CD44 Modulates Cell Migration and Invasion in Ewing Sarcoma Cells
Abstract
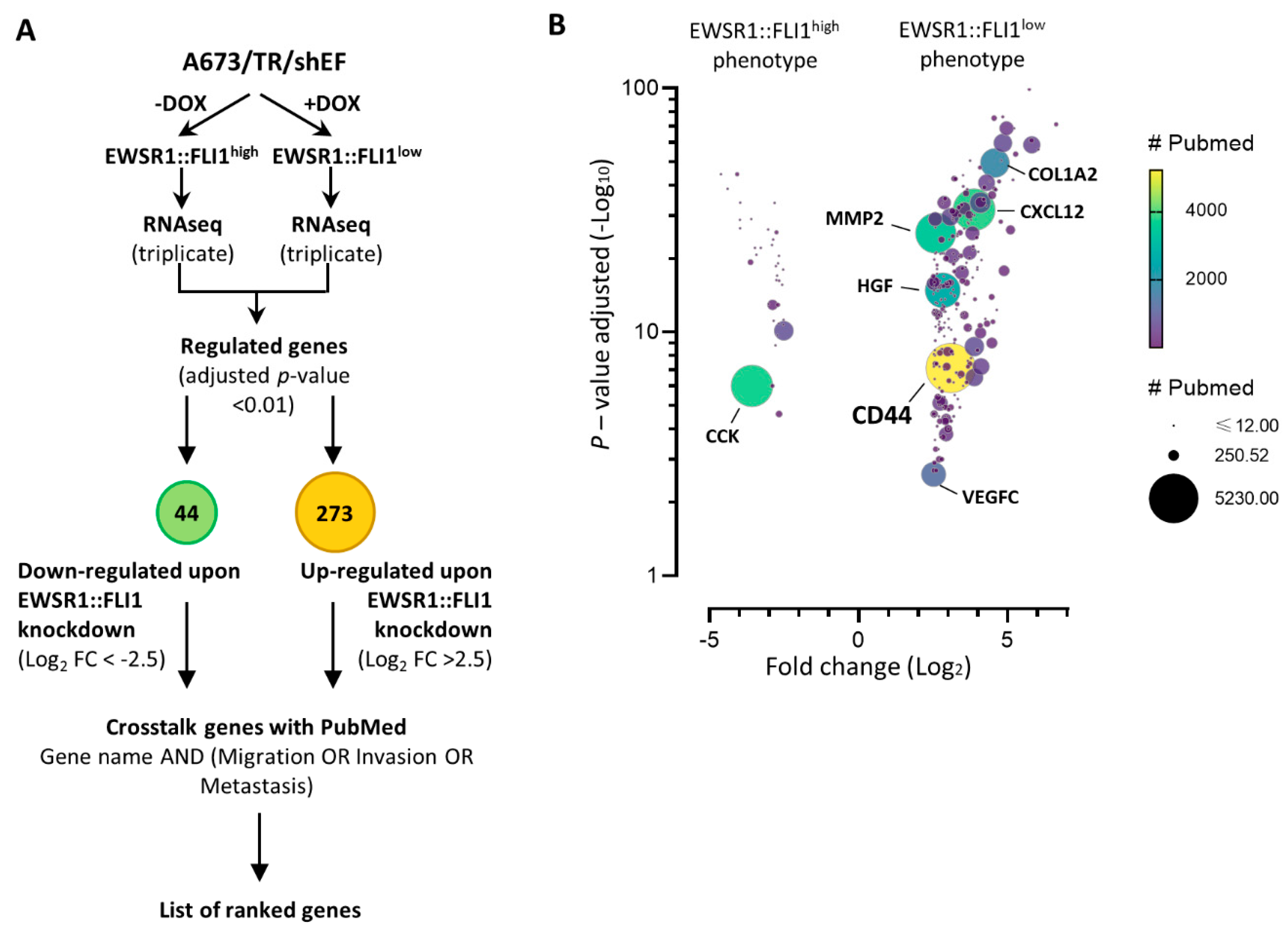
1. Introduction
2. Results
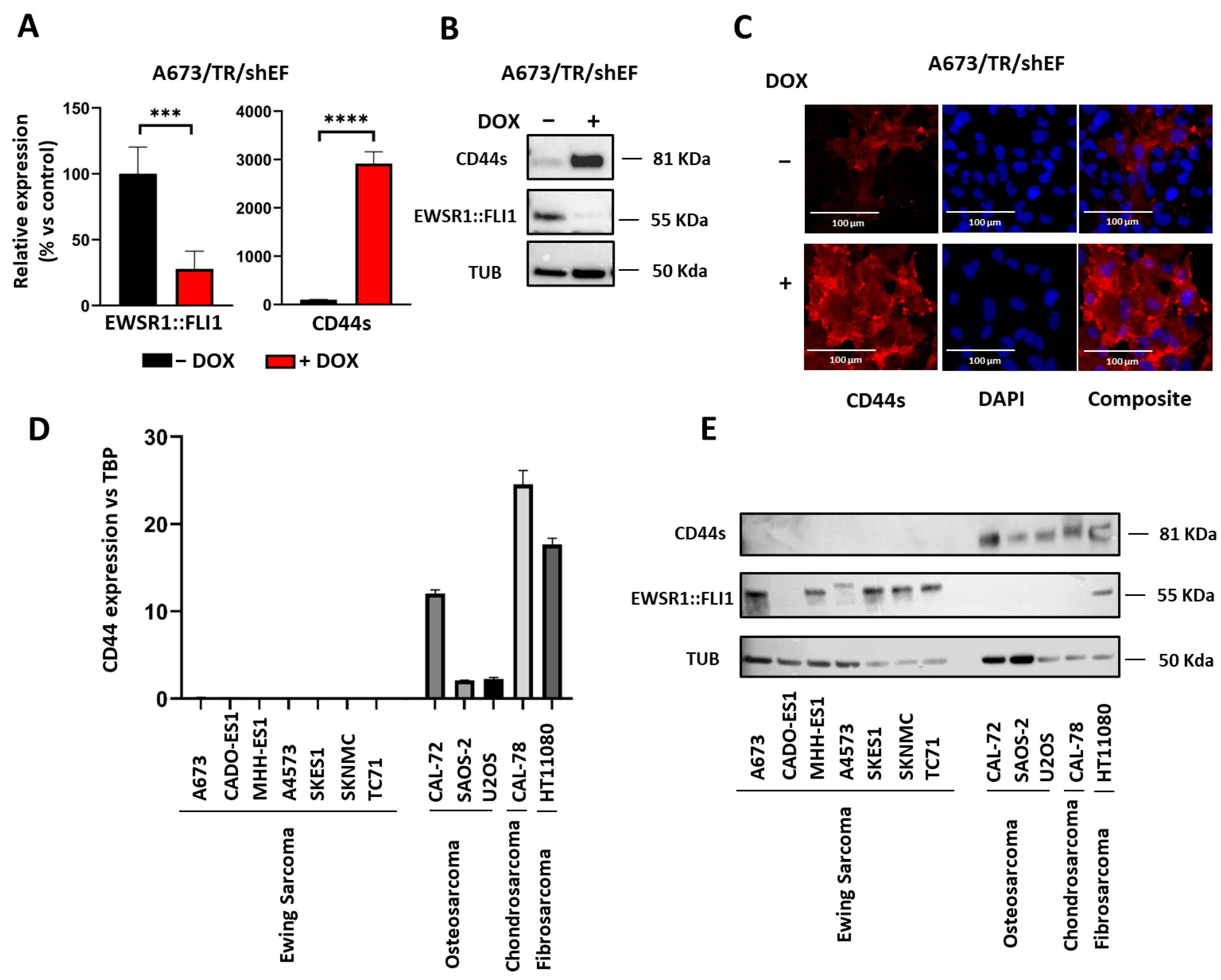
2.1. CD44s Is Upregulated upon EWSR1::FLI1 Knockdown
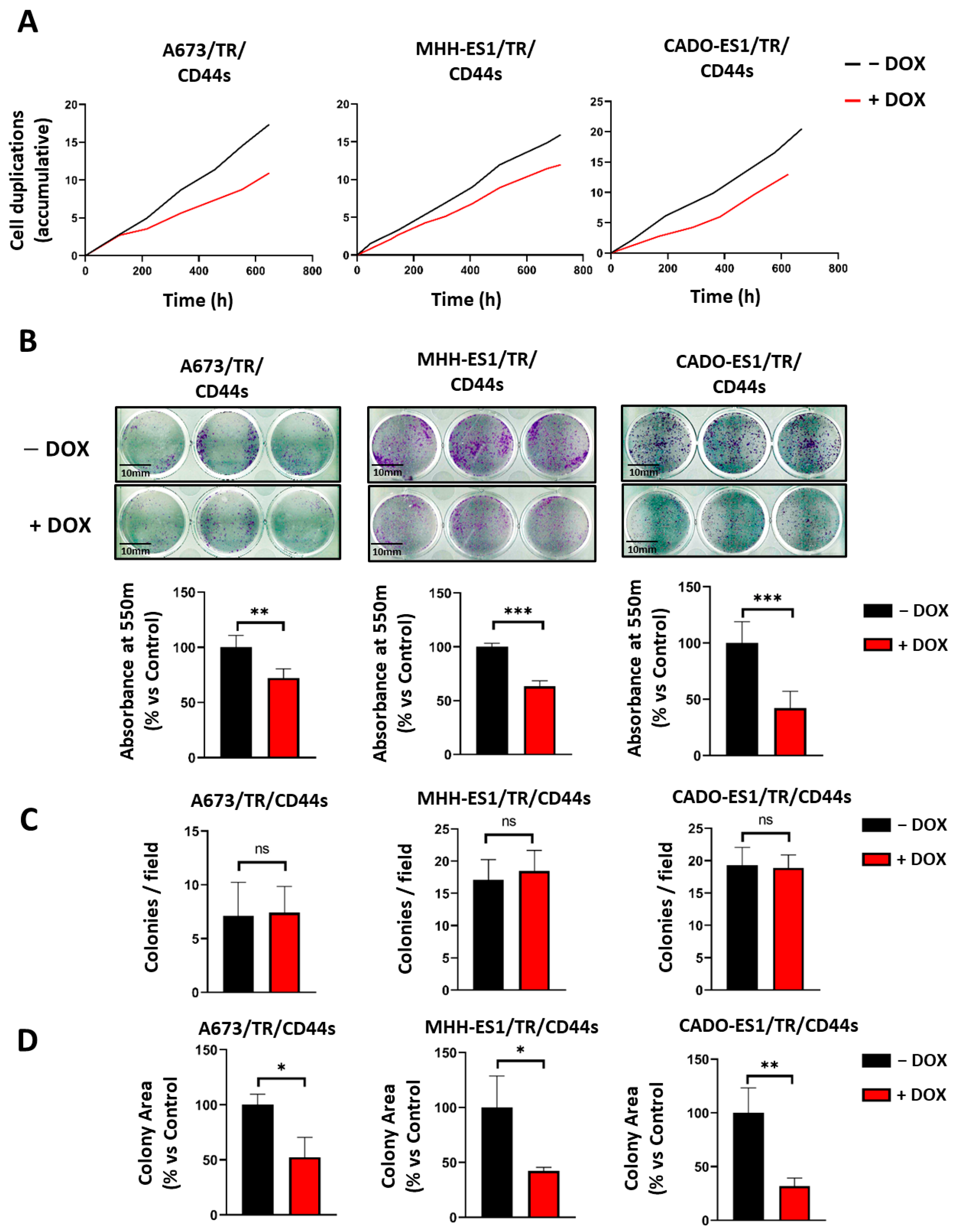
2.2. CD44s Expression Reduces Cell Proliferation without Impairing Clonogenicity
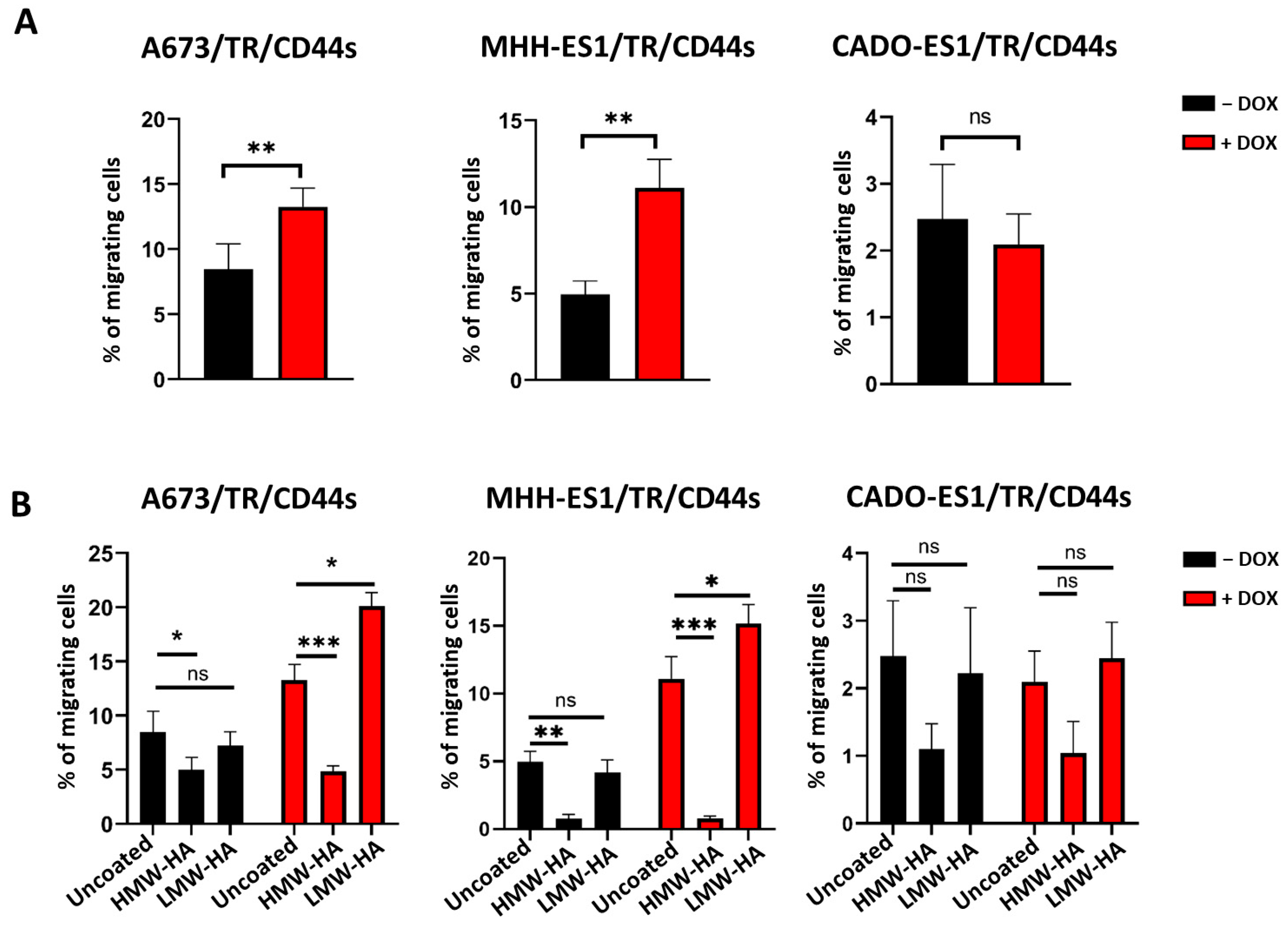
2.3. CD44s and LMW-HA Increase Cell Migration
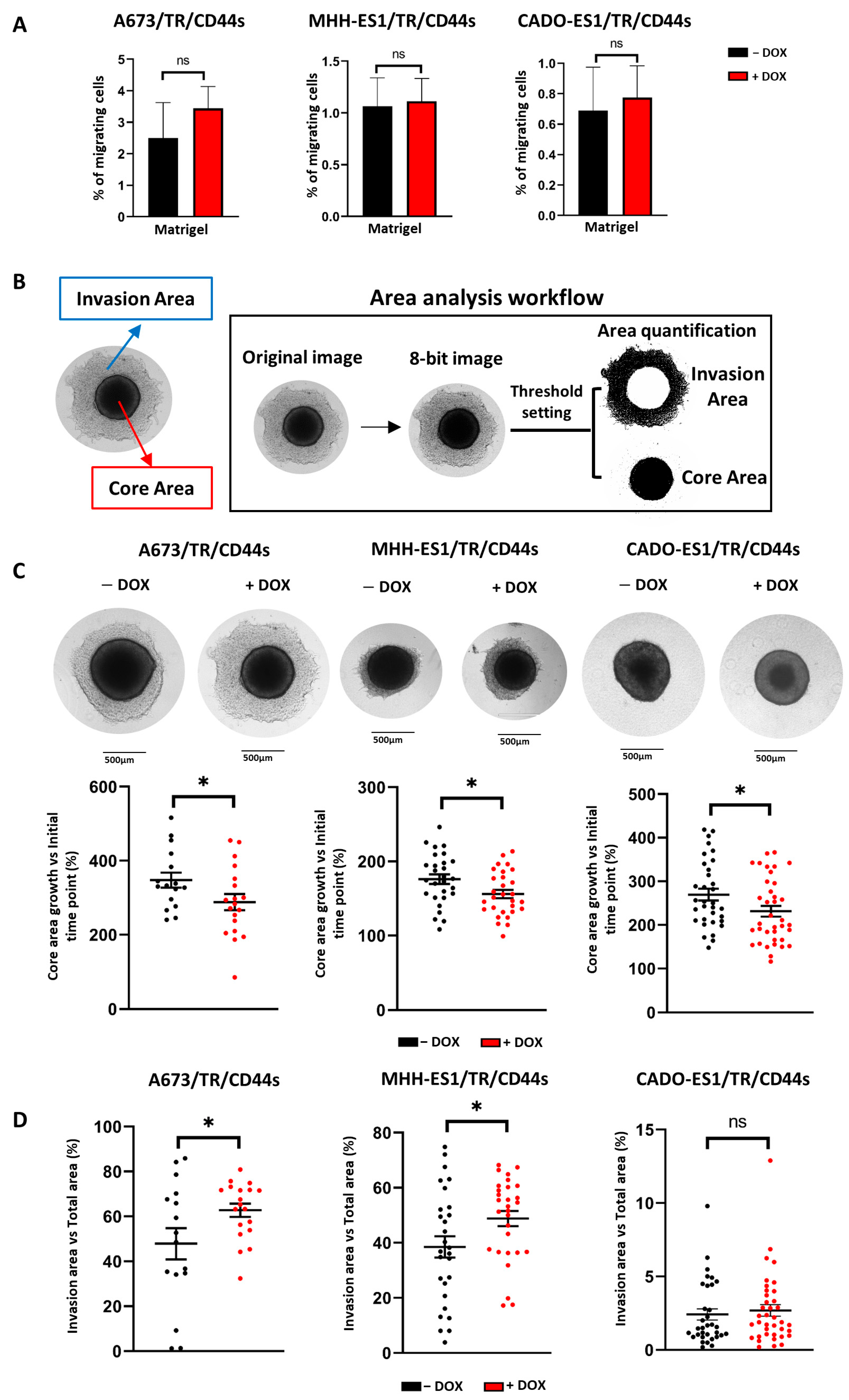
2.4. CD44s Expression Increases Cell Invasiveness
3. Discussion
4. Materials and Methods
4.1. Cell Lines
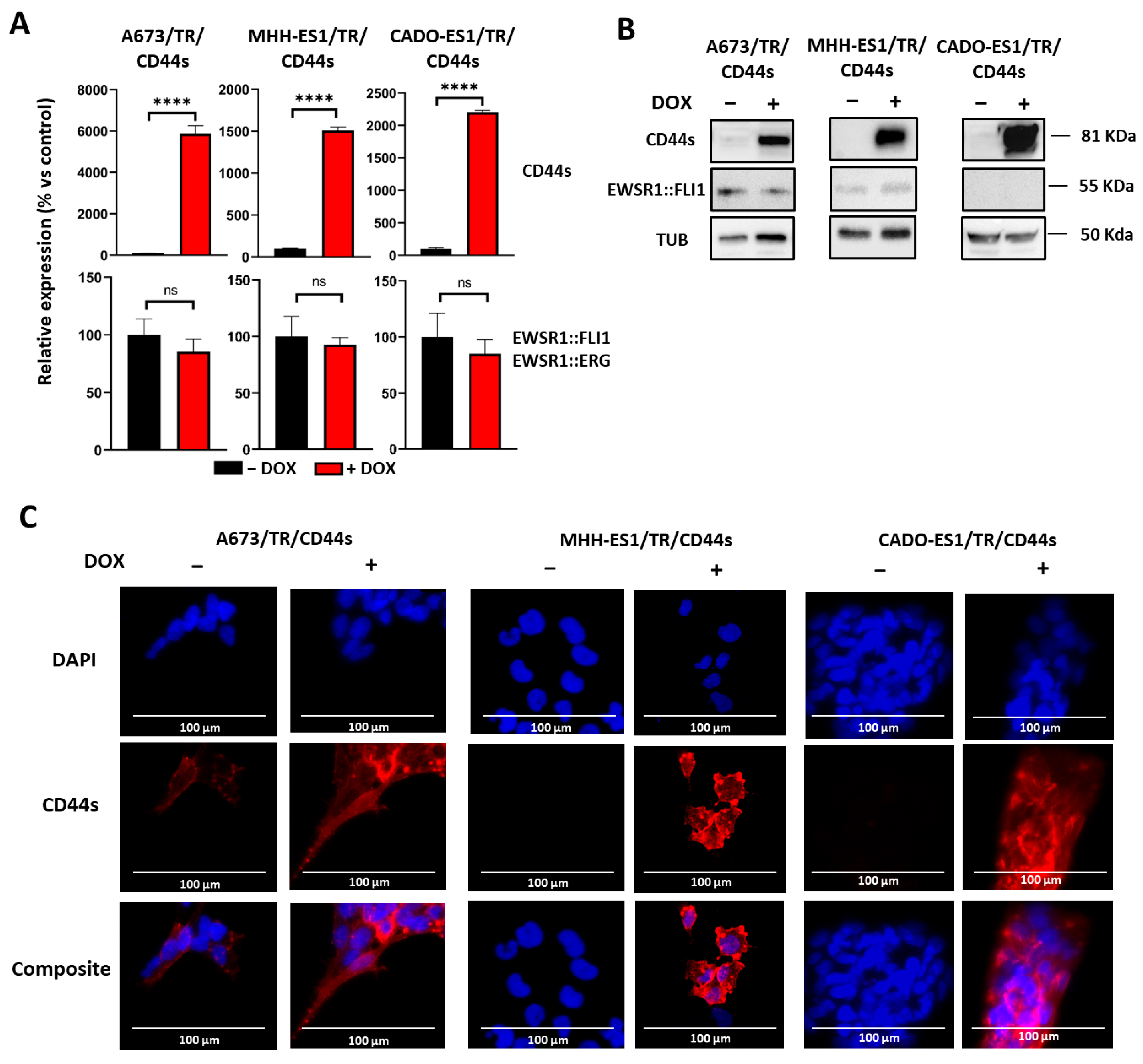
4.2. Establishment of Ewing Sarcoma Cell Lines Expressing the Doxycycline-Inducible CD44s Protein
4.3. Quantitative RT-PCR (RT-qPCR)
4.4. Western Blot Analysis and Antibodies
4.5. Immunofluorescence
4.6. Cell Proliferation Assay
4.7. Clonogenic Assay
4.8. Transwell Migration and Invasion Assay
4.9. Spheroid Invasion Assay
4.10. Transcriptomic Analysis (RNAseq)
4.11. Pubmed Terms Search
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grünewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; De Álava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Rev. Dis. Primers 2018, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Kauer, M.; Ban, J.; Kofler, R.; Walker, B.; Davis, S.; Meltzer, P.; Kovar, H. A molecular function map of Ewing’s sarcoma. PLoS ONE 2009, 4, e5415. [Google Scholar] [CrossRef]
- Riggi, N.; Knoechel, B.; Gillespie, S.M.; Rheinbay, E.; Boulay, G.; Suva, M.L.; Rossetti, N.E.; Boonseng, W.E.; Oksuz, O.; Cook, E.B.; et al. EWS-FLI1 Utilizes Divergent Chromatin Remodeling Mechanisms to Directly Activate or Repress Enhancer Elements in Ewing Sarcoma. Cancer Cell 2014, 26, 668–681. [Google Scholar] [CrossRef]
- Franzetti, G.-A.; Laud-Duval, K.; Van Der Ent, W.; Brisac, A.; Irondelle, M.; Aubert, S.; Dirksen, U.; Bouvier, C.; De Pinieux, G.; Snaar-Jagalska, E.; et al. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene 2017, 36, 3505–3514. [Google Scholar] [CrossRef]
