation. J. Assist. Reprod. Genet. 2016, 33, 1017–1025. [Google Scholar] [CrossRef]
- Beraldo, F.H.; Soares, I.N.; Goncalves, D.F.; Fan, J.; Thomas, A.A.; Santos, T.G.; Mohammad, A.H.; Roffé, M.; Calder, M.D.; Nikolova, S.; et al. Stress-Inducible Phosphoprotein 1 Has Unique Cochaperone Activity during Development and Regulates Cellular Response to Ischemia via the Prion Protein. FASEB J. 2013, 27, 3594–3607. [Google Scholar] [CrossRef]
- Sugino, N.; Shimamura, K.; Takiguchi, S.; Ono, M.; Nakata, M.; Nakamura, Y.; Uda, T.; Kato, H. Changes in Activity of Superoxide Dismutase in the Human Endometrium throughout the Menstrual Cycle and in Early Pregnancy. Hum Reprod. 1996, 11, 1073–1078. [Google Scholar] [CrossRef]
- Combelles, C.M.H.; Holick, E.A.; Racowsky, C. Release of Superoxide Dismutase-1 by Day 3 Embryos of Varying Quality and Implantation Potential. J. Assist. Reprod. Genet. 2012, 29, 305–311. [Google Scholar] [CrossRef]
- Ribeiro, J.C.; Braga, P.C.; Martins, A.D.; Silva, B.M.; Alves, M.G.; Oliveira, P.F. Antioxidants Present in Reproductive Tract Fluids and Their Relevance for Fertility. Antioxidants 2021, 10, 1441. [Google Scholar] [CrossRef]
- Smits, K.; Willems, S.; Van Steendam, K.; Van De Velde, M.; De Lange, V.; Ververs, C.; Roels, K.; Govaere, J.; Van Nieuwerburgh, F.; Peelman, L.; et al. Proteins Involved in Embryo- Maternal Interaction around the Signalling of Maternal Recognition of Pregnancy in the Horse. Sci. Rep. 2018, 8, 5249. [Google Scholar] [CrossRef]
- Pierzchała, D.; Liput, K.; Korwin-Kossakowska, A.; Ogłuszka, M.; Poławska, E.; Nawrocka, A.; Urbański, P.; Ciepłoch, A.; Juszczuk-Kubiak, E.; Lepczyński, A.; et al. Molecular Characterisation of Uterine Endometrial Proteins during Early Stages of Pregnancy in Pigs by Maldi Tof/Tof. Int. J. Mol. Sci. 2021, 22, 6720. [Google Scholar] [CrossRef] [PubMed]
- Schliffka, M.F.; Tortorelli, A.F.; Özgüç, Ö.; de Plater, L.; Polzer, O.; Pelzer, D.; Maître, J.L. Multiscale Analysis of Single and Double Maternal-Zygotic Myh9 and Myh10 Mutants during Mouse Preimplantation Development. Elife 2021, 10, e68536. [Google Scholar] [CrossRef] [PubMed]
- Schulz, L.C.; Bahr, J.M. Glucose-6-Phosphate Isomerase is Necessary for Embryo Implantation in the Domestic Ferret. Proc. Natl. Acad. Sci. USA 2003, 100, 8561–8566. [Google Scholar] [CrossRef]
- Bondza, P.K.; Metz, C.N.; Akoum, A. Postgestational Effects of Macrophage Migration Inhibitory Factor on Embryonic Implantation in Mice. Fertil. Steril. 2008, 90, 1433–1443. [Google Scholar] [CrossRef]
- Loones, M.T.; Rallu, M.; Mezger, V.; Morange, M. HSP Gene Expression and HSF2 in Mouse Development. Cell. Mol. Life Sci. 1997, 53, 179–190. [Google Scholar] [CrossRef]
- Chin, P.Y.; MacPherson, A.M.; Thompson, J.G.; Lane, M.; Robertson, S.A. Stress Response Genes are Suppressed in Mouse Preimplantation Embryos by Granulocyte-Macrophage Colony-Stimulating Factor (GM-CSF). Hum. Reprod. 2009, 24, 2997–3009. [Google Scholar] [CrossRef] [PubMed]
- Samakovli, D.; Tichá, T.; Vavrdová, T.; Závorková, N.; Pecinka, A.; Ovecka, M.; Samaj, J. HEAT SHOCK PROTEIN 90 Proteins and YODA Regulate Main Body Axis Formation during Early Embryogenesis. Plant Physiol. 2021, 186, 1526–1544. [Google Scholar] [CrossRef]
