How Can the Microbiome Induce Carcinogenesis and Modulate Drug Resistance in Cancer Therapy?
Abstract
1. Introduction
2. Drug Resistance in Cancer
Cancer and Infectious Diseases
3. The Human Microbiome
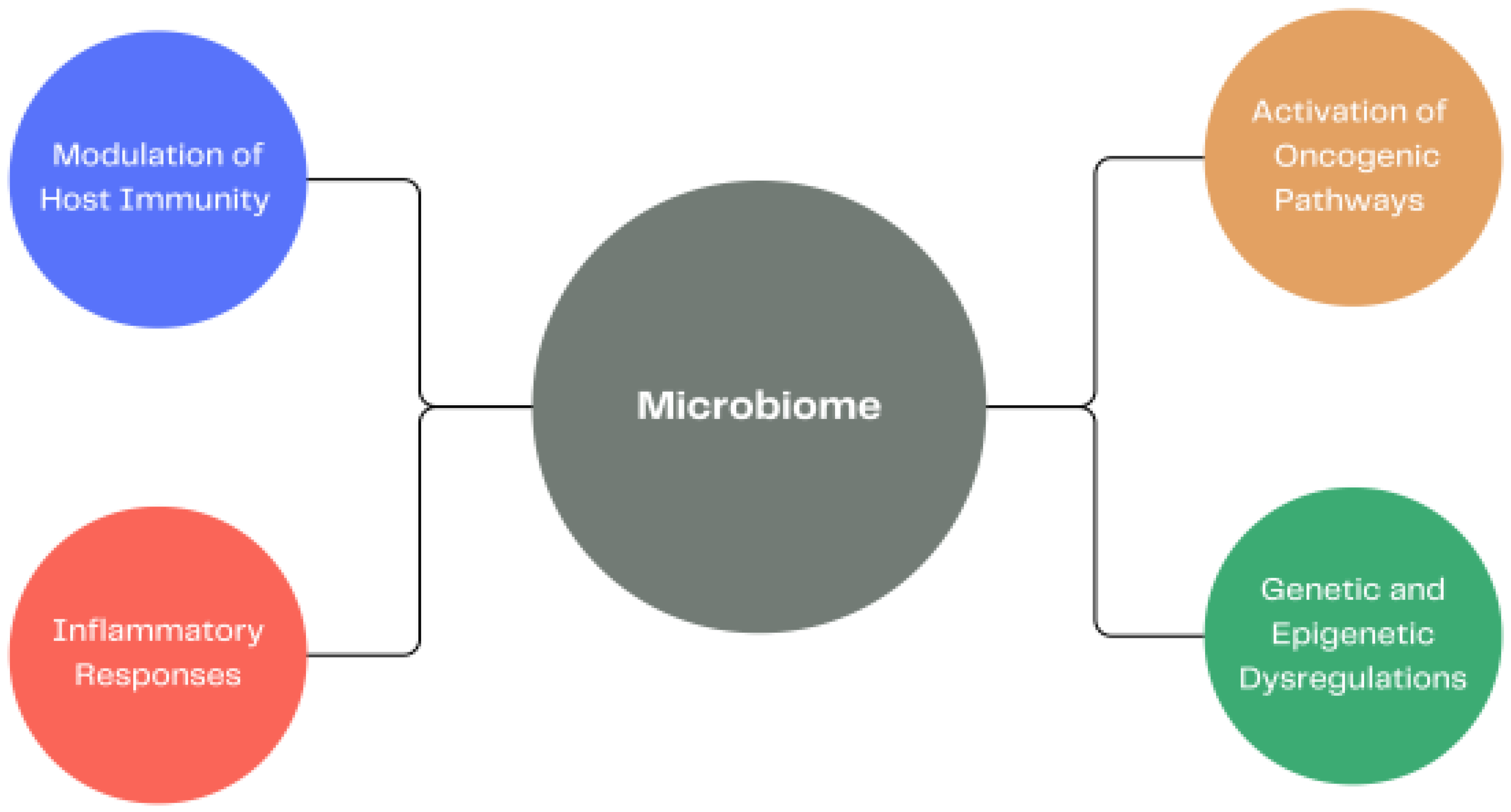
3.1. Altered Human Microbiome and Carcinogenesis
3.2. The Importance of the Gut Microbiome
Varied Modulatory Influences of the Gut Microbiome
4. Examples of Microbes and Their Association with Several Cancer Types
4.1. Colorectal Cancer
4.2. Breast Cancer
4.3. Gastric Cancer
Helicobacter Pylori and Gastric Carcinogenesis
5. Influence of Intratumoral Microbiota
6. Epigenetics
Non-Mutational Epigenetic Reprogramming
7. Epigenetic Regulation of CRC
8. Drug Resistance and Toxicity in Cancer Induced by Microbiome
8.1. Chemotherapy
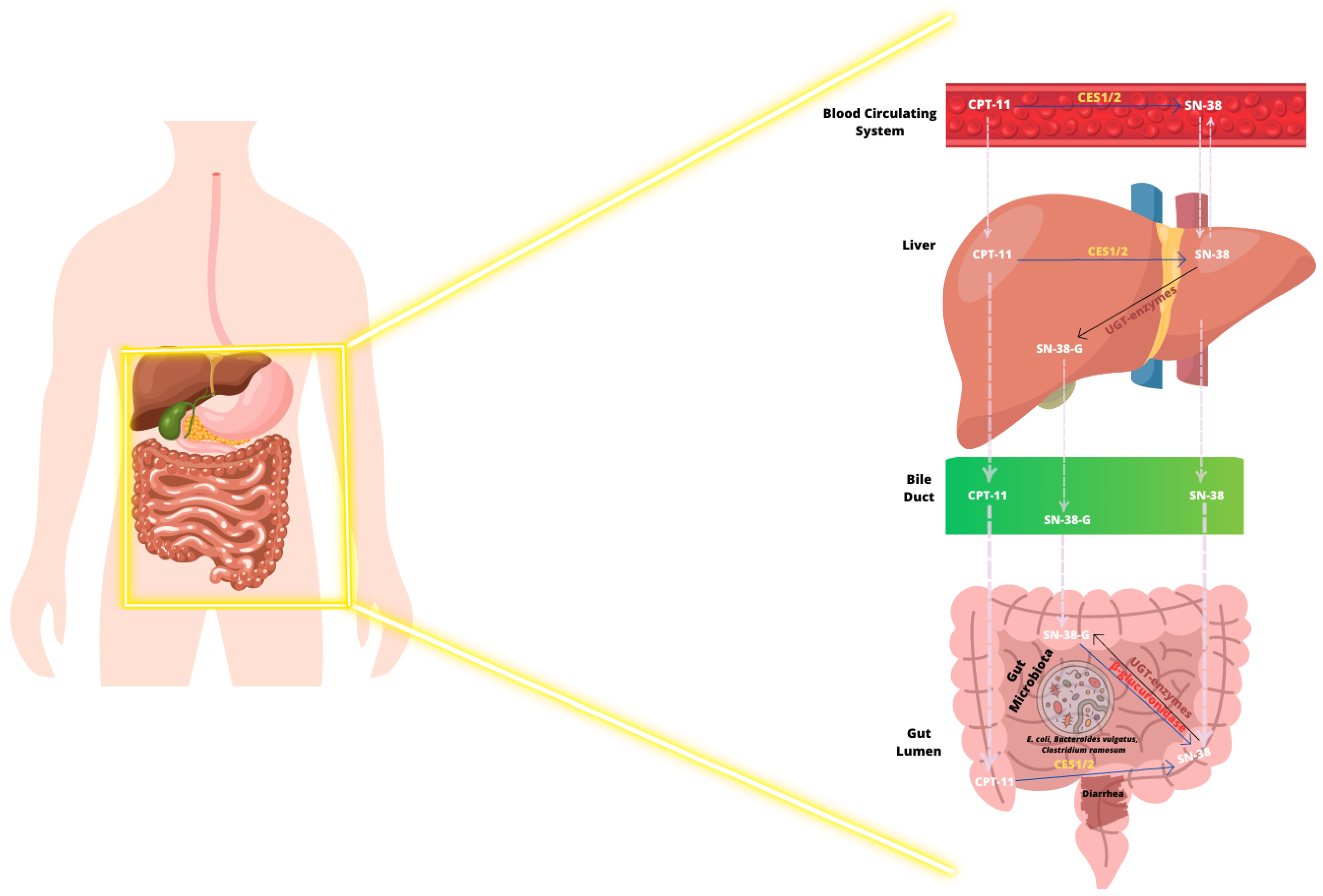
Irinotecan and Gut Microbiome
8.2. Immunotherapy
8.2.1. Immunotherapy in Advanced Melanoma and the Gut Microbiome
8.2.2. Microbiome and CAR-T Therapy
9. Allogeneic Hematopoietic Stem Cell Transplantation
9.1. Fecal Microbiota Transplantation as a Preventive Approach in Allo-HSCT
9.2. Colonization Screening to Guide Antibiotic Therapy in Allo-HSCT
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Senga, S.S.; Grose, R.P. Hallmarks of cancer-the new testament. Open Biol. 2021, 11, 200358. [Google Scholar] [CrossRef]
- Fernández-Figueroa, E.A.; Lino-Silva, S.; Peña-Velasco, J.E.; Rangel-Escareño, C. Pharmaco-Geno-Proteo-Metabolomics and Translational Research in Cancer. In Advances in Experimental Medicine and Biology; Ruiz-Garcia, E., Astudillo-de la Vega, H., Eds.; Springer Nature: Cham, Switzerland, 2019; Volume 1168, pp. 1–7. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics. CA Cancer J Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhu, W.; Wu, W. MicroRNAs Change the Landscape of Cancer Resistance. In Methods in Molecular Biology, 2nd ed.; Wu, W., Ed.; Humana Press: New York, NY, USA, 2018; Volume 1699, pp. 83–89. [Google Scholar] [CrossRef]
- Boveri, T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J. Cell Sci. 2008, 121, 1–84. [Google Scholar] [CrossRef]
- Waddington, C.H. The epigenotype. Endeavour 1942, 1, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.; Vogelstein, B. Hypomethylation distinguishes genes of some human cancers from their normal counter-parts. Nature 1983, 301, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Holliday, R. The inheritance of epigenetic defects. Science 1987, 238, 163–170. [Google Scholar] [CrossRef]
- Greger, V.; Passarge, E.; Höpping, W.; Messmer, E.; Horsthemke, B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum. Genet. 1989, 83, 155–158. [Google Scholar] [CrossRef]
- Saito, Y.; Liang, G.; Egger, G.; Friedman, J.M.; Chuang, J.C.; Coetzee, G.A.; Jones, P.A. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 2006, 9, 435–443. [Google Scholar] [CrossRef]
- Ross, J.P.; Rand, K.N.; Molloy, P.L. Hypomethylation of repeated DNA sequences in cancer. Epigenomics 2010, 2, 245–269. [Google Scholar] [CrossRef]
- Capurso, G.; Lahner, E. The interaction between smoking, alcohol and the gut microbiome. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 579–588. [Google Scholar] [CrossRef]
- Ogino, S.; Nowak, J.A.; Hamada, T.; Phipps, A.I.; Peters, U.; Milner, D.A., Jr.; Giovannucci, E.L.; Nishihara, R.; Giannakis, M.; Garrett, W.S.; et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut 2018, 67, 1168–1180. [Google Scholar] [CrossRef]
- Chen, J.; Domingue, J.C.; Sears, C.L. Microbiota dysbiosis in select human cancers: Evidence of association and causality. Semin. Immunol. 2017, 32, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef] [PubMed]
- Rajpoot, M.; Sharma, A.K.; Sharma, A.; Gupta, G.K. Understanding the microbiome: Emerging biomarkers for exploiting the microbiota for personalized medicine against cancer. Semin. Cancer Biol. 2018, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Healey, G.R.; Murphy, R.; Brough, L.; Butts, C.A.; Coad, J. Interindividual variability in gut microbiota and host response to dietary interventions. Nutr. Rev. 2017, 75, 1059–1080. [Google Scholar] [CrossRef]
- Vatanen, T.; Franzosa, E.A.; Schwager, R.; Tripathi, S.; Arthur, T.D.; Vehik, K.; Lernmark, Å.; Hagopian, W.A.; Rewers, M.J.; She, J.X.; et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018, 562, 589–594. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Sagaert, X.; Topal, B.; Haustermans, K.; Prenen, H. Gastric cancer. Lancet 2016, 388, 2654–2664. [Google Scholar] [CrossRef]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Yarchoan, R.; Uldrick, T.S. HIV-Associated Cancers and Related Diseases. N. Engl. J. Med. 2018, 378, 1029–1041. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef]
- Serrano, B.; Brotons, M.; Bosch, F.X.; Bruni, L. Epidemiology and burden of HPV-related disease. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 47, 14–26. [Google Scholar] [CrossRef]
- Ishitsuka, K.; Tamura, K. Human T-cell leukaemia virus type I and adult T-cell leukaemia-lymphoma. Lancet Oncol. 2014, 15, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, P.; Eslami, H.; Kafil, H.S. Carcinogenesis mechanisms of Fusobacterium nucleatum. Biomed. Pharmacother. 2017, 89, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Mima, K.; Sukawa, Y.; Nishihara, R.; Qian, Z.R.; Yamauchi, M.; Inamura, K.; Kim, S.A.; Masuda, A.; Nowak, J.A.; Nosho, K.; et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015, 1, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Akshintala, V.S.; Talukdar, R.; Singh, V.K.; Goggins, M. The Gut Microbiome in Pancreatic Disease. Clin. Gastroenterol. Hepatol. 2019, 17, 290–295. [Google Scholar] [CrossRef]
- Ren, Z.; Li, A.; Jiang, J.; Zhou, L.; Yu, Z.; Lu, H.; Xie, H.; Chen, X.; Shao, L.; Zhang, R.; et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019, 68, 1014–1023. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef]
- Garrett, W.S. Cancer and the microbiota. Science 2015, 348, 80–86. [Google Scholar] [CrossRef]
- Rajagopala, S.V.; Vashee, S.; Oldfield, L.M.; Suzuki, Y.; Venter, J.C.; Telenti, A.; Nelson, K.E. The Human Microbiome and Cancer. Cancer Prev. Res. 2017, 10, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Lertpiriyapong, K.; Whary, M.T.; Muthupalani, S.; Lofgren, J.L.; Gamazon, E.R.; Feng, Y.; Ge, Z.; Wang, T.C.; Fox, J.G. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut 2014, 63, 54–63. [Google Scholar] [CrossRef]
