Understanding the Molecular Basis of the Multiple Mitochondrial Dysfunctions Syndrome 2: The Disease-Causing His96Arg Mutation of BOLA3
Abstract
1. Introduction
2. Results
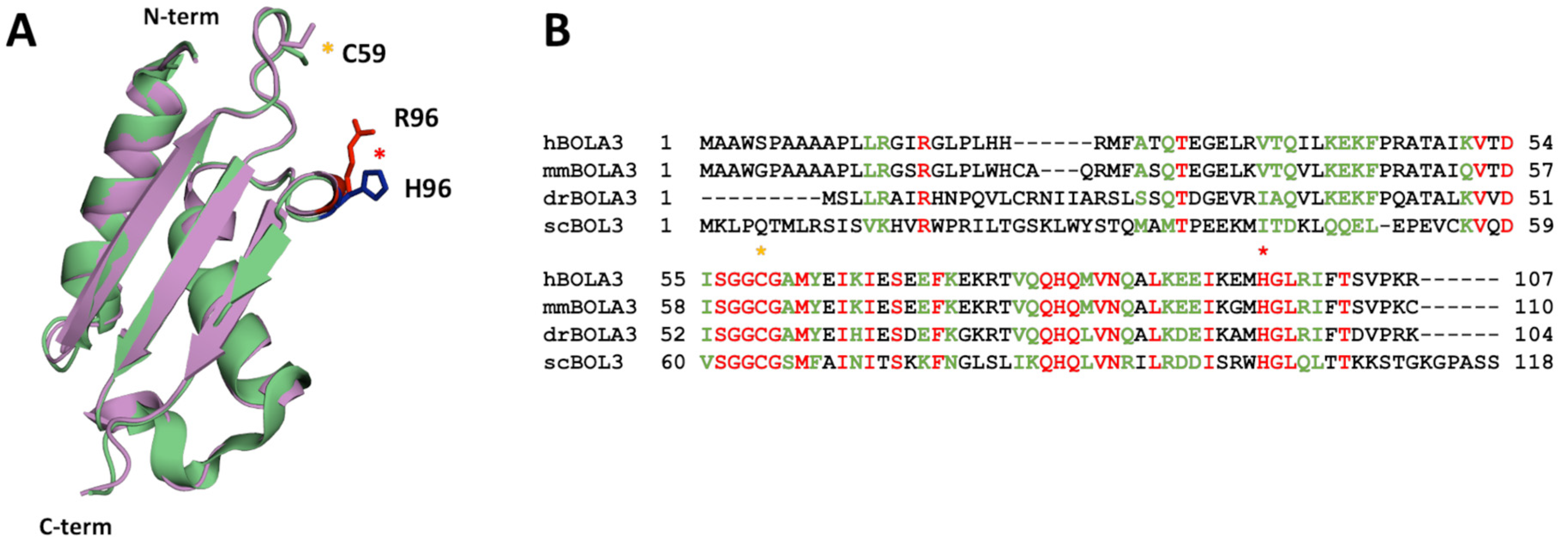
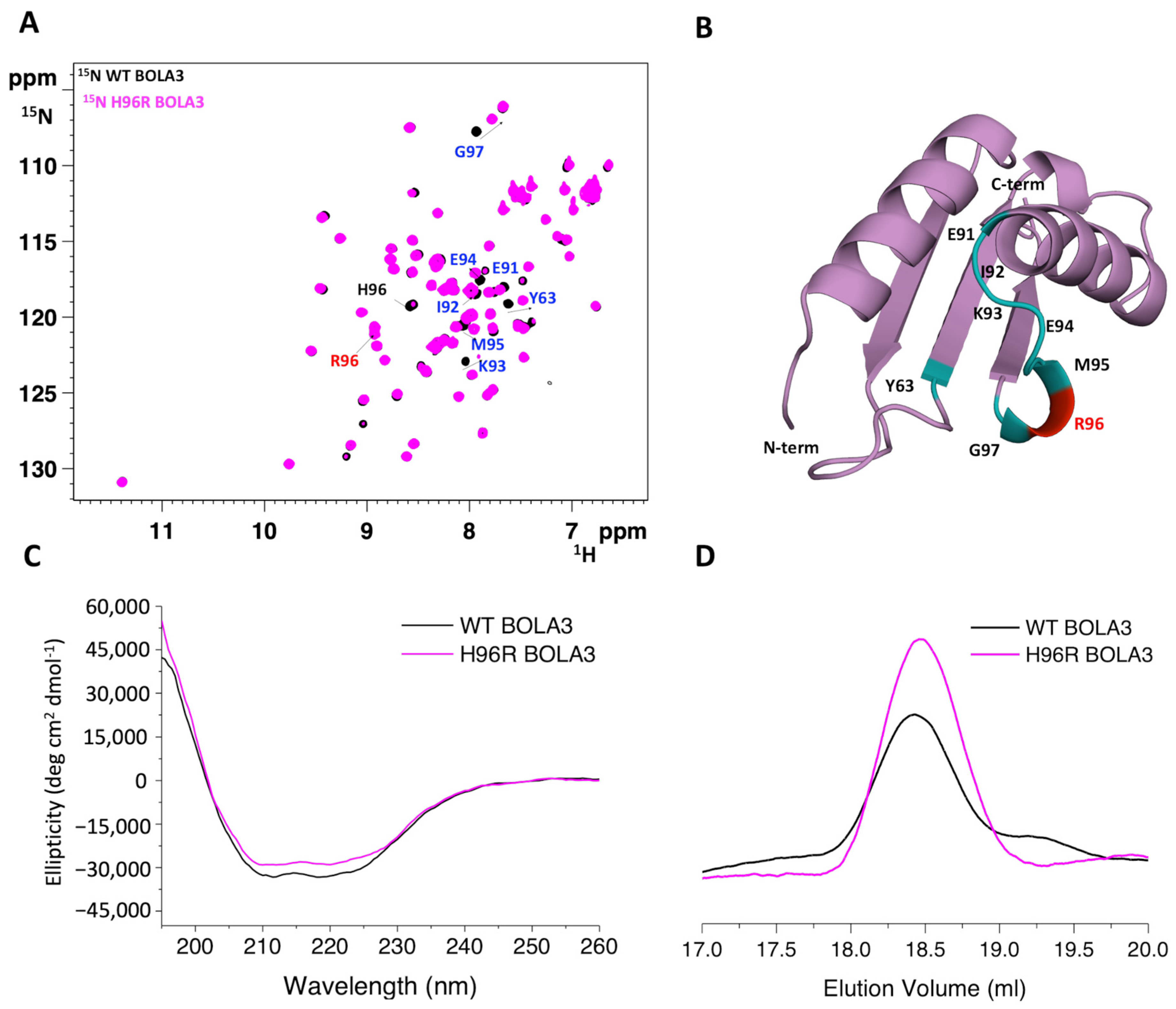
2.1. The His96Arg Substitution Does Not Affect the Structure of BOLA3
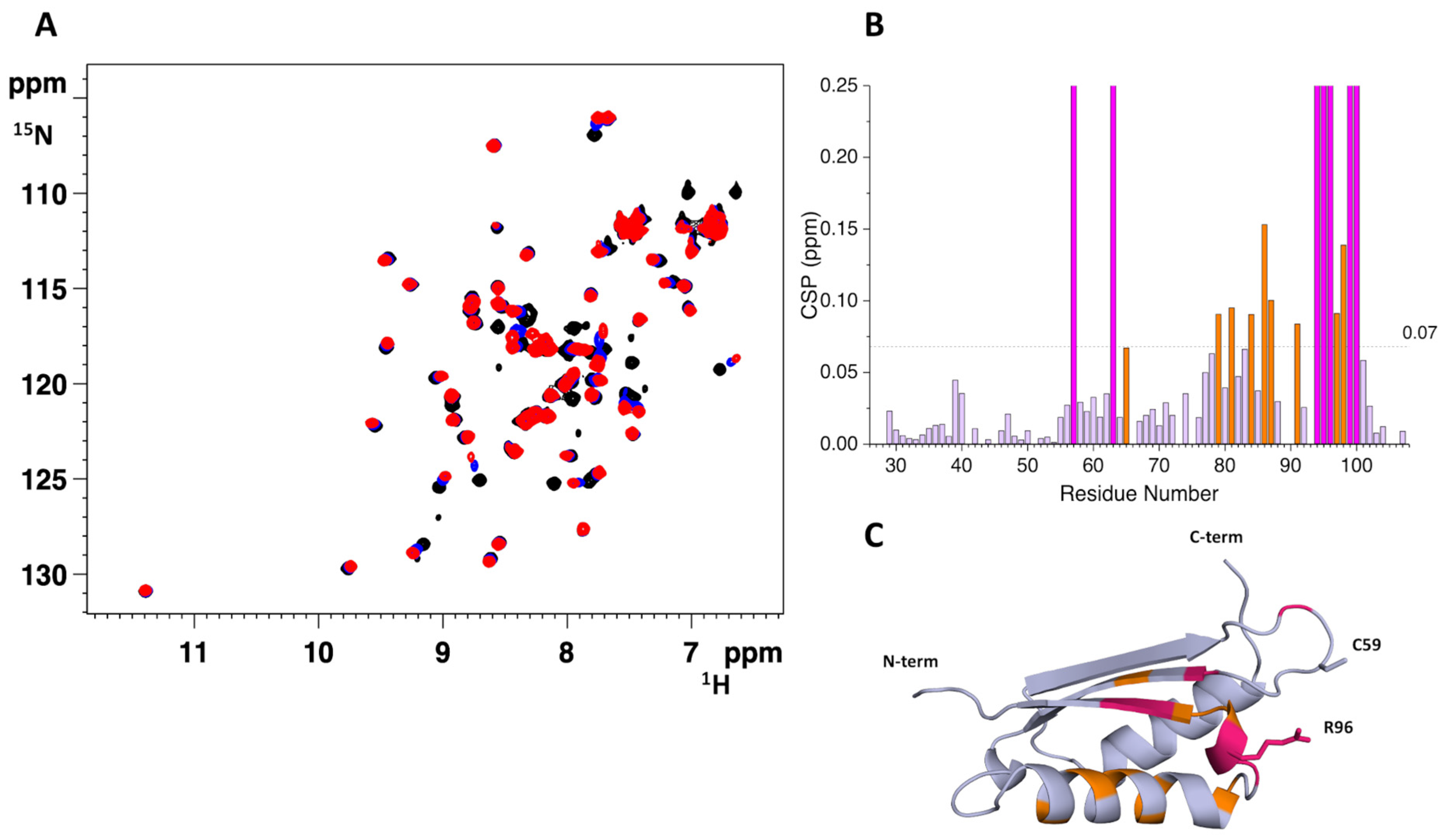
2.2. The His96Arg Substitution Does Not Impair the Interaction between BOLA3 and GLRX5
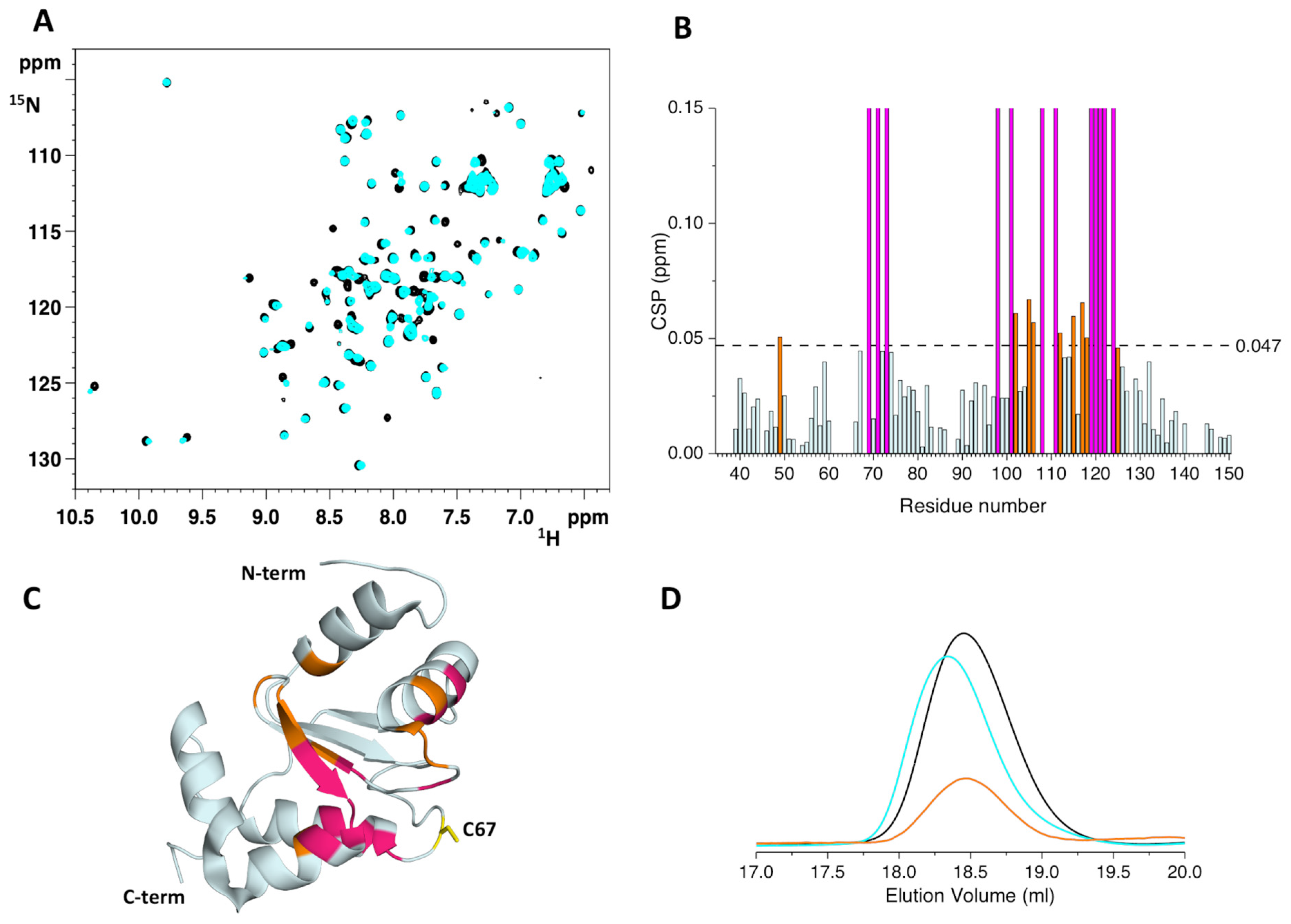
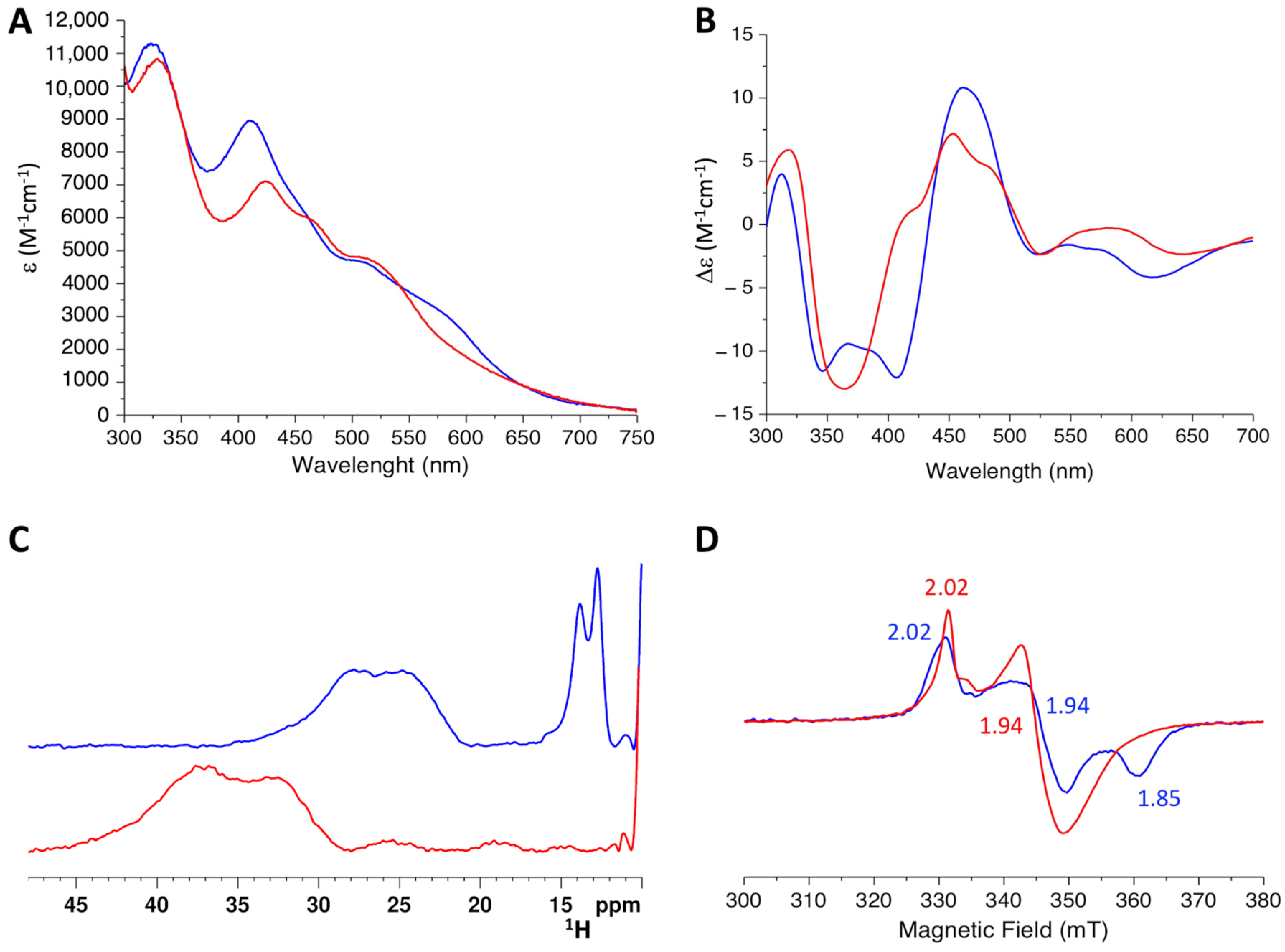
2.3. The His96Arg Substitution Does Not Impair the Formation of the BOLA3-[2Fe-2S]-GLRX5 Heterodimeric Complex
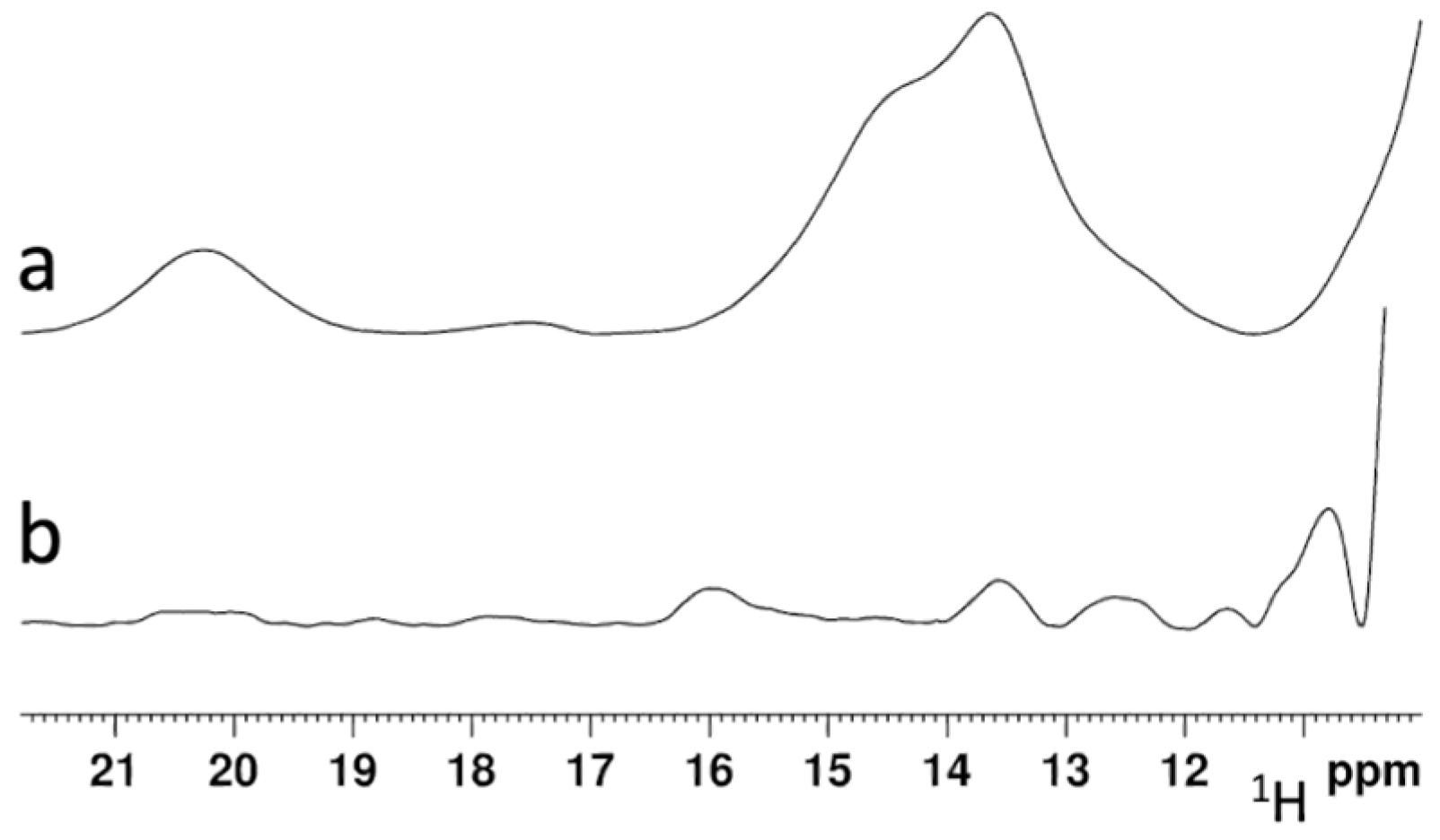
2.4. His96Arg Mutation Impairs the [2Fe-2S] Cluster Chaperone Activity of the BOLA3-[2Fe-2S]-GLRX5 Complex
3. Discussion
4. Materials and Methods
4.1. Proteins Expression and Purification
4.2. In Vitro Chemical Reconstitution of Apo Complex