- Chen, X.; Chen, X. PRM-Based Quantitative Proteomics Analysis of Altered HSPs Expression in Villi and Decidua of Early Missed Abortion Patients. Res. Square 2022, preprint. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, J.; Wang, Y.; Zhao, W.; Wang, W.; Cui, J.; Yang, J.; Yue, Y.; Zhang, S.; Chu, M.; et al. A Proteomic Atlas of Ligand-Receptor Interactions at the Ovine Maternal-Fetal Interface Reveals the Role of Histone Lactylation in Uterine Remodeling. J. Biol. Chem. 2022, 298, 101456. [Google Scholar] [CrossRef]
- Fisher, S.J.; Francisco, S.; Francisco, S.; Francisco, S.; Francisco, S.; Francisco, S.; Francisco, S.; Francisco, S.; Francisco, S. Tumour Exosome Integrins Determine Organotropic Metastasis. Nature 2016, 213, 329–335. [Google Scholar] [CrossRef]
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic Profiling of Extracellular Vesicles Allows for Human Breast Cancer Subtyping. Commun. Biol. 2019, 2, 325. [Google Scholar] [CrossRef]
- Segura-Benítez, M.; Carbajo-García, M.C.; Corachán, A.; Faus, A.; Pellicer, A.; Ferrero, H. Proteomic Analysis of Extracellular Vesicles Secreted by Primary Human Epithelial Endometrial Cells Reveals Key Proteins Related to Embryo Implantation. Reprod. Biol. Endocrinol. 2022, 20, 3. [Google Scholar] [CrossRef]
- Truong, T.; Gardner, D.K. Antioxidants Improve IVF Outcome and Subsequent Embryo Development in the Mouse. Hum. Reprod. 2017, 32, 2404–2413. [Google Scholar] [CrossRef]
- Van Winkle, L.J. Amino Acid Transport and Metabolism Regulate Early Embryo Development: Species Differences, Clinical Significance, and Evolutionary Implications. Cells 2021, 10, 3154. [Google Scholar] [CrossRef]
- Huo, P.; Zhu, Y.; Liang, C.; Yao, J.; Le, J.; Qin, L.; Lei, X.; Zhang, S. Non-Invasive Amino Acid Profiling of Embryo Culture Medium Using HPLC Correlates with Embryo Implantation Potential in Women Undergoing in Vitro Fertilization. Front. Physiol. 2020, 11, 405. [Google Scholar] [CrossRef]
- Rebecca, L.; Krisher, R.S.P. A Role for the Warburg Effect in Preimplantation Embryo Development: Metabolic Modification to Support Rapid Cell Proliferation. Mol. Repro. Dev. 2012, 23, 311–320. [Google Scholar] [CrossRef]
- Wang, X.; Matsumoto, H.; Zhao, X.; Das, S.K.; Paria, B.C. Embryonic Signals Direct the Formation of Tight Junctional Permeability Barrier in the Decidualizing Stroma during Embryo Implantation. J. Cell Sci. 2004, 117, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Whitby, S.; Zhou, W.; Dimitriadis, E. Alterations in Epithelial Cell Polarity during Endometrial Receptivity: A Systematic Review. Front. Endocrinol. 2020, 11, 596324. [Google Scholar] [CrossRef] [PubMed]
- Jee, B.; Dhar, R.; Singh, S.; Karmakar, S. Heat Shock Proteins and Their Role in Pregnancy: Redefining the Function of “Old Rum in a New Bottle”. Front. Cell Dev. Biol. 2021, 9, 648463. [Google Scholar] [CrossRef] [PubMed]
- Okan, A.; Demir, N.; Sozen, B. Unfolded Protein Response Triggers Differential Apoptotic Mechanisms in Ovaries and Early Embryos Exposed to Maternal Type 1 Diabetes. Sci. Rep. 2021, 11, 12759. [Google Scholar] [CrossRef] [PubMed]