- Sevcikova, A.; Izoldova, N.; Stevurkova, V.; Kasperova, B.; Chovanec, M.; Ciernikova, S.; Mego, M. The Impact of the Microbiome on Resistance to Cancer Treatment with Chemotherapeutic Agents and Immunotherapy. Int. J. Mol. Sci. 2022, 23, 488. [Google Scholar] [CrossRef] [PubMed]
- Goodman, L.S.; Wintrobe, M.M. Nitrogen mustard therapy; use of methyl-bis (beta-chloroethyl) amine hydrochloride and tris (beta-chloroethyl) amine hydrochloride for Hodgkin’s disease, lymphosarcoma, leukemia and certain allied and miscellaneous disorders. J. Am. Med. Assoc. 1946, 132, 126–132. [Google Scholar] [CrossRef]
- Farber, S.; Diamond, L.K. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N. Engl. J. Med. 1948, 238, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Crofton, J. Chemotherapy of pulmonary tuberculosis. Br. Med. J. 1959, 1, 1610–1614. [Google Scholar] [CrossRef] [PubMed]
- Bonadonna, G.; Brusamolino, E.; Valagussa, P.; Rossi, A.; Brugnatelli, L.; Brambilla, C.; De Lena, M.; Tancini, G.; Bajetta, E.; Musumeci, R.; et al. Combination chemotherapy as an adjuvant treatment in operable breast cancer. N. Engl. J. Med. 1976, 294, 405–410. [Google Scholar] [CrossRef]
- DeVita, V.T.; Simon, R.M.; Hubbard, S.M.; Young, R.C.; Berard, C.W.; Moxley, J.H., 3rd; Frei, E., 3rd; Carbone, P.P.; Canellos, G.P. Curability of advanced Hodgkin’s disease with chemotherapy. Long-term follow-up of MOPP-treated patients at the National Cancer Institute. Ann. Intern. Med. 1980, 92, 587–595. [Google Scholar] [CrossRef]
- Bosl, G.J.; Gluckman, R.; Geller, N.L.; Golbey, R.B.; Whitmore, W.F.; Herr, H.; Sogani, P.; Morse, M.; Martini, N.; Bains, M. VAB-6: An effective chemotherapy regimen for patients with germ-cell tumors. J. Clin. Oncol. 1986, 4, 1493–1499. [Google Scholar] [CrossRef]
- Hryniuk, W.; Bush, H. The importance of dose intensity in chemotherapy of metastatic breast cancer. J. Clin. Oncol. 1984, 2, 1281–1288. [Google Scholar] [CrossRef]
- Sternberg, C.N.; de Mulder, P.H.; Schornagel, J.H.; Théodore, C.; Fossa, S.D.; van Oosterom, A.T.; Witjes, F.; Spina, M.; van Groeningen, C.J.; de Balincourt, C.; et al. Randomized phase III trial of high-dose-intensity methotrexate, vinblastine, doxorubicin, and cisplatin (MVAC) chemotherapy and recombinant human granulocyte colony-stimulating factor versus classic MVAC in advanced urothelial tract tumors: European Organization for Research and Treatment of Cancer Protocol no. 30924. J. Clin. Oncol. 2001, 19, 2638–2646. [Google Scholar] [CrossRef]
- Citron, M.L.; Berry, D.A.; Cirrincione, C.; Hudis, C.; Winer, E.P.; Gradishar, W.J.; Davidson, N.E.; Martino, S.; Livingston, R.; Ingle, J.N.; et al. Randomized trial of dose-dense versus conventionally scheduled and sequential versus concurrent combination chemotherapy as postoperative adjuvant treatment of node-positive primary breast cancer: First report of Intergroup Trial C9741/Cancer and Leukemia Group B Trial 9741. J. Clin. Oncol. 2003, 21, 1431–1439. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Leach, D.R.; Krummel, M.F.; Allison, J.P. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996, 271, 1734–1736. [Google Scholar] [CrossRef] [PubMed]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.S.; Sawyers, C.L. Converting cancer therapies into cures: Lessons from infectious diseases. Cell 2012, 148, 1089–1098. [Google Scholar] [CrossRef]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Mishra, A.; Lai, G.C.; Yao, L.J.; Aung, T.T.; Shental, N.; Rotter-Maskowitz, A.; Shepherdson, E.; Singh, G.S.; Pai, R.; Shanti, A.; et al. Microbial exposure during early human development primes fetal immune cells. Cell 2021, 184, 3394–3409.e20. [Google Scholar] [CrossRef]
- Mueller, N.T.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The infant microbiome development: Mom matters. Trends Mol. Med. 2015, 21, 109–117. [Google Scholar] [CrossRef]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef] [PubMed]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Link, C.D. Is There a Brain Microbiome? Neurosci. Insights 2021, 16, 26331055211018709. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Mahurkar, A.; Rahnavard, G.; Crabtree, J.; Orvis, J.; Hall, A.B.; Brady, A.; Creasy, H.H.; McCracken, C.; Giglio, M.G.; et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017, 550, 61–66. [Google Scholar] [CrossRef]
- Amato, K.R.; Arrieta, M.C.; Azad, M.B.; Bailey, M.T.; Broussard, J.L.; Bruggeling, C.E.; Claud, E.C.; Costello, E.K.; Davenport, E.R.; Dutilh, B.E.; et al. The human gut microbiome and health inequities. Proc. Natl. Acad. Sci. USA 2021, 118, e2017947118. [Google Scholar] [CrossRef]
- Shapira, I.; Sultan, K.; Lee, A.; Taioli, E. Evolving concepts: How diet and the intestinal microbiome act as modulators of breast malignancy. ISRN Oncol. 2013, 2013, 693920. [Google Scholar] [CrossRef]
- Thomas, S.; Izard, J.; Walsh, E.; Batich, K.; Chongsathidkiet, P.; Clarke, G.; Sela, D.A.; Muller, A.J.; Mullin, J.M.; Albert, K.; et al. The Host Microbiome Regulates and Maintains Human Health: A Primer and Perspective for Non-Microbiologists. Cancer Res. 2017, 77, 1783–1812. [Google Scholar] [CrossRef]
- Dzutsev, A.; Badger, J.H.; Perez-Chanona, E.; Roy, S.; Salcedo, R.; Smith, C.K.; Trinchieri, G. Microbes and Cancer. Annu. Rev. Immunol. 2017, 35, 199–228. [Google Scholar] [CrossRef]
- Helmink, B.A.; Khan, M.A.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef]
- Wei, M.Y.; Shi, S.; Liang, C.; Meng, Q.C.; Hua, J.; Zhang, Y.Y.; Liu, J.; Zhang, B.; Xu, J.; Yu, X.J. The microbiota and microbiome in pancreatic cancer: More influential than expected. Mol. Cancer 2019, 18, 97. [Google Scholar] [CrossRef]
- Wang, G.; He, X.; Wang, Q. Intratumoral bacteria are an important “accomplice” in tumor development and metastasis. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188846. [Google Scholar] [CrossRef] [PubMed]
- de Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef] [PubMed]
- Niller, H.H.; Minarovits, J. Patho-epigenetics of Infectious Diseases Caused by Intracellular Bacteria. In Patho-Epigenetics of Infectious Disease; Advances in Experimental Medicine and Biology; Minarovits, J., Niller, H., Eds.; Springer Nature Switzerland AG: Cham, Switzerland, 2016; Volume 879, pp. 107–130. [Google Scholar] [CrossRef]
