Microtubule-Associated Serine/Threonine (MAST) Kinases in Development and Disease
Abstract
1. Introduction
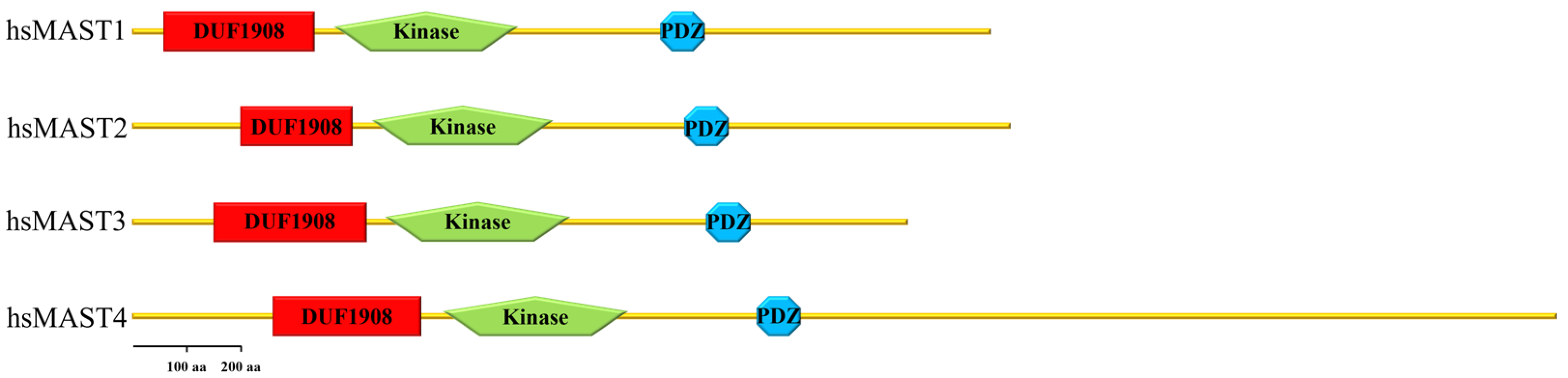
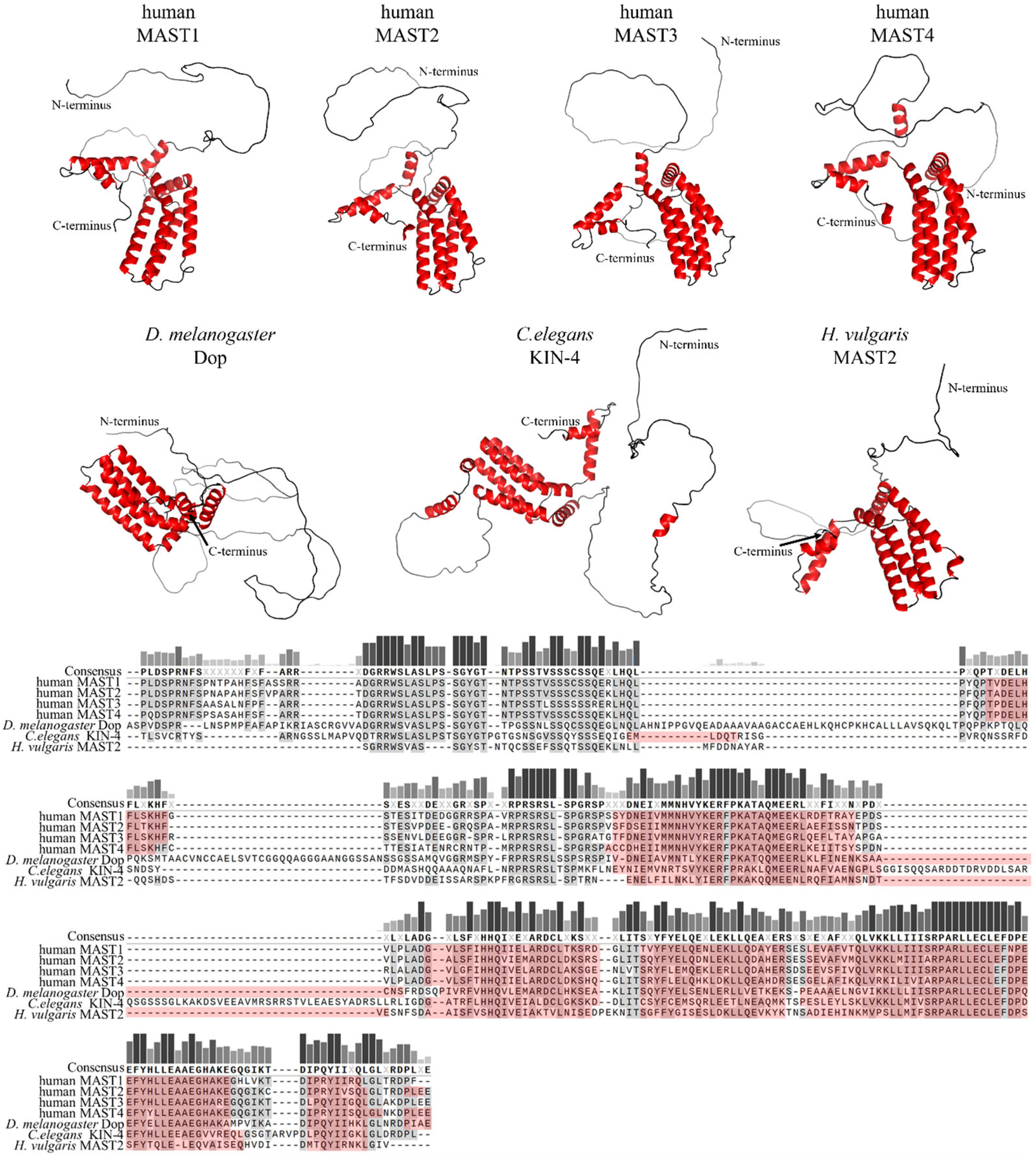
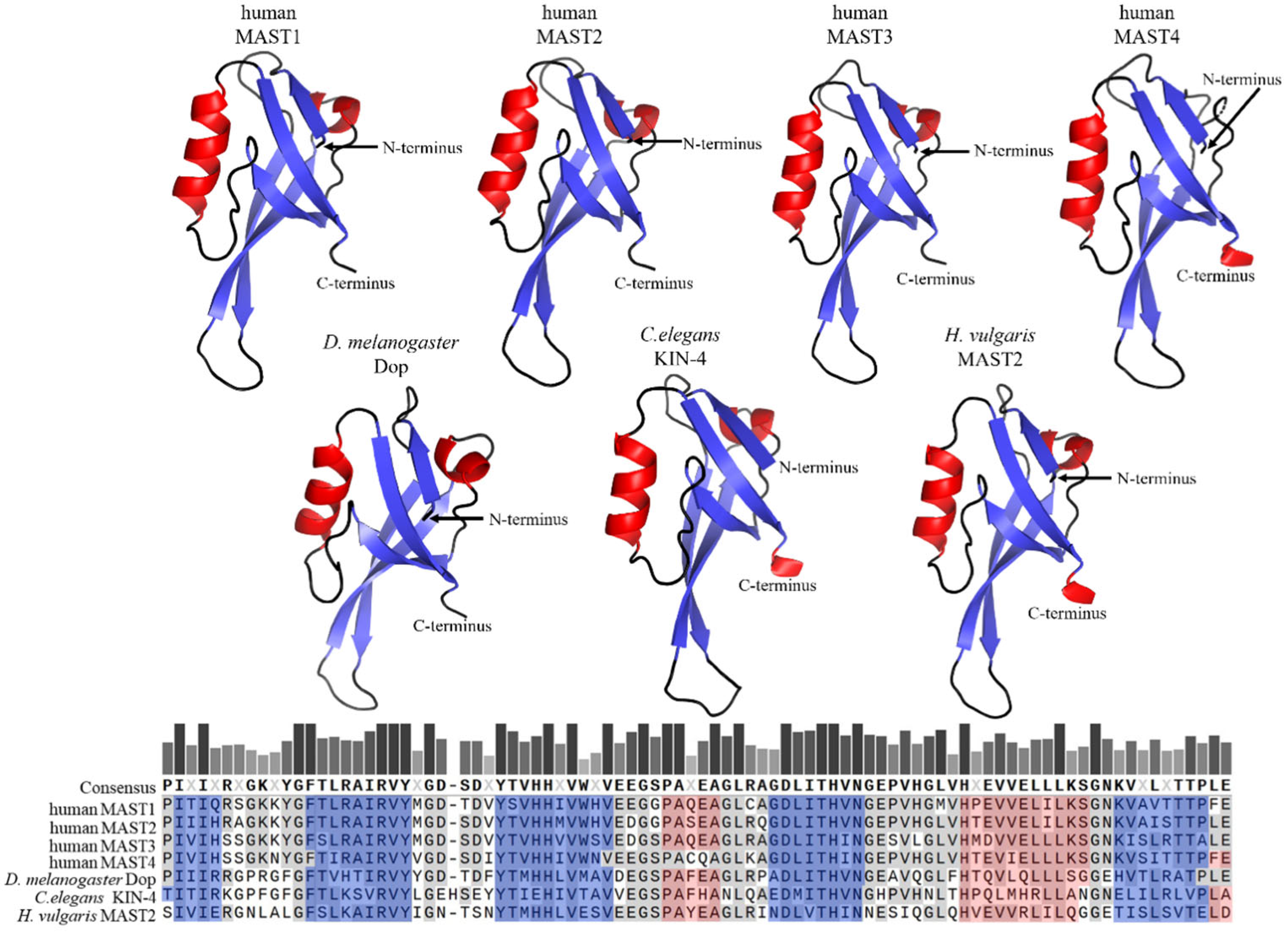
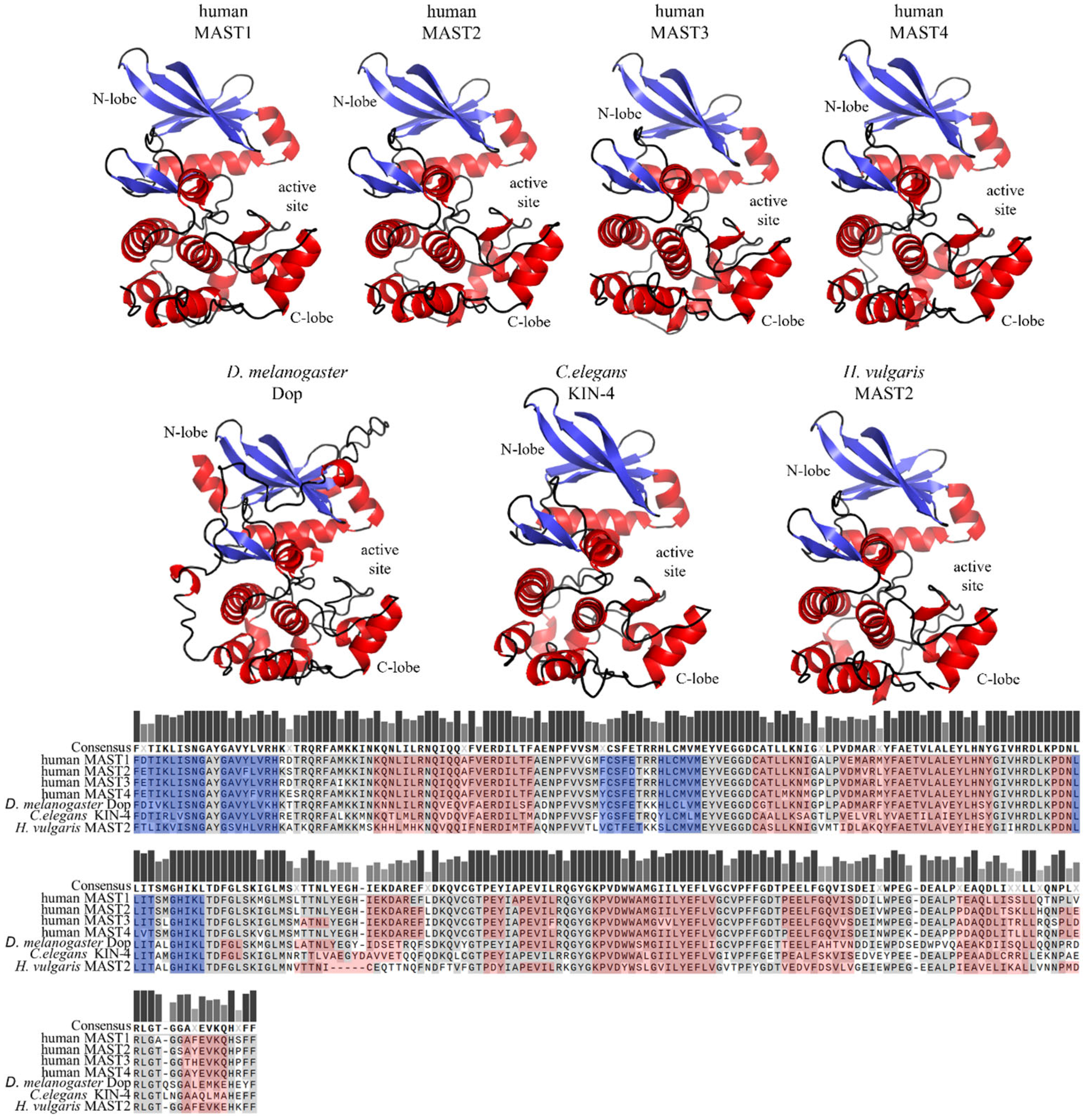
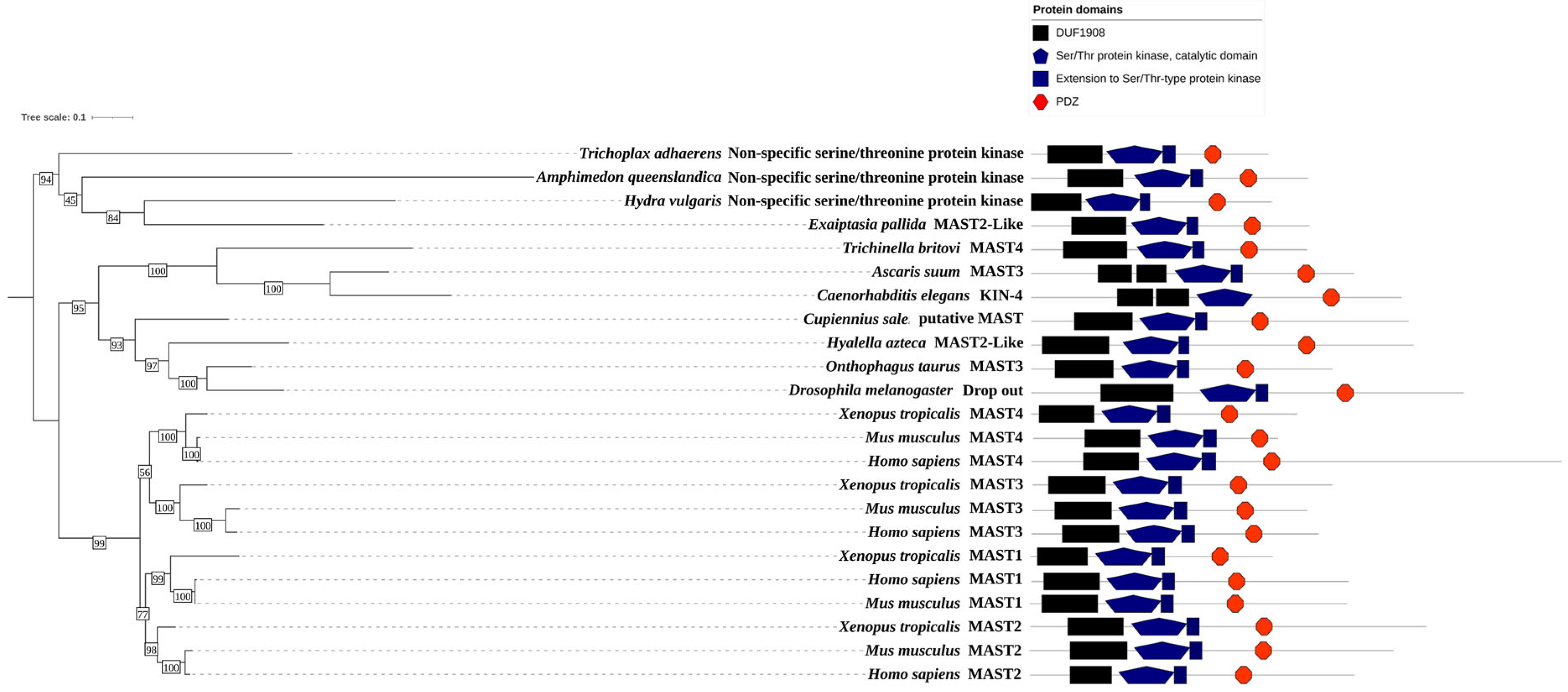
2. Domain Composition of MAST Kinases
3. Substrates and Interactors of MAST Kinases
| MASTK | Interactor | Phosphosite 1 | Interaction Domain 2 | References |
|---|---|---|---|---|
| MAST1 | USP1 | - | PDZ | [44] |
| MAST1 | Cdh1 | - | PDZ | [44] |
| MAST1 | CHIP | - | K317, K545 | [45] |
| MAST1 | MEK | S221 | - | [46] |
| MAST1 | c-Raf | - | - | [46] |
| MAST1 | HSP90 | - | - | [45] |
| MAST1 MAST2 | SNTB2 | - | PDZ | [36] |
| MAST1 MAST2 MAST3 | PTEN | C-term. | PDZ | [26] |
| MAST1 MAST2 | MAPs MAPs | - | KD + aa 948–1212 | [11,47] |
| MAST2 | TRAF6 | - | N-term. | [38] |
| MAST2 | RABV-G | - | PDZ | [21] |
| MAST2 | CFTR | - | PDZ | [48] |
| MAST2 | NHE3 | n.d. | PDZ | [34] |
| MAST2 | PCLKC | - | PDZ | [37] |
| MAST2 | 14-3-3 | - | - | [41] |
| MAST3 | ARPP-16 | S46 | - | [49] |
| MAST4 | Sox9 | S494 | - | [50] |
4. MAST Kinases in Human Disease
4.1. Association of MAST Kinase Mutations in Cancer
4.2. MAST Kinases in Neuronal Diseases
4.3. MAST Kinases in Cystic Fibrosis and Diarrhea
4.4. MAST Kinases in Male Fertility
4.5. MAST Kinases in Inflammation
| Disease Subgroup 1 | MASTK | Disease 2 | Cause 3 | References |
|---|---|---|---|---|
| Infertility | MAST2 | Nonobstructive azoospermia | Gene Duplication | [83] |
| Cancer | MAST4 | Multiple myeloma bone disease | Overexpressed | [30] |
| Acral melanoma | Various deleterious mutations | [85] | ||
| Ductal carcinoma in-situ Invasive breast cancer | Upregulated | [86] | ||
| MAST3 | Prostate cancer | Gene conversion | [87] | |
| MAST2 | Cutaneous melanoma | Translocation | [88] | |
| MAST2 | Esophageal cancer Pancreatic cancer Sarcomas | Overexpressed | [65] | |
| MAST2 | Liver cancer | Overexpression | [67] | |
| MAST2 | Chronic myeloid leukemia | Insertion of exon 8 in a BCR-ABL1 fusion gene | [89] | |
| MAST2 | Breast cancer | Translocation | [64] | |
| MAST2 | Breast cancer | Gene fusion | [60] | |
| MAST1 | ||||
| MAST1 | Breast cancer | High levels of DNA methylation | [90] | |
| MAST1 | Non-small-cell lung cancer | Upregulated | [91] | |
| MAST1 | Pheochromocytoma, paraganglioma | Overexpression by hypomethylation | [92] | |
| MAST1 | Hepatocellular carcinoma | Upregulated circRNA | [66] | |
| MAST1 | Uterine corpus endometrial carcinoma | Upregulated | [93] | |
| MAST1 | Lung cancer | S81Y C291F V316E | [94] | |
| Cardiovascular Diseases | MAST2 | Venous thrombosis | R89Q | [95] |
| Neuronal diseases | MAST3 | Developmental and epileptic encephalopathy | S101F S104L G515S L516P | [96] |
| MAST3 | Developmental and epileptic encephalopathy | G510S G515S | [97] | |
| MAST4 | Juvenile myoclonic epilepsy (JME) | T347M | [98] | |
| Childhood absence epilepsy | P1201R | |||
| MAST1 | Intellectual disability, speech delay, hypotonia, facial dysmorphism, autism | S93L | [99] | |
| MAST1 | Cerebral palsy, intellectual disability | P500L | [100] | |
| MAST1 | Intellectual disability | P1177R | [101] | |
| MAST1 | Neurologic abnormalities, developmental disability, mental retardation | Deletion | [102] | |
| MAST1 | Intellectual disability | L1180R | [103] | |
| MAST1 | Congenital bilateral Perisylvian syndrome | Deletion of Q223 to D230 | [104] | |
| MAST1 | Mega-corpus-callosum syndrome with cortical malformations without cerebellar Hypoplasia | G522E | [105] | |
| MAST1 | Mega-corpus-callosum syndrome with cerebellar hypoplasia and cortical malformations | L278del E194del K276del G517S E697del | [47] | |
| Inflammatory bowel disease | MAST3 | Crohn’s disease (CD) and ulcerative colitis (UC) | S861G | [40] |
| Others | MAST4 | Asthma (horses) | Overexpressed | [106] |
| MAST3 | Hepatic steatosis | Intronic variant | [107] | |
| MAST3 | Rheumatoid arthritis | Overexpressed in fibroblast-like synovial cells | [108] | |
| MAST2 | Type 2 diabetes mellitus | A1463T | [39] | |
| MAST2 | Rabies infection | Viral glycoprotein prevents complex formation (MAST2-PDZ and PTEN) and promotes neuronal survival | [21,33,63,109,110] |
5. MAST Kinases in Model Organisms
6. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Richardson, C.J.; Gao, Q.; Mitsopoulous, C.; Zvelebil, M.; Pearl, L.H.; Pearl, F.M. MoKCa database--mutations of kinases in cancer. Nucleic Acids Res. 2009, 37, D824–D831. [Google Scholar] [CrossRef][Green Version]
- Hanks, S.K.; Quinn, A.M.; Hunter, T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 1988, 241, 42–52. [Google Scholar] [CrossRef]
- Hanks, S.K.; Hunter, T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995, 9, 576–596. [Google Scholar] [CrossRef]
- Pearce, L.R.; Komander, D.; Alessi, D.R. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010, 11, 9–22. [Google Scholar] [CrossRef]
- Hanks, S.K. Genomic analysis of the eukaryotic protein kinase superfamily: A perspective. Genome Biol. 2003, 4, 111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Johnson, D.A.; Akamine, P.; Radzio-Andzelm, E.; Madhusudan, M.; Taylor, S.S. Dynamics of cAMP-dependent protein kinase. Chem. Rev. 2001, 101, 2243–2270. [Google Scholar] [CrossRef] [PubMed]
- Kannan, N.; Haste, N.; Taylor, S.S.; Neuwald, A.F. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc. Natl. Acad. Sci. USA 2007, 104, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Knighton, D.R.; Zheng, J.H.; Ten Eyck, L.F.; Xuong, N.H.; Taylor, S.S.; Sowadski, J.M. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 1991, 253, 414–420. [Google Scholar] [CrossRef]
- Taylor, S.S.; Soberg, K.; Kobori, E.; Wu, J.; Pautz, S.; Herberg, F.W.; Skalhegg, B.S. The Tails of Protein Kinase A. Mol. Pharmacol. 2022, 101, 219–225. [Google Scholar] [CrossRef]
- Walden, P.D.; Cowan, N.J. A novel 205-kilodalton testis-specific serine/threonine protein kinase associated with microtubules of the spermatid manchette. Mol. Cell Biol. 1993, 13, 7625–7635. [Google Scholar] [PubMed]
- Walden, P.D.; Millette, C.F. Increased activity associated with the MAST205 protein kinase complex during mammalian spermiogenesis. Biol. Reprod. 1996, 55, 1039–1044. [Google Scholar] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Monzon, V.; Paysan-Lafosse, T.; Wood, V.; Bateman, A. Reciprocal best structure hits: Using AlphaFold models to discover distant homologues. Bioinform. Adv. 2022, 2, vbac072. [Google Scholar]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Liu, X.; Fuentes, E.J. Emerging Themes in PDZ Domain Signaling: Structure, Function, and Inhibition. Int. Rev. Cell Mol. Biol. 2019, 343, 129–218. [Google Scholar]
- Ponting, C.P.; Phillips, C.; Davies, K.E.; Blake, D.J. PDZ domains: Targeting signalling molecules to sub-membranous sites. Bioessays 1997, 19, 469–479. [Google Scholar] [CrossRef]
- Songyang, Z.; Fanning, A.S.; Fu, C.; Xu, J.; Marfatia, S.M.; Chishti, A.H.; Crompton, A.; Chan, A.C.; Anderson, J.M.; Cantley, L.C. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 1997, 275, 73–77. [Google Scholar]
- Nourry, C.; Grant, S.G.; Borg, J.P. PDZ domain proteins: Plug and play! Sci. STKE 2003, 2003, RE7. [Google Scholar] [CrossRef]
- Terrien, E.; Chaffotte, A.; Lafage, M.; Khan, Z.; Prehaud, C.; Cordier, F.; Simenel, C.; Delepierre, M.; Buc, H.; Lafon, M.; et al. Interference with the PTEN-MAST2 interaction by a viral protein leads to cellular relocalization of PTEN. Sci. Signal 2012, 5, ra58. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Zheng, J.J. PDZ domains and their binding partners: Structure, specificity, and modification. Cell Commun. Signal 2010, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Hung, A.Y.; Sheng, M. PDZ domains: Structural modules for protein complex assembly. J. Biol. Chem. 2002, 277, 5699–5702. [Google Scholar] [CrossRef]
- Brenman, J.E.; Chao, D.S.; Gee, S.H.; McGee, A.W.; Craven, S.E.; Santillano, D.R.; Wu, Z.; Huang, F.; Xia, H.; Peters, M.F.; et al. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell 1996, 84, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.H.; Gujral, T.S.; Karp, E.S.; BuKhalid, R.; Grantcharova, V.P.; MacBeath, G. A systematic family-wide investigation reveals that ~30% of mammalian PDZ domains engage in PDZ-PDZ interactions. Chem. Biol. 2011, 18, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Valiente, M.; Andres-Pons, A.; Gomar, B.; Torres, J.; Gil, A.; Tapparel, C.; Antonarakis, S.E.; Pulido, R. Binding of PTEN to specific PDZ domains contributes to PTEN protein stability and phosphorylation by microtubule-associated serine/threonine kinases. J. Biol. Chem. 2005, 280, 28936–28943. [Google Scholar] [CrossRef]