- Ufer, C.; Wang, C.C. The Roles of Glutathione Peroxidases during Embryo Development. Front. Mol. Neurosci. 2011, 4, 11531. [Google Scholar] [CrossRef] [PubMed]
- Bedaiwy, M.; Agarwal, A.; Said, T.M.; Goldberg, J.M.; Sharma, R.K.; Worley, S.; Falcone, T. Role of Total Antioxidant Capacity in the Differential Growth of Human Embryos in Vitro. Fertil. Steril. 2006, 86, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Paszkowski, T.; Clarke, R.N. Antioxidative Capacity of Preimplantation Embryo Culture Medium Declines Following the Incubation of Poor Quality Embryos. Hum. Reprod. 1996, 11, 2493–2495. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Orsi, N.M.; Leese, H.J. Protection against Reactive Oxygen Species during Mouse Preimplantation Embryo Development: Role of EDTA, Oxygen Tension, Catalase, Superoxide Dismutase and Pyruvate. Mol. Reprod. Dev. 2001, 53, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Riou, C.; Brionne, A.; Cordeiro, L.; Harichaux, G.; Gargaros-ratajczak, A.; Labas, V.; Gautron, J.; Gérard, N.; Riou, C.; Brionne, A.; et al. Avian Uterine Fluid Proteome: Exosomes and Biological Processes Potentially Involved in Sperm Survival. Mol. Reprod. Dev. 2020, 87, 454–470. [Google Scholar] [CrossRef]
- Achache, H.; Revel, A. Endometrial Receptivity Markers, the Journey to Successful Embryo Implantation. Hum. Reprod. 2006, 12, 731–746. [Google Scholar] [CrossRef]
- O’Connor, B.B.; Pope, B.D.; Peters, M.M.; Ris-Stalpers, C.; Parker, K.K. The Role of Extracellular Matrix in Normal and Pathological Pregnancy: Future Applications of Microphysiological Systems in Reproductive Medicine. Exp. Biol. Med. 2020, 245, 1163–1174. [Google Scholar] [CrossRef]
- Kaloglu, C.; Onarlioglu, B. Extracellular Matrix Remodelling in Rat Endometrium during Early Pregnancy: The Role of Fibronectin and Laminin. Tissue Cell 2010, 42, 301–306. [Google Scholar] [CrossRef]
- Whitby, S.; Salamonsen, L.A.; Evans, J. The Endometrial Polarity Paradox: Differential Regulation of Polarity within Secretory-Phase Human Endometrium. Endocrinology 2018, 159, 506–518. [Google Scholar] [CrossRef]
- Yuan, J.; Cha, J.; Deng, W.; Bartos, A.; Sun, X.; Ho, H.Y.H.; Borg, J.P.; Yamaguchi, T.P.; Yang, Y.; Dey, S.K.; et al. Planar Cell Polarity Signaling in the Uterus Directs Appropriate Positioning of the Crypt for Embryo Implantation. Proc. Natl. Acad. Sci. USA 2016, 113, E8079–E8088. [Google Scholar] [CrossRef]
- Teh, W.T.; McBain, J.; Rogers, P. What is the Contribution of Embryo-Endometrial Asynchrony to Implantation Failure? J. Assist. Reprod. Genet. 2016, 33, 1419–1430. [Google Scholar] [CrossRef]
- Greening, D.W.; Nguyen, H.P.T.; Elgass, K.; Simpson, R.J.; Salamonsen, L.A. Human Endometrial Exosomes Contain Hormone-Specific Cargo Modulating Trophoblast Adhesive Capacity: Insights into Endometrial-Embryo Interactions. Biol. Reprod. 2016, 94, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.; Walker, K.J.; Bilandzic, M.; Kinnear, S.; Salamonsen, L.A. A Novel “Embryo-Endometrial” Adhesion Model Can Potentially Predict “Receptive” or “Non-Receptive” Endometrium. J. Assist. Reprod. Genet. 2020, 37, 5–16. [Google Scholar] [CrossRef]