- Bierne, H.; Hamon, M.; Cossart, P. Epigenetics and bacterial infections. Cold Spring Harb. Perspect. Med. 2012, 2, a010272. [Google Scholar] [CrossRef]
- Niller, H.H.; Banati, F.; Ay, E.; Minarovits, J. Epigenetic changes in virus-associated neoplasms. In Patho-Epigenetics of Disease; Springer: New York, NY, USA, 2012; pp. 179–225. [Google Scholar]
- Niller, H.H.; Banati, F.; Ay, E.; Minarovits, J. Microbe-induced epigenetic alterations. In Patho-Epigenetics of Disease; Springer: New York, NY, USA, 2012; pp. 419–455. [Google Scholar]
- Sears, C.L.; Garrett, W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014, 15, 317–328. [Google Scholar] [CrossRef]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Huber, A.; van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Okumura, S.; Konishi, Y.; Narukawa, M.; Sugiura, Y.; Yoshimoto, S.; Arai, Y.; Sato, S.; Yoshida, Y.; Tsuji, S.; Uemura, K.; et al. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat. Commun. 2021, 12, 5674. [Google Scholar] [CrossRef] [PubMed]
- Salvi, P.S.; Cowles, R.A. Butyrate and the Intestinal Epithelium: Modulation of Proliferation and Inflammation in Homeostasis and Disease. Cells 2021, 10, 1775. [Google Scholar] [CrossRef]
- Fessler, J.; Matson, V.; Gajewski, T.F. Exploring the emerging role of the microbiome in cancer immunotherapy. J. Immunother. Cancer 2019, 7, 108. [Google Scholar] [CrossRef]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Ding, T.; Schloss, P.D. Dynamics and associations of microbial community types across the human body. Nature 2014, 509, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Liu, M.; Fujimura, K.E.; Lyalina, S.; Nagarkar, D.R.; Charbit, B.; Bergstedt, J.; Patin, E.; Harrison, O.J.; Quintana-Murci, L.; et al. Gut microbiome stability and dynamics in healthy donors and patients with non-gastrointestinal cancers. J. Exp. Med. 2021, 218, e20200606. [Google Scholar] [CrossRef] [PubMed]
- Healy, C.M.; Moran, G.P. The microbiome and oral cancer: More questions than answers. Oral Oncol. 2019, 89, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Swaney, M.H.; Kalan, L.R. Living in Your Skin: Microbes, Molecules, and Mechanisms. Infect. Immun. 2021, 89, e00695-20. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.R.; Gabaldón, T. The Human Oral Microbiome in Health and Disease: From Sequences to Ecosystems. Microorganisms 2020, 8, 308. [Google Scholar] [CrossRef]
- Xu, J.; Peng, J.J.; Yang, W.; Fu, K.; Zhang, Y. Vaginal microbiomes and ovarian cancer: A review. Am. J. Cancer Res. 2020, 10, 743–756. [Google Scholar]
- Allen, J.; Sears, C.L. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: Contributions to colorectal cancer development. Genome Med. 2019, 11, 11. [Google Scholar] [CrossRef]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012, 22, 292–298. [Google Scholar] [CrossRef]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef]
- Flanagan, L.; Schmid, J.; Ebert, M.; Soucek, P.; Kunicka, T.; Liska, V.; Bruha, J.; Neary, P.; Dezeeuw, N.; Tommasino, M.; et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1381–1390. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Kauppila, J.H.; Karttunen, T.J.; Saarnio, J.; Nyberg, P.; Salo, T.; Graves, D.E.; Lehenkari, P.P.; Selander, K.S. Short DNA sequences and bacterial DNA induce esophageal, gastric, and colorectal cancer cell invasion. APMIS 2013, 121, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.L.; Freeman, D.J.; Pleasance, S.; Watson, P.; Moore, R.A.; Cochrane, K.; Allen-Vercoe, E.; Holt, R.A. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 2013, 1, 16. [Google Scholar] [CrossRef] [PubMed]
- Zackular, J.P.; Rogers, M.A.; Ruffin, M.T., 4th; Schloss, P.D. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. 2014, 7, 1112–1121. [Google Scholar] [CrossRef]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Böhm, J.; Brunetti, F.; Habermann, N.; et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef]
- Walker, A.W. Microbiota of the Human Body. In Microbiota of the Human Body: Implications in Health and Disease; Cohen, I.R., Lajtha, N.S., Lambris, J.D., Paoletti, R., Eds.; Springer Nature: Cham, Switzerland, 2016; Volume 902, pp. 5–32. [Google Scholar] [CrossRef]
- Elinav, E.; Nowarski, R.; Thaiss, C.A.; Hu, B.; Jin, C.; Flavell, R.A. Inflammation-induced cancer: Crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer 2013, 13, 759–771. [Google Scholar] [CrossRef]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial driver-passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef]
- Kean, J.M.; Rao, S.; Wang, M.; Garcea, R.L. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009, 5, e1000363. [Google Scholar] [CrossRef]
- Baez, C.F.; Brandão, R.; Villani, S.; Delbue, S. Human Polyomaviruses: The Battle of Large and Small Tumor Antigens. Virology 2017, 8, 1178122X17744785. [Google Scholar] [CrossRef]
- Saltzman, E.T.; Palacios, T.; Thomsen, M.; Vitetta, L. Intestinal Microbiome Shifts, Dysbiosis, Inflammation, and Non-alcoholic Fatty Liver Disease. Front. Microbiol. 2018, 9, 61. [Google Scholar] [CrossRef]
- Ahmad, A. Breast Cancer Statistics: Recent Trends; Springer: Cham, Switzerland, 2019; Volume 1152, pp. 1–7. [Google Scholar] [CrossRef]
- Kalinowski, L.; Saunus, J.M.; McCart, A.E.; Lakhani, S.R. Breast Cancer Heterogeneity in Primary and Metastatic Disease. In Breast Cancer Metastasis and Drug Resistance; Springer: Cham, Switzerland, 2019; Volume 1152, pp. 75–104. [Google Scholar] [CrossRef]
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef]
- Devoy, C.; Flores, Y.; Tangney, M. Understanding and harnessing triple-negative breast cancer-related microbiota in oncology. Front. Oncol. 2022, 12, 1020121. [Google Scholar] [CrossRef]
- Goedert, J.J.; Jones, G.; Hua, X.; Xu, X.; Yu, G.; Flores, R.; Falk, R.T.; Gail, M.H.; Shi, J.; Ravel, J.; et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: A population-based case-control pilot study. J. Natl. Cancer Inst. 2015, 107, djv147. [Google Scholar] [CrossRef] [PubMed]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, A.P.; Redinbo, M.R.; Bultman, S.J. The role of the microbiome in cancer development and therapy. CA Cancer J. Clin. 2017, 67, 326–344. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Douglass, J.; Prasath, V.; Neace, M.; Atrchian, S.; Manjili, M.H.; Shokouhi, S.; Habibi, M. The microbiome and breast cancer: A review. Breast Cancer Res. Treat. 2019, 178, 493–496. [Google Scholar] [CrossRef]