- Aynaud, M.-M.; Mirabeau, O.; Gruel, N.; Grossetête, S.; Boeva, V.; Durand, S.; Surdez, D.; Saulnier, O.; Zaïdi, S.; Gribkova, S.; et al. Transcriptional Programs Define Intratumoral Heterogeneity of Ewing Sarcoma at Single-Cell Resolution. Cell Rep. 2020, 30, 1767–1779.e6. [Google Scholar] [CrossRef] [PubMed]
- García-García, L.; Fernández-Tabanera, E.; Cervera, S.T.; Melero-Fernández De Mera, R.M.; Josa, S.; González-González, L.; Rodríguez-Martín, C.; Grünewald, T.G.P.; Alonso, J. The Transcription Factor FEZF1, a Direct Target of EWSR1-FLI1 in Ewing Sarcoma Cells, Regulates the Expression of Neural-Specific Genes. Cancers 2021, 13, 5668. [Google Scholar] [CrossRef]
- Mendiola, M.; Carrillo, J.; García, E.; Lalli, E.; Hernández, T.; De Alava, E.; Tirode, F.; Delattre, O.; García-Miguel, P.; López-Barea, F.; et al. The orphan nuclear receptor DAX1 is up-regulated by the EWS/FLI1 oncoprotein and is highly expressed in Ewing tumors. Int. J. Cancer 2006, 118, 1381–1389. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, J.; Garcia-Aragoncillo, E.; Azorin, D.; Agra, N.; Sastre, A.; Gonzalez-Mediero, I.; Garcia-Miguel, P.; Pestana, A.; Gallego, S.; Segura, D.; et al. Cholecystokinin down-regulation by RNA interference impairs Ewing tumor growth. Clin. Cancer Res. 2007, 13, 2429–2440. [Google Scholar] [CrossRef]
- Agra, N.; Cidre, F.; Garcia-Garcia, L.; de la Parra, J.; Alonso, J. Lysyl Oxidase Is Downregulated by the EWS/FLI1 Oncoprotein and Its Propeptide Domain Displays Tumor Supressor Activities in Ewing Sarcoma Cells. PLoS ONE 2013, 8, e66281. [Google Scholar] [CrossRef]
- Cidre-Aranaz, F.; Grünewald, T.G.P.; Surdez, D.; García-García, L.; Carlos Lázaro, J.; Kirchner, T.; González-González, L.; Sastre, A.; García-Miguel, P.; López-Pérez, S.E.; et al. EWS-FLI1-mediated suppression of the RAS-antagonist Sprouty 1 (SPRY1) confers aggressiveness to Ewing sarcoma. Oncogene 2017, 36, 766–776. [Google Scholar] [CrossRef]
- Hauer, K.; Calzada-Wack, J.; Steiger, K.; Grunewald, T.G.P.; Baumhoer, D.; Plehm, S.; Buch, T.; Da Costa, O.P.; Esposito, I.; Burdach, S.; et al. DKK2 Mediates Osteolysis, Invasiveness, and Metastatic Spread in Ewing Sarcoma. Cancer Res. 2013, 73, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef]