- Adey, N.B.; Huang, L.; Ormonde, P.A.; Baumgard, M.L.; Pero, R.; Byreddy, D.V.; Tavtigian, S.V.; Bartel, P.L. Threonine phosphorylation of the MMAC1/PTEN PDZ binding domain both inhibits and stimulates PDZ binding. Cancer Res. 2000, 60, 35–37. [Google Scholar]
- Lee, Y.R.; Chen, M.; Pandolfi, P.P. The functions and regulation of the PTEN tumour suppressor: New modes and prospects. Nat. Rev. Mol. Cell Biol. 2018, 19, 547–562. [Google Scholar] [CrossRef]
- Vazquez, F.; Ramaswamy, S.; Nakamura, N.; Sellers, W.R. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell Biol. 2000, 20, 5010–5018. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, F.; Zhang, D.; Huang, J.; Yang, Y.; Xu, J.; Gao, Y.; Ding, H.; Qu, Y.; Zhang, W.; et al. Estrogen-Responsive Gene MAST4 Regulates Myeloma Bone Disease. J. Bone Miner. Res. 2022, 37, 711–723. [Google Scholar] [CrossRef]
- Loh, S.H.; Francescut, L.; Lingor, P.; Bahr, M.; Nicotera, P. Identification of new kinase clusters required for neurite outgrowth and retraction by a loss-of-function RNA interference screen. Cell Death Differ. 2008, 15, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Gregorian, C.; Nakashima, J.; Dry, S.M.; Nghiemphu, P.L.; Smith, K.B.; Ao, Y.; Dang, J.; Lawson, G.; Mellinghoff, I.K.; Mischel, P.S.; et al. PTEN dosage is essential for neurofibroma development and malignant transformation. Proc. Natl. Acad. Sci. USA 2009, 106, 19479–19484. [Google Scholar] [CrossRef] [PubMed]
- Prehaud, C.; Wolff, N.; Terrien, E.; Lafage, M.; Megret, F.; Babault, N.; Cordier, F.; Tan, G.S.; Maitrepierre, E.; Menager, P.; et al. Attenuation of rabies virulence: Takeover by the cytoplasmic domain of its envelope protein. Sci. Signal 2010, 3, ra5. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lee, H.J.; Cooper, D.S.; Cebotaro, L.; Walden, P.D.; Choi, I.; Yun, C.C. Coexpression of MAST205 inhibits the activity of Na+/H+ exchanger NHE3. Am. J. Physiol. Renal Physiol. 2006, 290, F428–F437. [Google Scholar] [CrossRef]
- Alexander, R.T.; Grinstein, S. Tethering, recycling and activation of the epithelial sodium-proton exchanger, NHE3. J. Exp. Biol. 2009, 212 Pt 11, 1630–1637. [Google Scholar] [CrossRef]
- Lumeng, C.; Phelps, S.; Crawford, G.E.; Walden, P.D.; Barald, K.; Chamberlain, J.S. Interactions between beta 2-syntrophin and a family of microtubule-associated serine/threonine kinases. Nat. Neurosci. 1999, 2, 611–617. [Google Scholar] [CrossRef]
- Okazaki, N.; Takahashi, N.; Kojima, S.; Masuho, Y.; Koga, H. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation. Carcinogenesis 2002, 23, 1139–1148. [Google Scholar] [CrossRef]
- Xiong, H.; Li, H.; Chen, Y.; Zhao, J.; Unkeless, J.C. Interaction of TRAF6 with MAST205 regulates NF-kappaB activation and MAST205 stability. J. Biol. Chem. 2004, 279, 43675–43683. [Google Scholar] [CrossRef]
- Zhou, H.; Xiong, H.; Li, H.; Plevy, S.E.; Walden, P.D.; Sassaroli, M.; Prestwich, G.D.; Unkeless, J.C. Microtubule-associated serine/threonine kinase-205 kDa and Fc gamma receptor control IL-12 p40 synthesis and NF-kappa B activation. J. Immunol. 2004, 172, 2559–2568. [Google Scholar] [CrossRef]
- Labbe, C.; Goyette, P.; Lefebvre, C.; Stevens, C.; Green, T.; Tello-Ruiz, M.K.; Cao, Z.; Landry, A.L.; Stempak, J.; Annese, V.; et al. MAST3: A novel IBD risk factor that modulates TLR4 signaling. Genes. Immun. 2008, 9, 602–612. [Google Scholar] [CrossRef]
- Johnson, C.; Tinti, M.; Wood, N.T.; Campbell, D.G.; Toth, R.; Dubois, F.; Geraghty, K.M.; Wong, B.H.; Brown, L.J.; Tyler, J.; et al. Visualization and biochemical analyses of the emerging mammalian 14-3-3-phosphoproteome. Mol. Cell Proteom. 2011, 10, M110.005751. [Google Scholar] [CrossRef]
- Mackintosh, C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 2004, 381 Pt 2, 329–342. [Google Scholar] [CrossRef]
- Fu, H.; Subramanian, R.R.; Masters, S.C. 14-3-3 proteins: Structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 617–647. [Google Scholar] [CrossRef]
- Tyagi, A.; Kaushal, K.; Chandrasekaran, A.P.; Sarodaya, N.; Das, S.; Park, C.H.; Hong, S.H.; Kim, K.S.; Ramakrishna, S. CRISPR/Cas9-based genome-wide screening for deubiquitinase subfamily identifies USP1 regulating MAST1-driven cisplatin-resistance in cancer cells. Theranostics 2022, 12, 5949–5970. [Google Scholar] [CrossRef]