- Adlercreutz, H.; Martin, F.; Pulkkinen, M.; Dencker, H.; Rimér, U.; Sjöberg, N.O.; Tikkanen, M.J. Intestinal metabolism of estrogens. J. Clin. Endocrinol. Metab. 1976, 43, 497–505. [Google Scholar] [CrossRef]
- Org, E.; Mehrabian, M.; Parks, B.W.; Shipkova, P.; Liu, X.; Drake, T.A.; Lusis, A.J. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes 2016, 7, 313–322. [Google Scholar] [CrossRef]
- Xuan, C.; Shamonki, J.M.; Chung, A.; Dinome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial dysbiosis is associated with human breast cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef] [PubMed]
- Hieken, T.J.; Chen, J.; Hoskin, T.L.; Walther-Antonio, M.; Johnson, S.; Ramaker, S.; Xiao, J.; Radisky, D.C.; Knutson, K.L.; Kalari, K.R.; et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci. Rep. 2016, 6, 30751. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.A.; Bashir, M.; Rivas, M.N.; Duvall, K.; Sieling, P.A.; Pieber, T.R.; Vaishampayan, P.A.; Love, S.M.; Lee, D.J. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci. Rep. 2016, 6, 28061. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef]
- Santen, R.J. Benign Breast Disease in Women. In Endotext; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Hofland, J., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2018. [Google Scholar]
- Patel, S.H.; Vaidya, Y.H.; Patel, R.J.; Pandit, R.J.; Joshi, C.G.; Kunjadiya, A.P. Culture independent assessment of human milk microbial community in lactational mastitis. Sci. Rep. 2017, 7, 7804. [Google Scholar] [CrossRef]
- Hanshew, A.S.; Jetté, M.E.; Thibeault, S.L. Characterization and comparison of bacterial communities in benign vocal fold lesions. Microbiome 2014, 2, 43. [Google Scholar] [CrossRef]
- Amieva, M.; Peek, R.M. Pathobiology of Helicobacter pylori-Induced Gastric Cancer. Gastroenterology 2016, 150, 64–78. [Google Scholar] [CrossRef]
- Zamani, M.; Ebrahimtabar, F.; Zamani, V.; Miller, W.H.; Alizadeh-Navaei, R.; Shokri-Shirvani, J.; Derakhshan, M.H. Systematic review with meta-analysis: The worldwide prevalence of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 2018, 47, 868–876. [Google Scholar] [CrossRef]
- Perry, S.; de Jong, B.C.; Solnick, J.V.; Sanchez, M.; Yang, S.; Lin, P.L.; Hansen, L.M.; Talat, N.; Hill, P.C.; Hussain, R.; et al. Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS ONE 2010, 5, e8804. [Google Scholar] [CrossRef]
- Kira, J.I.; Isobe, N. Helicobacter pylori infection and demyelinating disease of the central nervous system. J. Neuroimmunol. 2019, 329, 14–19. [Google Scholar] [CrossRef]
- Piovani, D.; Danese, S.; Peyrin-Biroulet, L.; Nikolopoulos, G.K.; Lytras, T.; Bonovas, S. Environmental Risk Factors for Inflammatory Bowel Diseases: An Umbrella Review of Meta-analyses. Gastroenterology 2019, 157, 647–659.e4. [Google Scholar] [CrossRef] [PubMed]
- Compare, D.; Rocco, A.; Nardone, G. Risk factors in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2010, 14, 302–308. [Google Scholar] [PubMed]
- de Martel, C.; Ferlay, J.; Franceschi, S.; Vignat, J.; Bray, F.; Forman, D.; Plummer, M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012, 13, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Prz. Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef]
- Yamaoka, Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 629–641. [Google Scholar] [CrossRef]
- Hatakeyama, M. Helicobacter pylori CagA and gastric cancer: A paradigm for hit-and-run carcinogenesis. Cell Host Microbe 2014, 15, 306–316. [Google Scholar] [CrossRef]
- Ki, M.R.; Hwang, M.; Kim, A.Y.; Lee, E.M.; Lee, E.J.; Lee, M.M.; Sung, S.E.; Kim, S.H.; Lee, H.S.; Jeong, K.S. Role of vacuolating cytotoxin VacA and cytotoxin-associated antigen CagA of Helicobacter pylori in the progression of gastric cancer. Mol. Cell. Biochem. 2014, 396, 23–32. [Google Scholar] [CrossRef]
- Ansari, S.; Yamaoka, Y. Role of vacuolating cytotoxin A in Helicobacter pylori infection and its impact on gastric pathogenesis. Expert Rev. Anti Infect. Ther. 2020, 18, 987–996. [Google Scholar] [CrossRef]
- Umeda, M.; Murata-Kamiya, N.; Saito, Y.; Ohba, Y.; Takahashi, M.; Hatakeyama, M. Helicobacter pylori CagA causes mitotic impairment and induces chromosomal instability. J. Biol. Chem. 2009, 284, 22166–22172. [Google Scholar] [CrossRef]
- Huang, F.Y.; Chan, A.O.; Rashid, A.; Wong, D.K.; Cho, C.H.; Yuen, M.F. Helicobacter pylori induces promoter methylation of E-cadherin via interleukin-1β activation of nitric oxide production in gastric cancer cells. Cancer 2012, 118, 4969–4980. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S.; et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell 2019, 176, 998–1013.e16. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [PubMed]
- McAllister, F.; Khan, W.; Helmink, B.; Wargo, J.A. The Tumor Microbiome in Pancreatic Cancer: Bacteria and Beyond. Cancer Cell 2019, 36, 577–579. [Google Scholar] [CrossRef]
- Holliday, R. Epigenetics: A historical overview. Epigenetics 2006, 1, 76–80. [Google Scholar] [CrossRef]
- Baylin, S.B.; Ohm, J.E. Epigenetic gene silencing in cancer—A mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer 2006, 6, 107–116. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004, 429, 457–463. [Google Scholar] [CrossRef]
- Chen, K.; Zhao, B.S.; He, C. Nucleic Acid Modifications in Regulation of Gene Expression. Cell Chem. Biol. 2016, 23, 74–85. [Google Scholar] [CrossRef]
- Zhang, G.; Huang, H.; Liu, D.; Cheng, Y.; Liu, X.; Zhang, W.; Yin, R.; Zhang, D.; Zhang, P.; Liu, J.; et al. N6-methyladenine DNA modification in Drosophila. Cell 2015, 161, 893–906. [Google Scholar] [CrossRef]
- Jones, P.A.; Takai, D. The role of DNA methylation in mammalian epigenetics. Science 2001, 293, 1068–1070. [Google Scholar] [CrossRef] [PubMed]
- Frommer, M.; McDonald, L.E.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 1992, 89, 1827–1831. [Google Scholar] [CrossRef]
- Melnikov, A.A.; Gartenhaus, R.B.; Levenson, A.S.; Motchoulskaia, N.A.; Levenson, V.V. MSRE-PCR for analysis of gene-specific DNA methylation. Nucleic Acids Res. 2005, 33, e93. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, C. Coding or Noncoding, the Converging Concepts of RNAs. Front. Genet. 2019, 10, 496. [Google Scholar] [CrossRef] [PubMed]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef]