- Fernández-Tabanera, E.; Melero-Fernández de Mera, R.M.; Alonso, J. CD44 In Sarcomas: A Comprehensive Review and Future Perspectives. Front. Oncol. 2022, 12, 909450. [Google Scholar] [CrossRef]
- Hou, W.; Kong, L.; Hou, Z.; Ji, H. CD44 is a prognostic biomarker and correlated with immune infiltrates in gastric cancer. BMC Med. Genom. 2022, 15, 225. [Google Scholar] [CrossRef]
- Hassn Mesrati, M.; Syafruddin, S.E.; Mohtar, M.A.; Syahir, A. CD44: A Multifunctional Mediator of Cancer Progression. Biomolecules 2021, 11, 1850. [Google Scholar] [CrossRef] [PubMed]
- Roosta, Y.; Sanaat, Z.; Nikanfar, A.R.; Dolatkhah, R.; Fakhrjou, A. Predictive Value of CD44 for Prognosis in Patients with Breast Cancer. Asian Pac. J. Cancer Prev. 2020, 21, 2561–2567. [Google Scholar] [CrossRef]
- Zhang, Y.; Ding, C.; Wang, J.; Sun, G.; Cao, Y.; Xu, L.; Zhou, L.; Chen, X. Prognostic significance of CD44V6 expression in osteosarcoma: A meta-analysis. J. Orthop. Surg. Res. 2015, 10, 187. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Du, Y.; Lu, Y.; Luan, B.; Xu, C.; Yu, Y.; Zhao, H. CD44 Expression Predicts Prognosis of Ovarian Cancer Patients Through Promoting Epithelial-Mesenchymal Transition (EMT) by Regulating Snail, ZEB1, and Caveolin-1. Front. Oncol. 2019, 9, 802. [Google Scholar] [CrossRef]
- Paulis, Y.W.; Huijbers, E.J.; van der Schaft, D.W.; Soetekouw, P.M.; Pauwels, P.; Tjan-Heijnen, V.C.; Griffioen, A.W. CD44 enhances tumor aggressiveness by promoting tumor cell plasticity. Oncotarget 2015, 14, 19634–19646. [Google Scholar] [CrossRef]
- Ahmed, M.A.; Aleskandarany, M.A.; Rakha, E.A.; Moustafa, R.Z.; Benhasouna, A.; Nolan, C.; Green, A.R.; Ilyas, M.; Ellis, I.O. A CD44⁻/CD24⁺ phenotype is a poor prognostic marker in early invasive breast cancer. Breast Cancer Res. Treat. 2012, 133, 979–995. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, P.P.; Sackstein, R. CD44 and HCELL: Preventing hematogenous metastasis at step 1. FEBS Lett. 2011, 585, 3148–3158. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Feng, Y.; Shen, J.K.; Lin, M.; Choy, E.; Cote, G.M.; Harmon, D.C.; Mankin, H.J.; Hornicek, F.J.; Duan, Z. CD44 is a direct target of miR-199a-3p and contributes to aggressive progression in osteosarcoma. Sci. Rep. 2015, 5, 11365. [Google Scholar] [CrossRef]
- Guo, Q.; Yang, C.; Gao, F. The state of CD44 activation in cancer progression and therapeutic targeting. FEBS J. 2021, 289, 7970–7986. [Google Scholar] [CrossRef]
- Kong, L.; Ji, H.; Gan, X.; Cao, S.; Li, Z.; Jin, Y. Knockdown of CD44 inhibits proliferation, migration and invasion of osteosarcoma cells accompanied by downregulation of cathepsin S. J. Orthop. Surg. Res. 2022, 17, 154. [Google Scholar] [CrossRef]
- Gvozdenovic, A.; Arlt, M.J.E.; Campanile, C.; Brennecke, P.; Husmann, K.; Born, W.; Muff, R.; Fuchs, B. Silencing of CD44 Gene Expression in Human 143-B Osteosarcoma Cells Promotes Metastasis of Intratibial Tumors in SCID Mice. PLoS ONE 2013, 8, e60329. [Google Scholar] [CrossRef]
- Liu, T.; Yan, Z.; Liu, Y.; Choy, E.; Hornicek, F.J.; Mankin, H.; Duan, Z. CRISPR-Cas9-Mediated Silencing of CD44 in Human Highly Metastatic Osteosarcoma Cells. Cell. Physiol. Biochem. 2018, 46, 1218–1230. [Google Scholar] [CrossRef]
- Xiao, Z.; Wan, J.; Nur, A.A.; Dou, P.; Mankin, H.; Liu, T.; Ouyang, Z. Targeting CD44 by CRISPR-Cas9 in Multi-Drug Resistant Osteosarcoma Cells. Cell. Physiol. Biochem. 2018, 51, 1879–1893. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Ma, B.; Wang, Y.; Han, T.; Zheng, L.; Sun, C.; Liu, T.; Zhang, Y.; Qiu, X.; Fan, Q. miR-34a inhibits the metastasis of osteosarcoma cells by repressing the expression of CD44. Oncol. Rep. 2013, 29, 1027–1036. [Google Scholar] [CrossRef]
- Kogerman, P.; Sy, M.-S.; Culp, L.A. Counter-selection for over-expressed human CD44s in primary tumors versus lung metastases in a mouse fibrosarcoma model. Oncogene 1997, 15, 1407–1416. [Google Scholar] [CrossRef]