- Pan, C.; Chun, J.; Li, D.; Boese, A.C.; Li, J.; Kang, J.; Umano, A.; Jiang, Y.; Song, L.; Magliocca, K.R.; et al. Hsp90B enhances MAST1-mediated cisplatin resistance by protecting MAST1 from proteosomal degradation. J. Clin. Investig. 2019, 129, 4110–4123. [Google Scholar] [CrossRef]
- Jin, L.; Chun, J.; Pan, C.; Li, D.; Lin, R.; Alesi, G.N.; Wang, X.; Kang, H.B.; Song, L.; Wang, D.; et al. MAST1 Drives Cisplatin Resistance in Human Cancers by Rewiring cRaf-Independent MEK Activation. Cancer Cell 2018, 34, 315–330.e7. [Google Scholar] [CrossRef]
- Tripathy, R.; Leca, I.; van Dijk, T.; Weiss, J.; van Bon, B.W.; Sergaki, M.C.; Gstrein, T.; Breuss, M.; Tian, G.; Bahi-Buisson, N.; et al. Mutations in MAST1 Cause Mega-Corpus-Callosum Syndrome with Cerebellar Hypoplasia and Cortical Malformations. Neuron 2018, 100, 1354–1368.e5. [Google Scholar] [CrossRef]
- Ren, A.; Zhang, W.; Yarlagadda, S.; Sinha, C.; Arora, K.; Moon, C.S.; Naren, A.P. MAST205 competes with cystic fibrosis transmembrane conductance regulator (CFTR)-associated ligand for binding to CFTR to regulate CFTR-mediated fluid transport. J. Biol. Chem. 2013, 288, 12325–12334. [Google Scholar] [CrossRef]
- Andrade, E.C.; Musante, V.; Horiuchi, A.; Matsuzaki, H.; Brody, A.H.; Wu, T.; Greengard, P.; Taylor, J.R.; Nairn, A.C. ARPP-16 Is a Striatal-Enriched Inhibitor of Protein Phosphatase 2A Regulated by Microtubule-Associated Serine/Threonine Kinase 3 (Mast 3 Kinase). J. Neurosci. 2017, 37, 2709–2722. [Google Scholar] [CrossRef]
- Kim, P.; Park, J.; Lee, D.J.; Mizuno, S.; Shinohara, M.; Hong, C.P.; Jeong, Y.; Yun, R.; Park, H.; Park, S.; et al. Mast4 determines the cell fate of MSCs for bone and cartilage development. Nat. Commun. 2022, 13, 3960. [Google Scholar] [CrossRef]
- Girault, J.A.; Walaas, S.I.; Hemmings, H.C., Jr.; Greengard, P. ARPP-21, a cAMP-regulated phosphoprotein enriched in dopamine-innervated brain regions: Tissue distribution and regulation of phosphorylation in rat brain. Neuroscience 1990, 37, 317–325. [Google Scholar] [CrossRef]
- Horiuchi, A.; Williams, K.R.; Kurihara, T.; Nairn, A.C.; Greengard, P. Purification and cDNA cloning of ARPP-16, a cAMP-regulated phosphoprotein enriched in basal ganglia, and of a related phosphoprotein, ARPP-19. J. Biol. Chem. 1990, 265, 9476–9484. [Google Scholar] [CrossRef]
- Brene, S.; Lindefors, N.; Ehrlich, M.; Taubes, T.; Horiuchi, A.; Kopp, J.; Hall, H.; Sedvall, G.; Greengard, P.; Persson, H. Expression of mRNAs encoding ARPP-16/19, ARPP-21, and DARPP-32 in human brain tissue. J. Neurosci. 1994, 14 Pt 1, 985–998. [Google Scholar] [CrossRef]
- Musante, V.; Li, L.; Kanyo, J.; Lam, T.T.; Colangelo, C.M.; Cheng, S.K.; Brody, A.H.; Greengard, P.; Le Novere, N.; Nairn, A.C. Reciprocal regulation of ARPP-16 by PKA and MAST3 kinases provides a cAMP-regulated switch in protein phosphatase 2A inhibition. Elife 2017, 6, e24998. [Google Scholar] [CrossRef]
- Bell, D.M.; Leung, K.K.; Wheatley, S.C.; Ng, L.J.; Zhou, S.; Ling, K.W.; Sham, M.H.; Koopman, P.; Tam, P.P.; Cheah, K.S. SOX9 directly regulates the type-II collagen gene. Nat. Genet. 1997, 16, 174–178. [Google Scholar] [CrossRef]
- Bi, W.; Deng, J.M.; Zhang, Z.; Behringer, R.R.; de Crombrugghe, B. Sox9 is required for cartilage formation. Nat. Genet. 1999, 22, 85–89. [Google Scholar] [CrossRef]
- Lefebvre, V.; Huang, W.; Harley, V.R.; Goodfellow, P.N.; de Crombrugghe, B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol. Cell Biol. 1997, 17, 2336–2346. [Google Scholar] [CrossRef]
- Lee, S.J.; Park, J.; Lee, D.J.; Otsu, K.; Kim, P.; Mizuno, S.; Lee, M.J.; Kim, H.Y.; Harada, H.; Takahashi, S.; et al. Mast4 knockout shows the regulation of spermatogonial stem cell self-renewal via the FGF2/ERM pathway. Cell Death Differ. 2021, 28, 1441–1454. [Google Scholar] [CrossRef]
- Garland, P.; Quraishe, S.; French, P.; O’Connor, V. Expression of the MAST family of serine/threonine kinases. Brain Res. 2008, 1195, 12–19. [Google Scholar] [CrossRef]
- Robinson, D.R.; Kalyana-Sundaram, S.; Wu, Y.M.; Shankar, S.; Cao, X.; Ateeq, B.; Asangani, I.A.; Iyer, M.; Maher, C.A.; Grasso, C.S.; et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat. Med. 2011, 17, 1646–1651. [Google Scholar] [CrossRef]
- Wang, X.; Fredericksen, Z.S.; Vierkant, R.A.; Kosel, M.L.; Pankratz, V.S.; Cerhan, J.R.; Justenhoven, C.; Brauch, H.; Olson, J.E.; Couch, F.J. Association of genetic variation in mitotic kinases with breast cancer risk. Breast Cancer Res. Treat. 2010, 119, 453–462. [Google Scholar] [CrossRef]