- Angarica, V.E.; Del Sol, A. Bioinformatics Tools for Genome-Wide Epigenetic Research. Adv. Exp. Med. Biol. 2017, 978, 489–512. [Google Scholar] [CrossRef]
- Li, Y. Modern epigenetics methods in biological research. Methods 2021, 187, 104–113. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef]
- Flavahan, W.A.; Gaskell, E.; Bernstein, B.E. Epigenetic plasticity and the hallmarks of cancer. Science 2017, 357, eaal2380. [Google Scholar] [CrossRef]
- Jones, P.A.; Issa, J.P.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef]
- Huang, S. Tumor progression: Chance and necessity in Darwinian and Lamarckian somatic (mutationless) evolution. Prog. Biophys. Mol. Biol. 2012, 110, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Darwiche, N. Epigenetic mechanisms and the hallmarks of cancer: An intimate affair. Am. J. Cancer Res. 2020, 10, 1954–1978. [Google Scholar]
- Feng, Y.; Liu, X.; Pauklin, S. 3D chromatin architecture and epigenetic regulation in cancer stem cells. Protein Cell 2021, 12, 440–454. [Google Scholar] [CrossRef] [PubMed]
- Nam, A.S.; Chaligne, R.; Landau, D.A. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat. Rev. Genet. 2021, 22, 3–18. [Google Scholar] [CrossRef]
- Bitman-Lotan, E.; Orian, A. Nuclear organization and regulation of the differentiated state. Cell. Mol. Life Sci. 2021, 78, 3141–3158. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A landscape takes shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Chen, T. DNA Methylation Reprogramming during Mammalian Development. Genes 2019, 10, 257. [Google Scholar] [CrossRef]
- Hegde, A.N.; Smith, S.G. Recent developments in transcriptional and translational regulation underlying long-term synaptic plasticity and memory. Learn. Mem. 2019, 26, 307–317. [Google Scholar] [CrossRef]
- Kim, S.; Kaang, B.K. Epigenetic regulation and chromatin remodeling in learning and memory. Exp. Mol. Med. 2017, 49, e281. [Google Scholar] [CrossRef]
- Takane, K.; Midorikawa, Y.; Yagi, K.; Sakai, A.; Aburatani, H.; Takayama, T.; Kaneda, A. Aberrant promoter methylation of PPP1R3C and EFHD1 in plasma of colorectal cancer patients. Cancer Med. 2014, 3, 1235–1245. [Google Scholar] [CrossRef]
- Khare, S.; Verma, M. Epigenetics of colon cancer. Methods Mol. Biol. 2012, 863, 177–185. [Google Scholar] [CrossRef]
- Valenzuela, M.A.; Canales, J.; Corvalán, A.H.; Quest, A.F. Helicobacter pylori-induced inflammation and epigenetic changes during gastric carcinogenesis. World J. Gastroenterol. 2015, 21, 12742–12756. [Google Scholar] [CrossRef]
- Kumar, D.; Verma, M. Methods in cancer epigenetics and epidemiology. Methods Mol. Biol. 2009, 471, 273–288. [Google Scholar] [CrossRef]
- Deb, M.; Sengupta, D.; Kar, S.; Rath, S.K.; Roy, S.; Das, G.; Patra, S.K. Epigenetic drift towards histone modifications regulates CAV1 gene expression in colon cancer. Gene 2016, 581, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Deb, M.; Sengupta, D.; Rath, S.K.; Kar, S.; Parbin, S.; Shilpi, A.; Pradhan, N.; Bhutia, S.K.; Roy, S.; Patra, S.K. Clusterin gene is predominantly regulated by histone modifications in human colon cancer and ectopic expression of the nuclear isoform induces cell death. Biochim. Biophys. Acta 2015, 1852, 1630–1645. [Google Scholar] [CrossRef] [PubMed]
- Gezer, U.; Yörüker, E.E.; Keskin, M.; Kulle, C.B.; Dharuman, Y.; Holdenrieder, S. Histone Methylation Marks on Circulating Nucleosomes as Novel Blood-Based Biomarker in Colorectal Cancer. Int. J. Mol. Sci. 2015, 16, 29654–29662. [Google Scholar] [CrossRef]
- Benard, A.; Goossens-Beumer, I.J.; van Hoesel, A.Q.; de Graaf, W.; Horati, H.; Putter, H.; Zeestraten, E.C.; van de Velde, C.J.; Kuppen, P.J. Histone trimethylation at H3K4, H3K9 and H4K20 correlates with patient survival and tumor recurrence in early-stage colon cancer. BMC Cancer 2014, 14, 531. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.P.; Chiu, S.C.; Wu, C.C.; Lin, S.Z.; Kang, J.C.; Chen, Y.L.; Lin, P.C.; Pang, C.Y.; Harn, H.J. The association of methylation in the promoter of APC and MGMT and the prognosis of Taiwanese CRC patients. Genet. Test. Mol. Biomark. 2009, 13, 67–71. [Google Scholar] [CrossRef]
- Nazemalhosseini, M.E.; Kuppen, P.J.; Aghdaei, H.A.; Zali, M.R. The CpG island methylator phenotype (CIMP) in colorectal cancer. Gastroenterol. Hepatol. Bed Bench 2013, 6, 120–128. [Google Scholar]
- Ogino, S.; Cantor, M.; Kawasaki, T.; Brahmandam, M.; Kirkner, G.J.; Weisenberger, D.J.; Campan, M.; Laird, P.W.; Loda, M.; Fuchs, C.S. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut 2006, 55, 1000–1006. [Google Scholar] [CrossRef]
- Li, H.; Myeroff, L.; Kasturi, L.; Krumroy, L.; Schwartz, S.; Willson, J.K.; Stanbridge, E.; Casey, G.; Markowitz, S. Chromosomal autonomy of hMLH1 methylation in colon cancer. Oncogene 2002, 21, 1443–1449. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Zhang, X.R.; Park, J.L.; Kim, J.H.; Zhang, L.; Ma, J.L.; Liu, W.D.; Deng, D.J.; You, W.C.; Kim, Y.S.; et al. Genome-wide DNA methylation profiles altered by Helicobacter pylori in gastric mucosa and blood leukocyte DNA. Oncotarget 2016, 7, 37132–37144. [Google Scholar] [CrossRef] [PubMed]
- Papastergiou, V.; Karatapanis, S.; Georgopoulos, S.D. Helicobacter pylori and colorectal neoplasia: Is there a causal link? World J. Gastroenterol. 2016, 22, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Shmuely, H.; Melzer, E.; Braverman, M.; Domnitz, N.; Yahav, J. Helicobacter pylori infection is associated with advanced colorectal neoplasia. Scand. J. Gastroenterol. 2014, 49, 516–517. [Google Scholar] [CrossRef]
- Brew, R.; Erikson, J.S.; West, D.C.; Kinsella, A.R.; Slavin, J.; Christmas, S.E. Interleukin-8 as an autocrine growth factor for human colon carcinoma cells in vitro. Cytokine 2000, 12, 78–85. [Google Scholar] [CrossRef]
- Choi, W.; Lee, J.; Lee, J.Y.; Lee, S.M.; Kim, D.W.; Kim, Y.J. Classification of Colon Cancer Patients Based on the Methylation Patterns of Promoters. Genom. Inform. 2016, 14, 46–52. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Laghi, L.; Randolph, A.E.; Chauhan, D.P.; Marra, G.; Major, E.O.; Neel, J.V.; Boland, C.R. JC virus DNA is present in the mucosa of the human colon and in colorectal cancers. Proc. Natl. Acad. Sci. USA 1999, 96, 7484–7489. [Google Scholar] [CrossRef]