4.3. Biochemical and Spectroscopic UV-Vis, NMR and EPR Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Friedman, J.R.; Nunnari, J. Mitochondrial Form and Function. Nature 2014, 505, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Johannsen, D.L.; Ravussin, E. The Role of Mitochondria in Health and Disease. Curr. Opin. Pharmacol. 2009, 9, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Beinert, H.; Holm, R.H.; Münck, E. Iron-Sulfur Clusters: Nature’s Modular, Multipurpose Structures. Science 1997, 277, 653–659. [Google Scholar] [CrossRef]
- Lill, R. Function and Biogenesis of Iron-Sulphur Proteins. Nature 2009, 460, 831–838. [Google Scholar] [CrossRef]
- Rouault, T.A. The Indispensable Role of Mammalian Iron Sulfur Proteins in Function and Regulation of Multiple Diverse Metabolic Pathways. Biometals 2019, 32, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Ciofi-Baffoni, S.; Nasta, V.; Banci, L. Protein Networks in the Maturation of Human Iron–Sulfur Proteins. Metallomics 2018, 10, 49–72. [Google Scholar] [CrossRef]
- Lill, R.; Dutkiewicz, R.; Freibert, S.A.; Heidenreich, T.; Mascarenhas, J.; Netz, D.J.; Paul, V.D.; Pierik, A.J.; Richter, N.; Stümpfig, M.; et al. The Role of Mitochondria and the CIA Machinery in the Maturation of Cytosolic and Nuclear Iron-Sulfur Proteins. Eur. J. Cell Biol. 2015, 94, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Camponeschi, F.; Banci, L. Metal Cofactors Trafficking and Assembly in the Cell: A Molecular View. Pure Appl. Chem. 2019, 91, 231–245. [Google Scholar] [CrossRef]
- Lill, R.; Hoffmann, B.; Molik, S.; Pierik, A.J.; Rietzschel, N.; Stehling, O.; Uzarska, M.A.; Webert, H.; Wilbrecht, C.; Mühlenhoff, U. The Role of Mitochondria in Cellular Iron-Sulfur Protein Biogenesis and Iron Metabolism. Biochim. Biophys. Acta 2012, 1823, 1491–1508. [Google Scholar] [CrossRef]
- Lill, R.; Freibert, S.-A. Mechanisms of Mitochondrial Iron-Sulfur Protein Biogenesis. Annu. Rev. Biochem. 2020, 89, 471–499. [Google Scholar] [CrossRef]