- Labbe, C.; Boucher, G.; Foisy, S.; Alikashani, A.; Nkwimi, H.; David, G.; Beaudoin, M.; Goyette, P.; Charron, G.; Xavier, R.J.; et al. Genome-wide expression profiling implicates a MAST3-regulated gene set in colonic mucosal inflammation of ulcerative colitis patients. Inflamm. Bowel Dis. 2012, 18, 1072–1080. [Google Scholar] [CrossRef]
- Terrien, E.; Simenel, C.; Prehaud, C.; Buc, H.; Delepierre, M.; Lafon, M.; Wolff, N. 1H, 13C and 15N resonance assignments of the PDZ of microtubule-associated serine/threonine kinase 205 (MAST205) in complex with the C-terminal motif from the rabies virus glycoprotein. Biomol. NMR Assign. 2009, 3, 45–48. [Google Scholar] [CrossRef]
- Clay, M.R.; Varma, S.; West, R.B. MAST2 and NOTCH1 translocations in breast carcinoma and associated pre-invasive lesions. Hum. Pathol. 2013, 44, 2837–2844. [Google Scholar] [CrossRef]
- Eissmann, M.; Schwamb, B.; Melzer, I.M.; Moser, J.; Siele, D.; Kohl, U.; Rieker, R.J.; Wachter, D.L.; Agaimy, A.; Herpel, E.; et al. A functional yeast survival screen of tumor-derived cDNA libraries designed to identify anti-apoptotic mammalian oncogenes. PLoS ONE 2013, 8, e64873. [Google Scholar] [CrossRef]
- Yu, X.; Sheng, P.; Sun, J.; Zhao, X.; Zhang, J.; Li, Y.; Zhang, Y.; Zhang, W.; Wang, J.; Liu, K.; et al. The circular RNA circMAST1 promotes hepatocellular carcinoma cell proliferation and migration by sponging miR-1299 and regulating CTNND1 expression. Cell Death Dis. 2020, 11, 340. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, Y.; Jiang, P.; Fu, Z.; Liu, Y. High MAST2 mRNA expression and its role in diagnosis and prognosis of liver cancer. Sci. Rep. 2019, 9, 19865. [Google Scholar] [CrossRef]
- Sotelo, N.S.; Valiente, M.; Gil, A.; Pulido, R. A functional network of the tumor suppressors APC, hDlg, and PTEN, that relies on recognition of specific PDZ-domains. J. Cell Biochem. 2012, 113, 2661–2670. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, J.; Zhang, W.; Pan, L.; Zhang, D.; Zhao, P.; Wang, F.; Luo, H.; He, J.; Qin, Y.; et al. Young female patients with multiple myeloma have low occurrence of osteolytic lesion. Bone 2018, 110, 21–28. [Google Scholar] [CrossRef]
- Fuertes, M.A.; Castilla, J.; Alonso, C.; Perez, J.M. Cisplatin biochemical mechanism of action: From cytotoxicity to induction of cell death through interconnections between apoptotic and necrotic pathways. Curr. Med. Chem. 2003, 10, 257–266. [Google Scholar] [CrossRef]
- Kong, L.R.; Chua, K.N.; Sim, W.J.; Ng, H.C.; Bi, C.; Ho, J.; Nga, M.E.; Pang, Y.H.; Ong, W.R.; Soo, R.A.; et al. MEK Inhibition Overcomes Cisplatin Resistance Conferred by SOS/MAPK Pathway Activation in Squamous Cell Carcinoma. Mol. Cancer Ther. 2015, 14, 1750–1760. [Google Scholar] [CrossRef]
- Jaykumar, A.B.; Karra, A.S.; Cobb, M.H. Pulling a MAST1 on Cisplatin Resistance. Cancer Cell 2018, 34, 183–185. [Google Scholar] [CrossRef]
- Pan, C.; Kang, J.; Hwang, J.S.; Li, J.; Boese, A.C.; Wang, X.; Yang, L.; Boggon, T.J.; Chen, G.Z.; Saba, N.F.; et al. Cisplatin-mediated activation of glucocorticoid receptor induces platinum resistance via MAST1. Nat. Commun. 2021, 12, 4960. [Google Scholar] [CrossRef]
- Guggino, W.B.; Stanton, B.A. New insights into cystic fibrosis: Molecular switches that regulate CFTR. Nat. Rev. Mol. Cell Biol. 2006, 7, 426–436. [Google Scholar] [CrossRef]
- Guggino, W.B. The cystic fibrosis transmembrane regulator forms macromolecular complexes with PDZ domain scaffold proteins. Proc. Am. Thorac. Soc. 2004, 1, 28–32. [Google Scholar] [CrossRef]
- Broadbent, D.; Ahmadzai, M.M.; Kammala, A.K.; Yang, C.; Occhiuto, C.; Das, R.; Subramanian, H. Roles of NHERF Family of PDZ-Binding Proteins in Regulating GPCR Functions. Adv. Immunol. 2017, 136, 353–385. [Google Scholar]
- Li, C.; Naren, A.P. CFTR chloride channel in the apical compartments: Spatiotemporal coupling to its interacting partners. Integr. Biol. 2010, 2, 161–177. [Google Scholar] [CrossRef]
- Lamprecht, G.; Seidler, U. The emerging role of PDZ adapter proteins for regulation of intestinal ion transport. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 291, G766–G777. [Google Scholar] [CrossRef]
- Cheng, J.; Cebotaru, V.; Cebotaru, L.; Guggino, W.B. Syntaxin 6 and CAL mediate the degradation of the cystic fibrosis transmembrane conductance regulator. Mol. Biol. Cell 2010, 21, 1178–1187. [Google Scholar] [CrossRef]