- Bhagwat, S.R.; Redij, T.; Phalnikar, K.; Nayak, S.; Iyer, S.; Gadkar, S. Cell Surfactomes of Two Endometrial Epithelial Cell Lines That Differ in Their Adhesiveness to Embryonic Cells. Mol. Reprod. Dev. 2014, 340, 326–340. [Google Scholar] [CrossRef] [PubMed]
- Azizi, E.; Mofarahe, Z.S.; Naji, M. MicroRNAs, Small Regulatory Elements with Significant Effects on Human Implantation: A Review. J. Assist. Reprod. Genet. 2023, 40, 697–717. [Google Scholar] [CrossRef]
- Midekessa, G.; Godakumara, K.; Ord, J.; Viil, J.; Lättekivi, F.; Dissanayake, K.; Kopanchuk, S.; Rinken, A.; Andronowska, A.; Bhattacharjee, S.; et al. Zeta Potential of Extracellular Vesicles: Toward Understanding the Attributes That Determine Colloidal Stability. ACS Omega 2020, 5, 16701–16710. [Google Scholar] [CrossRef] [PubMed]
- Sticker, A.; Goeminne, L.; Martens, L.; Clement, L. Robust Summarization and Inference in Proteome-Wide Label-Free Quantification. Mol. Cell. Proteom. 2020, 19, 1209–1219. [Google Scholar] [CrossRef]
- Goeminne, L.J.E.; Sticker, A.; Martens, L.; Gevaert, K.; Clement, L. MSqRob Takes the Missing Hurdle: Uniting Intensity and Count-Based Proteomics. Anal. Chem. 2020, 92, 6278–6287. [Google Scholar] [CrossRef]

| KEGG Pathway | FDR | Fold Enrichment | Gene Names |
|---|---|---|---|
| Glutathione metabolism * | 4.0 × 10−1 | 6.6 | GSTP1, GSR, IDH1, PRDX6 |
| Biosynthesis of amino acids * | 4.40 × 10−1 | 6.6 | GOT1, IDH1, MAT2A, PGK1 |
| Metabolic pathways * | 6.90 × 10−1 | 24.6 | CNDP2, GPI, GOT1, GSTP1, GSR, HPRT1, ISYNA1, IDH1, LDHA, MIF, MAT2A, NIT2, PRDX6,PGK1, GALNT6 |
| Carbon metabolism * | 6.90 × 10−1 | 6.6 | GPI, GOT1, IDH1, PGK1, |
| Cysteine and methionine metabolism * | 7.40 × 10−1 | 4.9 | GOT1, LDHA, MAT2A |
| Glycolysis/Gluconeogenesis * | 9.4 × 10−1 | 4.9 | GPI, LDHA, PGK1 |
| Tight junction * | 9.4 × 10−1 | 6.6 | ACTB, MYH9, PCNA, PPP2R1A |
| Protein processing in endoplasmic reticulum * | 9.4 × 10−1 | 6.6 | RAD23B, CAPN1, HSP90AB1, LMAN2, |
| Peroxisome | 9.7 × 10−1 | 4.9 | IDH1, PRDX1, SOD1 |
| Phenylalanine metabolism | 1.0 × 10 | 3.3 | GOT1, MIF |
| 2-Oxocarboxylic acid metabolism | 1.0 × 10 | 3.3 | GOT1, IDH1 |
| UniProt Accession | Gene Name | Protein Description | Protein Fold Change | Function | References |
|---|---|---|---|---|---|
| P05997 | COL5A2 | Collagen alpha-2(V) chain | 5.28 | A main protein component of the extracellular matrix and upregulated in receptive endometrium and implantation sites. | [32,33] |
| P12004 | PCNA | Proliferating cell nuclear agent | 4.32 | Increased the expression of PCNA in stromal cells and myometrium with progressing gestation in rats and has a role in stromal cell proliferation. | [34] |
| P00390 | GSR | Glutathione reductase | 4.07 | Reported as a potential human cumulus cell quality marker for pregnancy prediction. Known to regulate glutathione (a major antioxidant) production in cells; hence, it has a role in embryo development. | [35,36] |
| P31153 | MAT2A | Methionine Adenosyltransferase 2A | 3.91 | Improves methionine-mediated DNA synthesis through the SAMTOR/mTORC1/S6K1/CAD pathway during human embryo implantation. Supports peri-conception embryo development in bovine. | [37,38] |