- Xu, H.; Niu, M.; Yuan, X.; Wu, K.; Liu, A. CD44 as a tumor biomarker and therapeutic target. Exp. Hematol. Oncol. 2020, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Wang, Q.; Su, Q.; Huang, H.; Luan, J.; Xu, X.; Wang, J. miR-373 Inhibits Glioma Cell U251 Migration and Invasion by Down-Regulating CD44 and TGFBR2. Cell Mol. Neurobiol. 2016, 36, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.; Oh, S.; Lee, K.M.; Yoo, S.A.; Shin, I. CD44 regulates cell proliferation, migration, and invasion via modulation of c-Src transcription in human breast cancer cells. Cell Signal 2015, 27, 1882–1894. [Google Scholar] [CrossRef]
- Yu, M.; He, P.; Liu, Y.; He, Y.; Du, Y.; Wu, M.; Zhang, G.; Yang, C.; Gao, F. Hyaluroan-regulated lymphatic permeability through S1P receptors is crucial for cancer metastasis. Med. Oncol. 2015, 32, 381. [Google Scholar] [CrossRef]
- Liu, M.; Tolg, C.; Turley, E. Dissecting the Dual Nature of Hyaluronan in the Tumor Microenvironment. Front. Immunol. 2019, 10, 947. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Azpurua, J.; Hine, C.; Vaidya, A.; Myakishev-Rempel, M.; Ablaeva, J.; Mao, Z.; Nevo, E.; Gorbunova, V.; Seluanov, A. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 2013, 499, 346–349. [Google Scholar] [CrossRef]
- Wolny, P.M.; Banerji, S.; Gounou, C.; Brisson, A.R.; Day, A.J.; Jackson, D.G.; Richter, R.P. Analysis of CD44-hyaluronan interactions in an artificial membrane system: Insights into the distinct binding properties of high and low molecular weight hyaluronan. J. Biol. Chem. 2010, 285, 30170–30180. [Google Scholar] [CrossRef]
- Yang, C.; Cao, M.; Liu, H.; He, Y.; Xu, J.; Du, Y.; Liu, Y.; Wang, W.; Cui, L.; Hu, J.; et al. The high and low molecular weight forms of hyaluronan have distinct effects on CD44 clustering. J. Biol. Chem. 2012, 287, 43094–43107. [Google Scholar] [CrossRef]
- Schwertfeger, K.L.; Cowman, M.K.; Telmer, P.G.; Turley, E.A.; McCarthy, J.B. Hyaluronan, Inflammation, and Breast Cancer Progression. Front. Immunol. 2015, 6, 236. [Google Scholar] [CrossRef]