- Farinha, C.M.; Matos, P.; Amaral, M.D. Control of cystic fibrosis transmembrane conductance regulator membrane trafficking: Not just from the endoplasmic reticulum to the Golgi. FEBS J. 2013, 280, 4396–4406. [Google Scholar] [CrossRef]
- Cheng, J.; Moyer, B.D.; Milewski, M.; Loffing, J.; Ikeda, M.; Mickle, J.E.; Cutting, G.R.; Li, M.; Stanton, B.A.; Guggino, W.B. A Golgi-associated PDZ domain protein modulates cystic fibrosis transmembrane regulator plasma membrane expression. J. Biol. Chem. 2002, 277, 3520–3529. [Google Scholar] [CrossRef]
- Cheng, J.; Wang, H.; Guggino, W.B. Modulation of mature cystic fibrosis transmembrane regulator protein by the PDZ domain protein CAL. J. Biol. Chem. 2004, 279, 1892–1898. [Google Scholar] [CrossRef]
- Huang, N.; Wen, Y.; Guo, X.; Li, Z.; Dai, J.; Ni, B.; Yu, J.; Lin, Y.; Zhou, W.; Yao, B.; et al. A Screen for Genomic Disorders of Infertility Identifies MAST2 Duplications Associated with Nonobstructive Azoospermia in Humans. Biol. Reprod. 2015, 93, 61. [Google Scholar] [CrossRef]
- Nagarajan, U.M.; Bushey, A.; Boss, J.M. Modulation of gene expression by the MHC class II transactivator. J. Immunol. 2002, 169, 5078–5088. [Google Scholar] [CrossRef]
- Lim, Y.; Lee, J.; Lee, D.Y. Is the survival rate for acral melanoma actually worse than other cutaneous melanomas? J. Dermatol. 2020, 47, 251–256. [Google Scholar] [CrossRef]
- Beretov, J.; Wasinger, V.C.; Millar, E.K.; Schwartz, P.; Graham, P.H.; Li, Y. Proteomic Analysis of Urine to Identify Breast Cancer Biomarker Candidates Using a Label-Free LC-MS/MS Approach. PLoS ONE 2015, 10, e0141876. [Google Scholar] [CrossRef]
- Jiang, Y.; Meyers, T.J.; Emeka, A.A.; Cooley, L.F.; Cooper, P.R.; Lancki, N.; Helenowski, I.; Kachuri, L.; Lin, D.W.; Stanford, J.L.; et al. Genetic Factors Associated with Prostate Cancer Conversion from Active Surveillance to Treatment. HGG Adv. 2022, 3, 100070. [Google Scholar] [CrossRef]
- Moran, J.M.T.; Le, L.P.; Nardi, V.; Golas, J.; Farahani, A.A.; Signorelli, S.; Onozato, M.L.; Foreman, R.K.; Duncan, L.M.; Lawrence, D.P.; et al. Identification of fusions with potential clinical significance in melanoma. Mod. Pathol. 2022, 35, 1837–1847. [Google Scholar] [CrossRef]
- Riva, E.; Manrique Arechavaleta, G.; De Almeida, C.; Costa, V.; Fernandez Del Campo, M.; Ifran Gonzalez, S.; Uriarte, R. A novel e8a2 BCR-ABL1 fusion with insertion of MAST2 exon 2 in a four-way translocation t (1;17;9;22) (p35;q24;q44;q11) in a patient with chronic myeloid leukemia. Leuk. Lymphoma 2016, 57, 203–205. [Google Scholar] [CrossRef]
- Mao, X.H.; Ye, Q.; Zhang, G.B.; Jiang, J.Y.; Zhao, H.Y.; Shao, Y.F.; Ye, Z.Q.; Xuan, Z.X.; Huang, P. Identification of differentially methylated genes as diagnostic and prognostic biomarkers of breast cancer. World J. Surg. Oncol. 2021, 19, 29. [Google Scholar] [CrossRef]
- Tan, B.; Zhang, J.; Wang, W.; Ma, H.; Yang, Y. E3 Ubiquitin Ligase CHIP Inhibits the Interaction between Hsp90beta and MAST1 to Repress Radiation Resistance in Non-Small-Cell Lung Cancer Stem Cells. Stem Cells Int. 2022, 2022, 2760899. [Google Scholar] [CrossRef]
- Oishi, T.; Iino, K.; Okawa, Y.; Kakizawa, K.; Matsunari, S.; Yamashita, M.; Taniguchi, T.; Maekawa, M.; Suda, T.; Oki, Y. DNA methylation analysis in malignant pheochromocytoma and paraganglioma. J. Clin. Transl. Endocrinol. 2017, 7, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Zhang, J.; Shi, R.; Yu, Z.; Chen, X.; Wang, H. Identification of an integrated kinase-related prognostic gene signature associated with tumor immune microenvironment in human uterine corpus endometrial carcinoma. Front. Oncol. 2022, 12, 944000. [Google Scholar] [CrossRef] [PubMed]
- Tomoshige, K.; Matsumoto, K.; Tsuchiya, T.; Oikawa, M.; Miyazaki, T.; Yamasaki, N.; Mishima, H.; Kinoshita, A.; Kubo, T.; Fukushima, K.; et al. Germline mutations causing familial lung cancer. J. Hum. Genet. 2015, 60, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Morange, P.E.; Peiretti, F.; Gourhant, L.; Proust, C.; Soukarieh, O.; Pulcrano-Nicolas, A.S.; Saripella, G.V.; Stefanucci, L.; Lacroix, R.; Ibrahim-Kosta, M.; et al. A rare coding mutation in the MAST2 gene causes venous thrombosis in a French family with unexplained thrombophilia: The Breizh MAST2 Arg89Gln variant. PLoS Genet. 2021, 17, e1009284. [Google Scholar] [CrossRef]