| O14980 | XPO1 | Exportin-1 | 3.86 | Linked with repeated implantation failure by affecting the proliferation and differentiation of endometrial stromal cells in humans. | [39] |
| P17931 | LGALS3 | Galectin-3 | 3.95 | Amplifies the inflammatory response; hence, it might play a role in early embryo implantation. Increased in the embryo implantation site and required for embryo implantation in mice. Increased LGALS3 induces endothelial cells morphogenesis and angiogenesis. Previously reported as being upregulated in Ishikawa cells treated with trophectoderm EVs (intracellularly). | [14,40,41,42,43] |
| P68036 | UBE2L3 | Ubiquitin-conjugating enzyme E2 L3 | 3.12 | Only expressed in the uterus during pregnancy, supports embryo survival, and increases implantation potential in mice. | [44] |
| Q13753 | LAMC2 | Laminin subunit gamma-2 | 2.82 | Interacting molecule in the embryo–endometrial interface. | [45] |
| P32119 | PRDX2 | Peroxiredoxin-2 | 2.82 | Regulates trophoblast cell proliferation and apoptosis during early pregnancy and is mediated by c-Myc. Downregulation is linked with recurrent miscarriages. Previously reported as being upregulated in Ishikawa cells treated with trophectoderm EVs (intracellularly). | [14,27] |
| P50990 | CCT8 | T-complex protein 1 subunit theta | 3.87 | Altered in females with endometriosis during the window of implantation in humans. | [46] |
| Q08431 | MFGE8 | Lactadherin | 2.75 | Expressed in embryo–maternal interface in humans and equine. Known to play a role in embryo attachment to the endometrial epithelial cells. Increased secretion has been linked with stimulation by hCG. Previously identified as being significantly upregulated in Ishikawa cells treated with trophectoderm EVs (intracellularly). | [14,47,48,49,50,51] |
| P30041 | PRDX6 | Peroxiredoxin 6 | 2.78 | Mediates antioxidant activity; hence, it is important for embryo development. Previously identified as changing intracellularly in Ishikawa cells treated with trophectoderm. | [14,52] |
| P00558 | PGK1 | Phosphoglycerate kinase 1 | 2.69 | Increased in in vitro decidualization in endometrial stromal cells by regulating angiogenesis and glycolysis. Deficiency leads to impaired decidualization. | [53] |
| P52434 | POLR2H | RNA polymerase II, I and III subunit H | 2.66 | Increased in endometrium in high fertility heifers in the midluteal phase of the estrous cycle. | [54] |
| Q8NCL4 | GALNT6 | Polypeptide N-acetylgalactosaminyltransferase 6 | 2.54 | Upregulated in human blastocyst-stage embryos; potentially involved in the synthesis of oncofetal fibronectin, thus facilitating embryo attachment to endometrium. | [55] |
| P00338 | LDHA | Lactate dehydrogenase A | 2.35 | Lactate dehydrogenase (LDH) isoform, LDHB (which favors pyruvate formation) is transformed to LDHA (which favors lactate formation) during the early phase of embryo implantation in blastocysts that can potentially support tissue invasion. Previously reported as being upregulated in Ishikawa cells treated with trophectoderm-derived EVs (intracellularly). | [14,31] |
| P31948 | STIP1 | Stress-inducible phosphoprotein 1 | 2.28 | Lack of STIP1 causes embryonic lethality in mice. | [56] |