- Lebigot, E.; Schiff, M.; Golinelli-Cohen, M.-P. A Review of Multiple Mitochondrial Dysfunction Syndromes, Syndromes Associated with Defective Fe-S Protein Maturation. Biomedicines 2021, 9, 989. [Google Scholar] [CrossRef]
- Stehling, O.; Wilbrecht, C.; Lill, R. Mitochondrial Iron-Sulfur Protein Biogenesis and Human Disease. Biochimie 2014, 100, 61–77. [Google Scholar] [CrossRef]
- Maio, N.; Rouault, T.A. Mammalian Iron Sulfur Cluster Biogenesis and Human Diseases. IUBMB Life 2022, 74, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Camponeschi, F.; Ciofi-Baffoni, S.; Calderone, V.; Banci, L. Molecular Basis of Rare Diseases Associated to the Maturation of Mitochondrial [4Fe-4S]-Containing Proteins. Biomolecules 2022, 12, 1009. [Google Scholar] [CrossRef] [PubMed]
- Wachnowsky, C.; Fidai, I.; Cowan, J.A. Iron–Sulfur Cluster Biosynthesis and Trafficking—Impact on Human Disease Conditions. Metallomics 2018, 10, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Mayr, J.A.; Feichtinger, R.G.; Tort, F.; Ribes, A.; Sperl, W. Lipoic