- Koyama, H.; Hibi, T.; Isogai, Z.; Yoneda, M.; Fujimori, M.; Amano, J.; Kawakubo, M.; Kannagi, R.; Kimata, K.; Taniguchi, S.I.; et al. Hyperproduction of Hyaluronan in Neu-Induced Mammary Tumor Accelerates Angiogenesis through Stromal Cell Recruitment. Am. J. Pathol. 2007, 170, 1086–1099. [Google Scholar] [CrossRef]
- Misra, S.; Heldin, P.; Hascall, V.C.; Karamanos, N.K.; Skandalis, S.S.; Markwald, R.R.; Ghatak, S. Hyaluronan-CD44 interactions as potential targets for cancer therapy. FEBS J. 2011, 278, 1429–1443. [Google Scholar] [CrossRef]
- Avigdor, A.; Goichberg, P.; Shivtiel, S.; Dar, A.; Peled, A.; Samira, S.; Kollet, O.; Hershkoviz, R.; Alon, R.; Hardan, I.; et al. CD44 and hyaluronic acid cooperate with SDF-1 in the trafficking of human CD34+ stem/progenitor cells to bone marrow. Blood 2004, 103, 2981–2989. [Google Scholar] [CrossRef] [PubMed]
- Khaldoyanidi, S.; Denzel, A.; Zöller, M. Requirement for CD44 in proliferation and homing of hematopoietic precursor cells. J. Leukoc. Biol. 1996, 60, 579–592. [Google Scholar] [CrossRef]
- Schwankhaus, N.; Gathmann, C.; Wicklein, D.; Riecken, K.; Schumacher, U.; Valentiner, U. Cell adhesion molecules in metastatic neuroblastoma models. Clin. Exp. Metastasis 2014, 31, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Valentiner, U.; Valentiner, F.U.; Schumacher, U. Expression of CD44 is associated with a metastatic pattern of human neuroblastoma cells in a SCID mouse xenograft model. Tumour Biol. 2008, 29, 152–160. [Google Scholar] [CrossRef]
- Richter, U.; Wicklein, D.; Geleff, S.; Schumacher, U. The interaction between CD44 on tumour cells and hyaluronan under physiologic flow conditions: Implications for metastasis formation. Histochem. Cell Biol. 2012, 137, 687–695. [Google Scholar] [CrossRef]
- Benien, P.; Swami, A. 3D tumor models: History, advances and future perspectives. Future Oncol. 2014, 10, 1311–1327. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Yoon, C.; Zhou, X.-H.; Zhou, Y.-C.; Zhou, W.-W.; Liu, H.; Yang, X.; Lu, J.; Lee, S.Y.; Huang, K. ERK1/2-Nanog signaling pathway enhances CD44(+) cancer stem-like cell phenotypes and epithelial-to-mesenchymal transition in head and neck squamous cell carcinomas. Cell Death Dis. 2020, 11, 266. [Google Scholar] [CrossRef]