- Lin, P.Y.; Fung, C.Y.; Chang, F.P.; Huang, W.S.; Chen, W.C.; Wang, J.Y.; Chang, D. Prevalence and genotype identification of human JC virus in colon cancer in Taiwan. J. Med. Virol. 2008, 80, 1828–1834. [Google Scholar] [CrossRef]
- Sinagra, E.; Raimondo, D.; Gallo, E.; Stella, M.; Cottone, M.; Orlando, A.; Rossi, F.; Orlando, E.; Messina, M.; Tomasello, G.; et al. Could JC virus provoke metastasis in colon cancer? World J. Gastroenterol. 2014, 20, 15745–15749. [Google Scholar] [CrossRef][Green Version]
- Theodoropoulos, G.; Panoussopoulos, D.; Papaconstantinou, I.; Gazouli, M.; Perdiki, M.; Bramis, J.; Lazaris, A. Assessment of JC polyoma virus in colon neoplasms. Dis. Colon Rectum 2005, 48, 86–91. [Google Scholar] [CrossRef]
- Farhana, L.; Banerjee, H.N.; Verma, M.; Majumdar, A.P. Role of Microbiome in Carcinogenesis Process and Epigenetic Regulation of Colorectal Cancer. Methods Mol. Biol. 2018, 1856, 35–55. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Lehouritis, P.; Cummins, J.; Stanton, M.; Murphy, C.T.; McCarthy, F.O.; Reid, G.; Urbaniak, C.; Byrne, W.L.; Tangney, M. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci. Rep. 2015, 5, 14554. [Google Scholar] [CrossRef] [PubMed]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Daillère, R.; Vétizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Von Bültzingslöwen, I.; Adlerberth, I.; Wold, A.E.; Dahlén, G.; Jontell, M. Oral and intestinal microflora in 5-fluorouracil treated rats, translocation to cervical and mesenteric lymph nodes and effects of probiotic bacteria. Oral Microbiol. Immunol. 2003, 18, 278–284. [Google Scholar] [CrossRef]
- Le Bastard, Q.; Ward, T.; Sidiropoulos, D.; Hillmann, B.M.; Chun, C.L.; Sadowsky, M.J.; Knights, D.; Montassier, E. Fecal microbiota transplantation reverses antibiotic and chemotherapy-induced gut dysbiosis in mice. Sci. Rep. 2018, 8, 6219. [Google Scholar] [CrossRef]
- Guthrie, L.; Gupta, S.; Daily, J.; Kelly, L. Human microbiome signatures of differential colorectal cancer drug metabolism. NPJ Biofilms Microbiomes 2017, 3, 27. [Google Scholar] [CrossRef]
- Montassier, E.; Gastinne, T.; Vangay, P.; Al-Ghalith, G.A.; des Varannes, S.; Massart, S.; Moreau, P.; Potel, G.; de La Cochetière, M.F.; Batard, E.; et al. Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment. Pharmacol. Ther. 2015, 42, 515–528. [Google Scholar] [CrossRef]
- Schluter, J.; Peled, J.U.; Taylor, B.P.; Markey, K.A.; Smith, M.; Taur, Y.; Niehus, R.; Staffas, A.; Dai, A.; Fontana, E.; et al. The gut microbiota is associated with immune cell dynamics in humans. Nature 2020, 588, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Cullin, N.; Azevedo, A.C.; Straussman, R.; Stein-Thoeringer, C.K.; Elinav, E. Microbiome and cancer. Cancer Cell 2021, 39, 1317–1341. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y. Eukaryotic DNA topoisomerase I: Genome gatekeeper and its intruders, camptothecins. Semin. Oncol. 1996, 1, 3–10. [Google Scholar]
- Hsiang, Y.H.; Liu, L.F. Identification of mammalian DNA topoisomerase I as an intracellular target of the anticancer drug camptothecin. Cancer Res. 1988, 48, 1722–1726. [Google Scholar]
- Yohe, M.E.; Heske, C.M.; Stewart, E.; Adamson, P.C.; Ahmed, N.; Antonescu, C.R.; Chen, E.; Collins, N.; Ehrlich, A.; Galindo, R.L.; et al. Insights into pediatric rhabdomyosarcoma research: Challenges and goals. Pediatr. Blood Cancer 2019, 66, e27869. [Google Scholar] [CrossRef]
- Bagatell, R.; London, W.B.; Wagner, L.M.; Voss, S.D.; Stewart, C.F.; Maris, J.M.; Kretschmar, C.; Cohn, S.L. Phase II study of irinotecan and temozolomide in children with relapsed or refractory neuroblastoma: A Children’s Oncology Group study. J. Clin. Oncol. 2011, 29, 208–213. [Google Scholar] [CrossRef]
- Di Desidero, T.; Antonelli, A.; Orlandi, P.; Ferrari, S.M.; Fioravanti, A.; Alì, G.; Fontanini, G.; Basolo, F.; Francia, G.; Bocci, G. Synergistic efficacy of irinotecan and sunitinib combination in preclinical models of anaplastic thyroid cancer. Cancer Lett. 2017, 411, 35–43. [Google Scholar] [CrossRef]
- Chen, Z.; Jiang, L. The clinical application of fruquintinib on colorectal cancer. Expert. Rev. Clin. Pharmacol. 2019, 12, 713–721. [Google Scholar] [CrossRef]
- Liu, Q.; Hua, S.; Wang, X.; Chen, F.; Gou, S. The introduction of immunosuppressor (TDO inhibitor) significantly improved the efficacy of irinotecan in treating hepatocellular carcinoma. Cancer Immunol. Immunother. 2021, 70, 497–508. [Google Scholar] [CrossRef]
- Kawato, Y.; Aonuma, M.; Hirota, Y.; Kuga, H.; Sato, K. Intracellular roles of SN-38, a metabolite of the camptothecin derivative CPT-11, in the antitumor effect of CPT-11. Cancer Res. 1991, 51, 4187–4191. [Google Scholar]
- Slatter, J.G.; Su, P.; Sams, J.P.; Schaaf, L.J.; Wienkers, L.C. Bioactivation of the anticancer agent CPT-11 to SN-38 by human hepatic microsomal carboxylesterases and the in vitro assessment of potential drug interactions. Drug Metab. Dispos. 1997, 25, 1157–1164. [Google Scholar] [PubMed]
- Rudakova, E.V.; Boltneva, N.P.; Makhaeva, G.F. Comparative analysis of esterase activities of human, mouse, and rat blood. Bull. Exp. Biol. Med. 2011, 152, 73–75. [Google Scholar] [CrossRef]
- Gupta, E.; Lestingi, T.M.; Mick, R.; Ramirez, J.; Vokes, E.E.; Ratain, M.J. Metabolic fate of irinotecan in humans: Correlation of glucuronidation with diarrhea. Cancer Res. 1994, 54, 3723–3725. [Google Scholar] [PubMed]
- Wallace, B.D.; Roberts, A.B.; Pollet, R.M.; Ingle, J.D.; Biernat, K.A.; Pellock, S.J.; Venkatesh, M.K.; Guthrie, L.; O’Neal, S.K.; Robinson, S.J.; et al. Structure and Inhibition of Microbiome β-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem. Biol. 2015, 22, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.D.; Wang, H.; Lane, K.T.; Scott, J.E.; Orans, J.; Koo, J.S.; Venkatesh, M.; Jobin, C.; Yeh, L.A.; Mani, S.; et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 2010, 330, 831–835. [Google Scholar] [CrossRef]