- Shu, L.; Xiao, N.; Qin, J.; Tian, Q.; Zhang, Y.; Li, H.; Liu, J.; Li, Q.; Gu, W.; Wang, P.; et al. The Role of Microtubule Associated Serine/Threonine Kinase 3 Variants in Neurodevelopmental Diseases: Genotype-Phenotype Association. Front. Mol. Neurosci. 2021, 14, 775479. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, E.; Christensen, K.R.; Bryant, E.; Schneider, A.; Rakotomamonjy, J.; Muir, A.M.; Giannelli, J.; Littlejohn, R.O.; Roeder, E.R.; Schmidt, B.; et al. Pathogenic MAST3 Variants in the STK Domain Are Associated with Epilepsy. Ann. Neurol. 2021, 90, 274–284. [Google Scholar] [CrossRef]
- Landoulsi, Z.; Laatar, F.; Noe, E.; Mrabet, S.; Ben Djebara, M.; Achaz, G.; Nava, C.; Baulac, S.; Kacem, I.; Gargouri-Berrechid, A.; et al. Clinical and genetic study of Tunisian families with genetic generalized epilepsy: Contribution of CACNA1H and MAST4 genes. Neurogenetics 2018, 19, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Bowling, K.M.; Thompson, M.L.; Amaral, M.D.; Finnila, C.R.; Hiatt, S.M.; Engel, K.L.; Cochran, J.N.; Brothers, K.B.; East, K.M.; Gray, D.E.; et al. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med. 2017, 9, 43. [Google Scholar] [CrossRef]
- McMichael, G.; Bainbridge, M.N.; Haan, E.; Corbett, M.; Gardner, A.; Thompson, S.; van Bon, B.W.; van Eyk, C.L.; Broadbent, J.; Reynolds, C.; et al. Whole-exome sequencing points to considerable genetic heterogeneity of cerebral palsy. Mol. Psychiatry 2015, 20, 176–182. [Google Scholar] [CrossRef]
- Gilissen, C.; Hehir-Kwa, J.Y.; Thung, D.T.; van de Vorst, M.; van Bon, B.W.; Willemsen, M.H.; Kwint, M.; Janssen, I.M.; Hoischen, A.; Schenck, A.; et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014, 511, 344–347. [Google Scholar] [CrossRef]
- Dolan, M.; Mendelsohn, N.J.; Pierpont, M.E.; Schimmenti, L.A.; Berry, S.A.; Hirsch, B. A novel microdeletion/microduplication syndrome of 19p13.13. Genet. Med. 2010, 12, 503–511. [Google Scholar] [CrossRef]
- Ben-Mahmoud, A.; Al-Shamsi, A.M.; Ali, B.R.; Al-Gazali, L. Evaluating the Role of MAST1 as an Intellectual Disability Disease Gene: Identification of a Novel De Novo Variant in a Patient with Developmental Disabilities. J. Mol. Neurosci. 2020, 70, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Hecher, L.; Johannsen, J.; Bierhals, T.; Buhk, J.H.; Hempel, M.; Denecke, J. The Clinical Picture of a Bilateral Perisylvian Syndrome as the Initial Symptom of Mega-Corpus-Callosum Syndrome due to a MAST1-Gene Mutation. Neuropediatrics 2020, 51, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Garcia, M.E.; Cotrina-Vinagre, F.J.; Gomez-Cano, M.L.A.; Martinez de Aragon, A.; Martin-Hernandez, E.; Martinez-Azorin, F. MAST1 variant causes mega-corpus-callosum syndrome with cortical malformations but without cerebellar hypoplasia. Am. J. Med. Genet. A 2020, 182, 1483–1490. [Google Scholar] [CrossRef] [PubMed]
- Mainguy-Seers, S.; Beaudry, F.; Fernandez-Prada, C.; Martin, J.G.; Lavoie, J.P. Neutrophil Extracellular Vesicles and Airway Smooth Muscle Proliferation in the Natural Model of Severe Asthma in Horses. Cells 2022, 11, 3347. [Google Scholar] [CrossRef]
- Haas, M.E.; Pirruccello, J.P.; Friedman, S.N.; Wang, M.; Emdin, C.A.; Ajmera, V.H.; Simon, T.G.; Homburger, J.R.; Guo, X.; Budoff, M.; et al. Machine learning enables new insights into genetic contributions to liver fat accumulation. Cell Genom. 2021, 1, 100066. [Google Scholar] [CrossRef]
- Xu, Q.; Yin, S.; Yao, Y.; Li, X.; Song, B.; Yang, Y.; Liu, Y.; Chen, R.; Li, J.; Ma, T.; et al. MAST3 modulates the inflammatory response and proliferation of fibroblast-like synoviocytes in rheumatoid arthritis. Int. Immunopharmacol. 2019, 77, 105900. [Google Scholar] [CrossRef]
- Khan, Z.; Terrien, E.; Delhommel, F.; Lefebvre-Omar, C.; Bohl, D.; Vitry, S.; Bernard, C.; Ramirez, J.; Chaffotte, A.; Ricquier, K.; et al. Structure-based optimization of a PDZ-binding motif within a viral peptide stimulates neurite outgrowth. J. Biol. Chem. 2019, 294, 13755–13768. [Google Scholar] [CrossRef]
- Delhommel, F.; Chaffotte, A.; Terrien, E.; Raynal, B.; Buc, H.; Delepierre, M.; Cordier, F.; Wolff, N. Deciphering the unconventional peptide binding to the PDZ domain of MAST2. Biochem. J. 2015, 469, 159–168. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Groza, T.; Gomez, F.L.; Mashhadi, H.H.; Munoz-Fuentes, V.; Gunes, O.; Wilson, R.; Cacheiro, P.; Frost, A.; Keskivali-Bond, P.; Vardal, B.; et al. The International Mouse Phenotyping Consortium: Comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res. 2023, 51, D1038–D1045. [Google Scholar] [CrossRef]