| P00441 | SOD1 | Superoxide dismutase 1 | 2.27 | Activity peaks in the midluteal phase of the menstrual cycle in humans. Released by human embryos and found in IVF-spent media, but its relation to implantation potential is not clear. Has been linked with fertility capacity in mice. Previously reported as being upregulated in Ishikawa cells treated with trophectoderm-derived EVs (intracellularly). | [14,57,58,59] |
| P09211 | GSTP1 | Glutathione S-transferase Pi | 2.11 | Increased in the uterine fluid of pregnant mares compared to cyclin mares. Reduce inflammation by reducing cyclooxygenase-2 (COX-2). Previously reported as being upregulated in Ishikawa cells treated with trophectoderm-derived EVs (intracellularly). | [14,60] |
| P17987 | TCP1 | T-complex 1 | 2.1 | Increased during pregnancy in horses and potentially play a role in the maternal recognition of pregnancy. Downregulated in pregnancy loss compared to healthy pregnancies. Necessary for folding newly synthesized proteins such as actin and tubulin. Previously reported as being upregulated in Ishikawa cells treated with trophectoderm-derived EVs (intracellularly). | [14,61] |
| P35579 | MYH9 | Myosin heavy chain 9 | 2.02 | Loss of MYH9 is lethal to embryos and plays a key role in cytokinesis in mice. | [62] |
| P06744 | GPI | Glucose-6-phosphate isomerase | 2.53 | Plays a role in glycolysis and is needed for embryo implantation in ferrets. | [63] |
| P14174 | MIF | Macrophage migration inhibitory factor | 2.43 | A pro-inflammatory cytokine that showed a slight increase in the secretory phase of the menstrual cycle in humans. | [28,64] |
| P08238 | HSP90AB1 | Heat shock protein HSP 90-beta | 2.18 | Downregulated in human villi and decidua of early missed abortion patients. Play roles in placental development and cell proliferation in early mouse embryo development. Previously reported as being upregulated in Ishikawa cells treated with trophectoderm-derived EVs (intracellularly). | [14,65,66,67,68,69] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muhandiram, S.; Dissanayake, K.; Orro, T.; Godakumara, K.; Kodithuwakku, S.; Fazeli, A. Secretory Proteomic Responses of Endometrial Epithelial Cells to Trophoblast-Derived Extracellular Vesicles. Int. J. Mol. Sci. 2023, 24, 11924. https://doi.org/10.3390/ijms241511924
Muhandiram S, Dissanayake K, Orro T, Godakumara K, Kodithuwakku S, Fazeli A. Secretory Proteomic Responses of Endometrial Epithelial Cells to Trophoblast-Derived Extracellular Vesicles. International Journal of Molecular Sciences. 2023; 24(15):11924. https://doi.org/10.3390/ijms241511924
Chicago/Turabian StyleMuhandiram, Subhashini, Keerthie Dissanayake, Toomos Orro, Kasun Godakumara, Suranga Kodithuwakku, and Alireza Fazeli. 2023. "Secretory Proteomic Responses of Endometrial Epithelial Cells to Trophoblast-Derived Extracellular Vesicles" International Journal of Molecular Sciences 24, no. 15: 11924. https://doi.org/10.3390/ijms241511924
APA StyleMuhandiram, S., Dissanayake, K., Orro, T., Godakumara, K., Kodithuwakku, S., & Fazeli, A. (2023). Secretory Proteomic Responses of Endometrial Epithelial Cells to Trophoblast-Derived Extracellular Vesicles. International Journal of Molecular Sciences, 24(15), 11924. https://doi.org/10.3390/ijms241511924