- Kodawara, T.; Higashi, T.; Negoro, Y.; Kamitani, Y.; Igarashi, T.; Watanabe, K.; Tsukamoto, H.; Yano, R.; Masada, M.; Iwasaki, H.; et al. The Inhibitory Effect of Ciprofloxacin on the β-Glucuronidase-mediated Deconjugation of the Irinotecan Metabolite SN-38-G. Basic Clin. Pharmacol. Toxicol. 2016, 118, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.; Liu, T.; Zhu, X.; Ahmad, S.; Williams, A.L.; Phan, A.T.; Zhao, H.; Scott, J.E.; Yeh, L.A.; Wong, S.T. Old drug new use—Amoxapine and its metabolites as potent bacterial β-glucuronidase inhibitors for alleviating cancer drug toxicity. Clin. Cancer Res. 2014, 20, 3521–3530. [Google Scholar] [CrossRef]
- Stringer, A.M.; Gibson, R.J.; Logan, R.M.; Bowen, J.M.; Yeoh, A.S.; Keefe, D.M. Faecal microflora and beta-glucuronidase expression are altered in an irinotecan-induced diarrhea model in rats. Cancer Biol. Ther. 2008, 7, 1919–1925. [Google Scholar] [CrossRef]
- Takasuna, K.; Hagiwara, T.; Hirohashi, M.; Kato, M.; Nomura, M.; Nagai, E.; Yokoi, T.; Kamataki, T. Inhibition of intestinal microflora beta-glucuronidase modifies the distribution of the active metabolite of the antitumor agent, irinotecan hydrochloride (CPT-11) in rats. Cancer Chemother. Pharmacol. 1998, 42, 280–286. [Google Scholar] [CrossRef]
- Kehrer, D.F.; Sparreboom, A.; Verweij, J.; de Bruijn, P.; Nierop, C.A.; van de Schraaf, J.; Ruijgrok, E.J.; de Jonge, M.J. Modulation of irinotecan-induced diarrhea by cotreatment with neomycin in cancer patients. Clin. Cancer Res. 2001, 7, 1136–1141. [Google Scholar]
- de Jong, F.A.; Kehrer, D.F.; Mathijssen, R.H.; Creemers, G.J.; de Bruijn, P.; van Schaik, R.H.; Planting, A.S.; van der Gaast, A.; Eskens, F.A.; Janssen, J.T.; et al. Prophylaxis of irinotecan-induced diarrhea with neomycin and potential role for UGT1A1*28 genotype screening: A double-blind, randomized, placebo-controlled study. Oncologist 2006, 11, 944–954. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Gao, R.; Lv, C.; Yu, Z.; Wang, H.; Geng, X.; Wang, Z.; Dou, W. Berberine Improves Irinotecan-Induced Intestinal Mucositis Without Impairing the Anti-colorectal Cancer Efficacy of Irinotecan by Inhibiting Bacterial β-glucuronidase. Front. Pharmacol. 2021, 12, 774560. [Google Scholar] [CrossRef]
- Leardini, D.; Venturelli, F.; Baccelli, F.; Cerasi, S.; Muratore, E.; Brigidi, P.; Pession, A.; Prete, A.; Masetti, R. Pharmacomicrobiomics in Pediatric Oncology: The Complex Interplay between Commonly Used Drugs and Gut Microbiome. Int. J. Mol. Sci. 2022, 23, 15387. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Hayase, E.; Jenq, R.R. Role of the intestinal microbiome and microbial-derived metabolites in immune checkpoint blockade immunotherapy of cancer. Genome Med. 2021, 13, 107. [Google Scholar] [CrossRef]
- Coutzac, C.; Jouniaux, J.M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L.; et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M.; et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2019, 381, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Ascierto, P.A.; Long, G.V.; Robert, C.; Brady, B.; Dutriaux, C.; Di Giacomo, A.M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; et al. Survival Outcomes in Patients With Previously Untreated BRAF Wild-Type Advanced Melanoma Treated With Nivolumab Therapy: Three-Year Follow-up of a Randomized Phase 3 Trial. JAMA Oncol. 2019, 5, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef]
- Amaria, R.N.; Reddy, S.M.; Tawbi, H.A.; Davies, M.A.; Ross, M.I.; Glitza, I.C.; Cormier, J.N.; Lewis, C.; Hwu, W.J.; Hanna, E.; et al. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat. Med. 2018, 24, 1649–1654. [Google Scholar] [CrossRef] [PubMed]
- Baruch, E.N.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021, 371, 602–609. [Google Scholar] [CrossRef] [PubMed]
- McQuade, J.L.; Ologun, G.O.; Arora, R.; Wargo, J.A. Gut Microbiome Modulation Via Fecal Microbiota Transplant to Augment Immunotherapy in Patients with Melanoma or Other Cancers. Curr. Oncol. Rep. 2020, 22, 74. [Google Scholar] [CrossRef] [PubMed]
- Dubin, K.; Callahan, M.K.; Ren, B.; Khanin, R.; Viale, A.; Ling, L.; No, D.; Gobourne, A.; Littmann, E.; Huttenhower, C.; et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 2016, 7, 10391. [Google Scholar] [CrossRef]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.A.; Thomas, A.M.; Bolte, L.A.; Björk, J.R.; de Ruijter, L.K.; Armanini, F.; Asnicar, F.; Blanco-Miguez, A.; Board, R.; Calbet-Llopart, N.; et al. Cross-cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma. Nat. Med. 2022, 28, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef] [PubMed]
- June, C.H.; Sadelain, M. Chimeric Antigen Receptor Therapy. N. Engl. J. Med. 2018, 379, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Ansell, S.M.; Lesokhin, A.M.; Borrello, I.; Halwani, A.; Scott, E.C.; Gutierrez, M.; Schuster, S.J.; Millenson, M.M.; Cattry, D.; Freeman, G.J.; et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med. 2015, 372, 311–319. [Google Scholar] [CrossRef]
- Kuehn, B.M. The Promise and Challenges of CAR-T Gene Therapy. JAMA 2017, 318, 2167–2169. [Google Scholar] [CrossRef]
- Stein-Thoeringer, C.K.; Saini, N.Y.; Zamir, E.; Blumenberg, V.; Schubert, M.L.; Mor, U.; Fante, M.A.; Schmidt, S.; Hayase, E.; Hayase, T.; et al. A non-antibiotic-disrupted gut microbiome is associated with clinical responses to CD19-CAR-T cell cancer immunotherapy. Nat. Med. 2023, 29, 906–916. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.; Dai, A.; Ghilardi, G.; Amelsberg, K.V.; Devlin, S.M.; Pajarillo, R.; Slingerland, J.B.; Beghi, S.; Herrera, P.S.; Giardina, P.; et al. Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat. Med. 2022, 28, 713–723. [Google Scholar] [CrossRef]
- Jenq, R.R.; van den Brink, M.R. Allogeneic haematopoietic stem cell transplantation: Individualized stem cell and immune therapy of cancer. Nat. Rev. Cancer 2010, 10, 213–221. [Google Scholar] [CrossRef]
- Zeiser, R.; Blazar, B.R. Acute Graft-versus-Host Disease—Biologic Process, Prevention, and Therapy. N. Engl. J. Med. 2017, 377, 2167–2179. [Google Scholar] [CrossRef]
- Shono, Y.; Ueha, S.; Wang, Y.; Abe, J.; Kurachi, M.; Matsuno, Y.; Sugiyama, T.; Nagasawa, T.; Imamura, M.; Matsushima, K. Bone marrow graft-versus-host disease: Early destruction of hematopoietic niche after MHC-mismatched hematopoietic stem cell transplantation. Blood 2010, 115, 5401–5411. [Google Scholar] [CrossRef] [PubMed]