- Yan, Y.; Zuo, X.; Wei, D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Trans. Med. 2015, 4, 1033–1043. [Google Scholar] [CrossRef]
- Tsukita, S.; Oishi, K.; Sato, N.; Sagara, J.; Kawai, A.; Tsukita, S. ERM Family Members as Molecular Linkers Between the Cell Surface Glycoprotein CD44 and Actin-Based Cytoskeletons. J. Cell Biol. 1994, 126, 391–401. [Google Scholar] [CrossRef]
- Matsui, Y.; Chansky, H.A.; Barahmand-Pour, F.; Zielinska-Kwiatkowska, A.; Tsumaki, N.; Myoui, A.; Yoshikawa, H.; Yang, L.; Eyre, D.R. COL11A2 Collagen Gene Transcription Is Differentially Regulated by EWS/ERG Sarcoma Fusion Protein and Wild-type ERG. J. Biol. Chem. 2003, 278, 11369–11375. [Google Scholar] [CrossRef] [PubMed]
- Cantile, M.; Marra, L.; Franco, R.; Ascierto, P.; Liguori, G.; De Chiara, A.; Botti, G. Molecular detection and targeting of EWSR1 fusion transcripts in soft tissue tumors. Med. Oncol. 2013, 30, 412. [Google Scholar] [CrossRef] [PubMed]
- Kodama, K.; Doi, O.; Higashiyama, M.; Yokouchi, H.; Tateishi, R.; Mori, Y. Differentiation of a Ewing’s Sarcoma Cell Line towards Neural and Mesenchymal Cell Lineages. Jpn. J. Cancer Res. 1994, 85, 335–338. [Google Scholar] [CrossRef]
- Hotfilder, M.; Mallela, N.; Seggewiß, J.; Dirksen, U.; Korsching, E. Defining a Characteristic Gene Expression Set Responsible for Cancer Stem Cell-Like Features in a Sub-Population of Ewing Sarcoma Cells CADO-ES1. Int. J. Mol. Sci. 2018, 19, 3908. [Google Scholar] [CrossRef] [PubMed]
- Kodama, K.; Doi, O.; Higashiyama, M.; Mori, Y.; Horai, T.; Tateishi, R.; Aoki, Y.; Misawa, S. Establishment and characterization of a new Ewing’s sarcoma cell line. Cancer Genet. Cytogenet. 1991, 57, 19–30. [Google Scholar] [CrossRef]
- Korsching, E.; Matschke, J.; Hotfilder, M. Splice variants denote differences between a cancer stem cell side population of EWSR1-ERG-based Ewing sarcoma cells, its main population and EWSR1-FLI-based cells. Int. J. Mol. Med. 2022, 49, 39. [Google Scholar] [CrossRef]