- Baldarelli, R.M.; Smith, C.M.; Finger, J.H.; Hayamizu, T.F.; McCright, I.J.; Xu, J.; Shaw, D.R.; Beal, J.S.; Blodgett, O.; Campbell, J.; et al. The mouse Gene Expression Database (GXD): 2021 update. Nucleic Acids Res. 2021, 49, D924–D931. [Google Scholar] [CrossRef]
- Mannsfeldt, A.G.; Carroll, P.; Stucky, C.L.; Lewin, G.R. Stomatin, a MEC-2 like protein, is expressed by mammalian sensory neurons. Mol. Cell Neurosci. 1999, 13, 391–404. [Google Scholar] [CrossRef]
- Nurrish, S.; Segalat, L.; Kaplan, J.M. Serotonin inhibition of synaptic transmission: Galpha(0) decreases the abundance of UNC-13 at release sites. Neuron 1999, 24, 231–242. [Google Scholar] [CrossRef]
- Miller, K.G.; Emerson, M.D.; Rand, J.B. Goalpha and diacylglycerol kinase negatively regulate the Gqalpha pathway in C. elegans. Neuron 1999, 24, 323–333. [Google Scholar] [CrossRef]
- Nakano, S.; Ikeda, M.; Tsukada, Y.; Fei, X.; Suzuki, T.; Niino, Y.; Ahluwalia, R.; Sano, A.; Kondo, R.; Ihara, K.; et al. Presynaptic MAST kinase controls opposing postsynaptic responses to convey stimulus valence in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2020, 117, 1638–1647. [Google Scholar] [CrossRef] [PubMed]
- An, S.W.A.; Choi, E.S.; Hwang, W.; Son, H.G.; Yang, J.S.; Seo, K.; Nam, H.J.; Nguyen, N.T.H.; Kim, E.J.E.; Suh, B.K.; et al. KIN-4/MAST kinase promotes PTEN-mediated longevity of Caenorhabditis elegans via binding through a PDZ domain. Aging Cell 2019, 18, e12906. [Google Scholar] [CrossRef] [PubMed]
- Hain, D.; Langlands, A.; Sonnenberg, H.C.; Bailey, C.; Bullock, S.L.; Muller, H.A. The Drosophila MAST kinase Drop out is required to initiate membrane compartmentalisation during cellularisation and regulates dynein-based transport. Development 2014, 141, 2119–2130. [Google Scholar] [CrossRef]
- Galewsky, S.; Schulz, R.A. Drop out: A third chromosome maternal-effect locus required for formation of the Drosophila cellular blastoderm. Mol. Reprod. Dev. 1992, 32, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, A.; Mazumdar, M. How one becomes many: Blastoderm cellularization in Drosophila melanogaster. Bioessays 2002, 24, 1012–1022. [Google Scholar] [CrossRef]
- Sokac, A.M.; Biel, N.; De Renzis, S. Membrane-actin interactions in morphogenesis: Lessons learned from Drosophila cellularization. Semin. Cell Dev. Biol. 2023, 133, 107–122. [Google Scholar] [CrossRef]
- Schmidt, A.; Grosshans, J. Dynamics of cortical domains in early Drosophila development. J. Cell Sci. 2018, 131, jcs212795. [Google Scholar] [CrossRef]
- Gross, S.P.; Welte, M.A.; Block, S.M.; Wieschaus, E.F. Dynein-mediated cargo transport in vivo. A switch controls travel distance. J. Cell Biol. 2000, 148, 945–956. [Google Scholar] [CrossRef]
- Meyer, W.J.; Schreiber, S.; Guo, Y.; Volkmann, T.; Welte, M.A.; Muller, H.A. Overlapping functions of argonaute proteins in patterning and morphogenesis of Drosophila embryos. PLoS Genet. 2006, 2, e134. [Google Scholar] [CrossRef]
- Welte, M.A.; Gross, S.P.; Postner, M.; Block, S.M.; Wieschaus, E.F. Developmental regulation of vesicle transport in Drosophila embryos: Forces and kinetics. Cell 1998, 92, 547–557. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rumpf, M.; Pautz, S.; Drebes, B.; Herberg, F.W.; Müller, H.-A.J. Microtubule-Associated Serine/Threonine (MAST) Kinases in Development and Disease. Int. J. Mol. Sci. 2023, 24, 11913. https://doi.org/10.3390/ijms241511913
Rumpf M, Pautz S, Drebes B, Herberg FW, Müller H-AJ. Microtubule-Associated Serine/Threonine (MAST) Kinases in Development and Disease. International Journal of Molecular Sciences. 2023; 24(15):11913. https://doi.org/10.3390/ijms241511913
Chicago/Turabian StyleRumpf, Marie, Sabine Pautz, Benedikt Drebes, Friedrich W. Herberg, and Hans-Arno J. Müller. 2023. "Microtubule-Associated Serine/Threonine (MAST) Kinases in Development and Disease" International Journal of Molecular Sciences 24, no. 15: 11913. https://doi.org/10.3390/ijms241511913
APA StyleRumpf, M., Pautz, S., Drebes, B., Herberg, F. W., & Müller, H.-A. J. (2023). Microtubule-Associated Serine/Threonine (MAST) Kinases in Development and Disease. International Journal of Molecular Sciences, 24(15), 11913. https://doi.org/10.3390/ijms241511913