Acid Biosynthesis Defects. J. Inherit. Metab. Dis. 2014, 37, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Melber, A.; Na, U.; Vashisht, A.; Weiler, B.D.; Lill, R.; Wohlschlegel, J.A.; Winge, D.R. Role of Nfu1 and Bol3 in Iron-Sulfur Cluster Transfer to Mitochondrial Clients. eLife 2016, 5, e15991. [Google Scholar] [CrossRef] [PubMed]
- Uzarska, M.A.; Nasta, V.; Weiler, B.D.; Spantgar, F.; Ciofi-Baffoni, S.; Saviello, M.R.; Gonnelli, L.; Mühlenhoff, U.; Banci, L.; Lill, R. Mitochondrial Bol1 and Bol3 Function as Assembly Factors for Specific Iron-Sulfur Proteins. eLife 2016, 5, e16673. [Google Scholar] [CrossRef] [PubMed]
- Nasta, V.; Giachetti, A.; Ciofi-Baffoni, S.; Banci, L. Structural Insights into the Molecular Function of Human [2Fe-2S] BOLA1-GRX5 and [2Fe-2S] BOLA3-GRX5 Complexes. Biochim. Biophys. Acta 2017, 1861, 2119–2131. [Google Scholar] [CrossRef]
- Sen, S.; Rao, B.; Wachnowsky, C.; Cowan, J.A. Cluster Exchange Reactivity of [2Fe-2S] Cluster-Bridged Complexes of BOLA3 with Monothiol Glutaredoxins. Metallomics 2018, 10, 1282–1290. [Google Scholar] [CrossRef]
- Nasta, V.; Suraci, D.; Gourdoupis, S.; Ciofi-Baffoni, S.; Banci, L. A Pathway for Assembling [4Fe-4S]2+ Clusters in Mitochondrial Iron-Sulfur Protein Biogenesis. FEBS J. 2020, 287, 2312–2327. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Singh, A.; Maio, N.; Rouault, T.A. Assembly of the [4Fe-4S] Cluster of NFU1 Requires the Coordinated Donation of Two [2Fe-2S] Clusters from the Scaffold Proteins, ISCU2 and ISCA1. Hum. Mol. Genet. 2020, 29, 3165–3182. [Google Scholar] [CrossRef] [PubMed]
- Kohda, M.; Tokuzawa, Y.; Kishita, Y.; Nyuzuki, H.; Moriyama, Y.; Mizuno, Y.; Hirata, T.; Yatsuka, Y.; Yamashita-Sugahara, Y.; Nakachi, Y.; et al. A Comprehensive Genomic Analysis Reveals the Genetic Landscape of Mitochondrial Respiratory Chain Complex Deficiencies. PLoS Genet. 2016, 12, e1005679. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, M.; Inaba, Y.; Motobayashi, M.; Hara, Y.; Numata, R.; Amano, Y.; Shingu, K.; Yamamoto, Y.; Murayama, K.; Ohtake, A.; et al. An Infant Case of Diffuse Cerebrospinal Lesions and Cardiomyopathy Caused by a BOLA3 Mutation. Brain Dev. 2018, 40, 484–488. [Google Scholar] [CrossRef]
- Imai-Okazaki, A.; Kishita, Y.; Kohda, M.; Mizuno, Y.; Fushimi, T.; Matsunaga, A.; Yatsuka, Y.; Hirata, T.; Harashima, H.; Takeda, A.; et al. Cardiomyopathy in Children with Mitochondrial Disease: Prognosis and Genetic Background. Int. J. Cardiol. 2019, 279, 115–121. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and Sequence Analysis Tools Services from EMBL-EBI in 2022. Nucleic Acids Res 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Banci, L.; Brancaccio, D.; Ciofi-Baffoni, S.; Del Conte, R.; Gadepalli, R.; Mikolajczyk, M.; Neri, S.; Piccioli, M.; Winkelmann, J. [2Fe-2S] Cluster Transfer in Iron-Sulfur Protein Biogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 6203–6208. [Google Scholar] [CrossRef]