- Guzmán, C.; Bagga, M.; Kaur, A.; Westermarck, J.; Abankwa, D. ColonyArea: An ImageJ Plugin to Automatically Quantify Colony Formation in Clonogenic Assays. PLoS ONE 2014, 9, e92444. [Google Scholar] [CrossRef]
- Montagnac, G.; Meas-Yedid, V.; Irondelle, M.; Castro-Castro, A.; Franco, M.; Shida, T.; Nachury, M.V.; Benmerah, A.; Olivo-Marin, J.-C.; Chavrier, P. αTAT1 catalyses microtubule acetylation at clathrin-coated pits. Nature 2013, 502, 567–570. [Google Scholar] [CrossRef]
- Kelm, J.M.; Timmins, N.E.; Brown, C.J.; Fussenegger, M.; Nielsen, L.K. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 2003, 83, 173–180. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Tabanera, E.; García-García, L.; Rodríguez-Martín, C.; Cervera, S.T.; González-González, L.; Robledo, C.; Josa, S.; Martínez, S.; Chapado, L.; Monzón, S.; et al. CD44 Modulates Cell Migration and Invasion in Ewing Sarcoma Cells. Int. J. Mol. Sci. 2023, 24, 11774. https://doi.org/10.3390/ijms241411774
Fernández-Tabanera E, García-García L, Rodríguez-Martín C, Cervera ST, González-González L, Robledo C, Josa S, Martínez S, Chapado L, Monzón S, et al. CD44 Modulates Cell Migration and Invasion in Ewing Sarcoma Cells. International Journal of Molecular Sciences. 2023; 24(14):11774. https://doi.org/10.3390/ijms241411774
Chicago/Turabian StyleFernández-Tabanera, Enrique, Laura García-García, Carlos Rodríguez-Martín, Saint T. Cervera, Laura González-González, Cristina Robledo, Santiago Josa, Selene Martínez, Luis Chapado, Sara Monzón, and et al. 2023. "CD44 Modulates Cell Migration and Invasion in Ewing Sarcoma Cells" International Journal of Molecular Sciences 24, no. 14: 11774. https://doi.org/10.3390/ijms241411774
APA StyleFernández-Tabanera, E., García-García, L., Rodríguez-Martín, C., Cervera, S. T., González-González, L., Robledo, C., Josa, S., Martínez, S., Chapado, L., Monzón, S., Melero-Fernández de Mera, R. M., & Alonso, J. (2023). CD44 Modulates Cell Migration and Invasion in Ewing Sarcoma Cells. International Journal of Molecular Sciences, 24(14), 11774. https://doi.org/10.3390/ijms241411774