- Shimoji, S.; Hashimoto, D.; Tsujigiwa, H.; Miyawaki, K.; Kato, K.; Takahashi, S.; Ogasawara, R.; Jiromaru, T.; Iwasaki, H.; Miyamoto, T.; et al. Graft-versus-host disease targets ovary and causes female infertility in mice. Blood 2017, 129, 1216–1225. [Google Scholar] [CrossRef] [PubMed]
- Hartrampf, S.; Dudakov, J.A.; Johnson, L.K.; Smith, O.M.; Tsai, J.; Singer, N.V.; West, M.L.; Hanash, A.M.; Albert, M.H.; Liu, B.; et al. The central nervous system is a target of acute graft versus host disease in mice. Blood 2013, 121, 1906–1910. [Google Scholar] [CrossRef]
- Zama, D.; Biagi, E.; Masetti, R.; Gasperini, P.; Prete, A.; Candela, M.; Brigidi, P.; Pession, A. Gut microbiota and hematopoietic stem cell transplantation: Where do we stand? Bone Marrow Transplant. 2017, 52, 7–14. [Google Scholar] [CrossRef]
- Paulos, C.M.; Wrzesinski, C.; Kaiser, A.; Hinrichs, C.S.; Chieppa, M.; Cassard, L.; Palmer, D.C.; Boni, A.; Muranski, P.; Yu, Z.; et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J. Clin. Investig. 2007, 117, 2197–2204. [Google Scholar] [CrossRef]
- Peuker, K.; Muff, S.; Wang, J.; Künzel, S.; Bosse, E.; Zeissig, Y.; Luzzi, G.; Basic, M.; Strigli, A.; Ulbricht, A.; et al. Epithelial calcineurin controls microbiota-dependent intestinal tumor development. Nat. Med. 2016, 22, 506–515. [Google Scholar] [CrossRef]
- Zitvogel, L.; Ayyoub, M.; Routy, B.; Kroemer, G. Microbiome and Anticancer Immunosurveillance. Cell 2016, 165, 276–287. [Google Scholar] [CrossRef]
- Roy, S.; Trinchieri, G. Microbiota: A key orchestrator of cancer therapy. Nat. Rev. Cancer 2017, 17, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Daillère, R.; Roberti, M.P.; Routy, B.; Kroemer, G. Anticancer effects of the microbiome and its products. Nat. Rev. Microbiol. 2017, 15, 465–478. [Google Scholar] [CrossRef]
- Blazar, B.R.; Murphy, W.J.; Abedi, M. Advances in graft-versus-host disease biology and therapy. Nat. Rev. Immunol. 2012, 12, 443–458. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Knight, R. Species divergence and the measurement of microbial diversity. FEMS Microbiol. Rev. 2008, 32, 557–578. [Google Scholar] [CrossRef] [PubMed]
- Taur, Y.; Jenq, R.R.; Perales, M.A.; Littmann, E.R.; Morjaria, S.; Ling, L.; No, D.; Gobourne, A.; Viale, A.; Dahi, P.B.; et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014, 124, 1174–1182. [Google Scholar] [CrossRef]
- Jenq, R.R.; Taur, Y.; Devlin, S.M.; Ponce, D.M.; Goldberg, J.D.; Ahr, K.F.; Littmann, E.R.; Ling, L.; Gobourne, A.C.; Miller, L.C.; et al. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol. Blood Marrow Transplant. 2015, 21, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Peled, J.U.; Gomes, A.L.C.; Devlin, S.M.; Littmann, E.R.; Taur, Y.; Sung, A.D.; Weber, D.; Hashimoto, D.; Slingerland, A.E.; Slingerland, J.B.; et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N. Engl. J. Med. 2020, 382, 822–834. [Google Scholar] [CrossRef] [PubMed]
- Pession, A.; Zama, D.; Muratore, E.; Leardini, D.; Gori, D.; Guaraldi, F.; Prete, A.; Turroni, S.; Brigidi, P.; Masetti, R. Fecal Microbiota Transplantation in Allogeneic Hematopoietic Stem Cell Transplantation Recipients: A Systematic Review. J. Pers. Med. 2021, 11, 100. [Google Scholar] [CrossRef] [PubMed]
- Cammarota, G.; Ianiro, G.; Tilg, H.; Rajili’c-Stojanovi’c, M.; Kump, P.; Satokari, R.; Sokol, H.; Arkkila, P.; Pintus, C.; Hart, A.; et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut 2017, 66, 569–580. [Google Scholar] [CrossRef]
- Ianiro, G.; Bibbò, S.; Scaldaferri, F.; Gasbarrini, A.; Cammarota, G. Fecal microbiota transplantation in inflammatory bowel disease: Beyond the excitement. Medicine 2014, 93, e97. [Google Scholar] [CrossRef]
- Wardill, H.R.; Secombe, K.R.; Bryant, R.V.; Hazenberg, M.D.; Costello, S.P. Adjunctive fecal microbiota transplantation in supportive oncology: Emerging indications and considerations in immunocompromised patients. eBioMedicine 2019, 44, 730–740. [Google Scholar] [CrossRef]
- Andersen, S.; Staudacher, H.; Weber, N.; Kennedy, G.; Varelias, A.; Banks, M.; Bauer, J. Pilot study investigating the effect of enteral and parenteral nutrition on the gastrointestinal microbiome post-allogeneic transplantation. Br. J. Haematol. 2020, 188, 570–581. [Google Scholar] [CrossRef]
- Mao, D.; Jiang, Q.; Sun, Y.; Mao, Y.; Guo, L.; Zhang, Y.; Man, M.; Ouyang, G.; Sheng, L. Treatment of intestinal graft-versus-host disease with unrelated donor fecal microbiota transplantation capsules: A case report. Medicine 2020, 99, e22129. [Google Scholar] [CrossRef] [PubMed]
- Innes, A.J.; Mullish, B.H.; Fernando, F.; Adams, G.; Marchesi, J.R.; Apperley, J.F.; Brannigan, E.; Davies, F.; Pavlů, J. Faecal microbiota transplant: A novel biological approach to extensively drug-resistant organism-related non-relapse mortality. Bone Marrow Transplant. 2017, 52, 1452–1454. [Google Scholar] [CrossRef] [PubMed]
- Fabozzi, F.; Trovato, C.M.; Diamanti, A.; Mastronuzzi, A.; Zecca, M.; Tripodi, S.I.; Masetti, R.; Leardini, D.; Muratore, E.; Barat, V.; et al. Management of Nutritional Needs in Pediatric Oncology: A Consensus Statement. Cancers 2022, 14, 3378. [Google Scholar] [CrossRef] [PubMed]

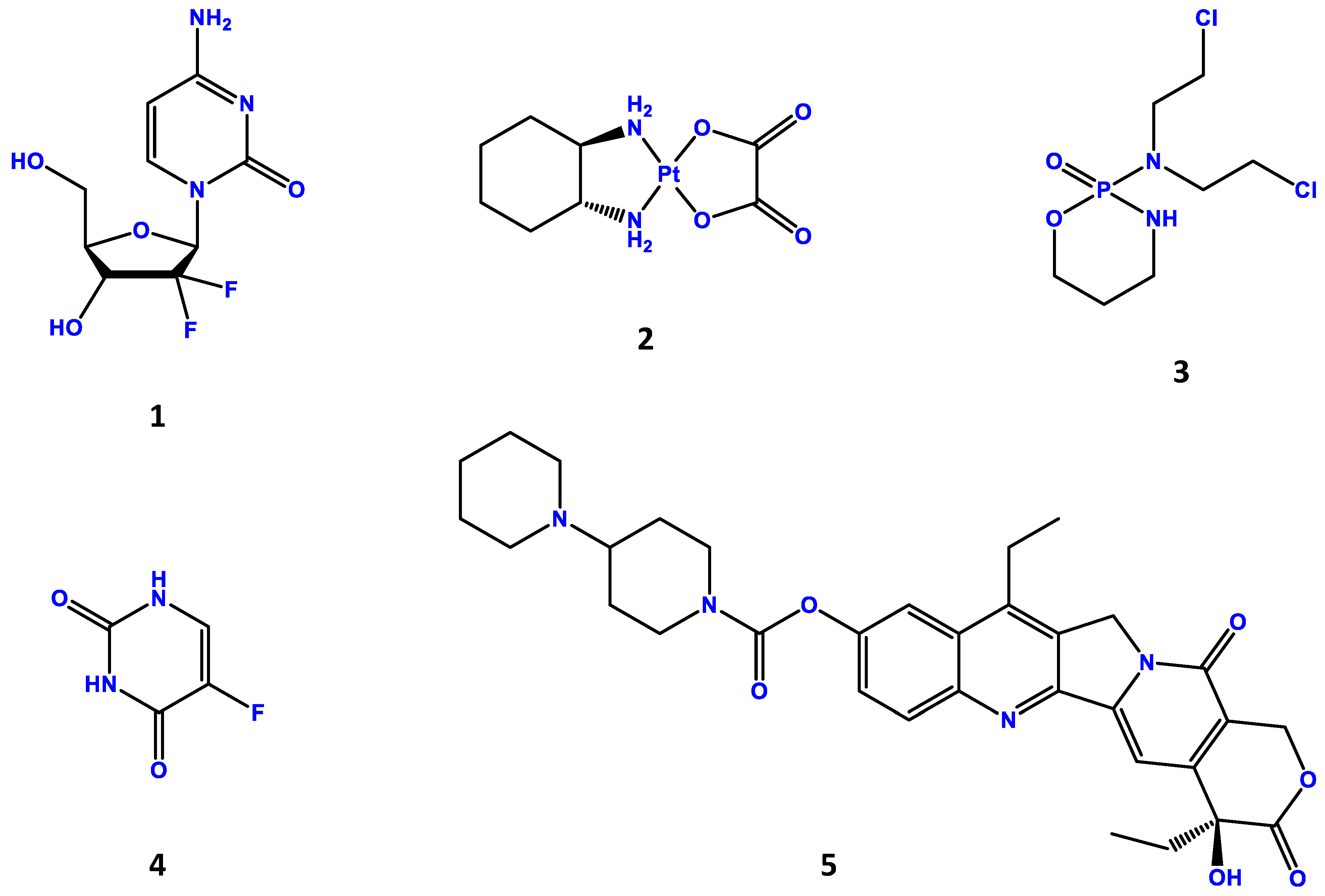
| Authors | Microbiome Features |
|---|---|
| Routy et al. [221] | ↑ Akkermansia muciniphila, Alistipes and Firmicutes |
| Gopalakrishnan et al. [219] | ↑ Faecalibacterium prausnitzii |
| Matson et al. [220] | ↑ Group of eight species driven by Bifidobacterium longum |
| Frankel et al. [232] | ↑ Bacteroides caccae |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendes, I.; Vale, N. How Can the Microbiome Induce Carcinogenesis and Modulate Drug Resistance in Cancer Therapy? Int. J. Mol. Sci. 2023, 24, 11855. https://doi.org/10.3390/ijms241411855
Mendes I, Vale N. How Can the Microbiome Induce Carcinogenesis and Modulate Drug Resistance in Cancer Therapy? International Journal of Molecular Sciences. 2023; 24(14):11855. https://doi.org/10.3390/ijms241411855
Chicago/Turabian StyleMendes, Inês, and Nuno Vale. 2023. "How Can the Microbiome Induce Carcinogenesis and Modulate Drug Resistance in Cancer Therapy?" International Journal of Molecular Sciences 24, no. 14: 11855. https://doi.org/10.3390/ijms241411855
APA StyleMendes, I., & Vale, N. (2023). How Can the Microbiome Induce Carcinogenesis and Modulate Drug Resistance in Cancer Therapy? International Journal of Molecular Sciences, 24(14), 11855. https://doi.org/10.3390/ijms241411855