- Saudino, G.; Suraci, D.; Nasta, V.; Ciofi-Baffoni, S.; Banci, L. Molecular Basis of Multiple Mitochondrial Dysfunctions Syndrome 2 Caused by CYS59TYR BOLA3 Mutation. Int. J. Mol. Sci. 2021, 22, 4848. [Google Scholar] [CrossRef]
- Banci, L.; Camponeschi, F.; Ciofi-Baffoni, S.; Muzzioli, R. Elucidating the Molecular Function of Human BOLA2 in GRX3-Dependent Anamorsin Maturation Pathway. J. Am. Chem. Soc. 2015, 137, 16133–16143. [Google Scholar] [CrossRef]
- Li, H.; Mapolelo, D.T.; Randeniya, S.; Johnson, M.K.; Outten, C.E. Human Glutaredoxin 3 Forms [2Fe-2S]-Bridged Complexes with Human BolA2. Biochemistry 2012, 51, 1687–1696. [Google Scholar] [CrossRef]
- Camponeschi, F.; Piccioli, M.; Banci, L. The Intriguing mitoNEET: Functional and Spectroscopic Properties of a Unique [2Fe-2S] Cluster Coordination Geometry. Molecules 2022, 27, 8218. [Google Scholar] [CrossRef] [PubMed]
- Noodleman, L.; Baerends, E.J. Electronic Structure, Magnetic Properties, ESR, and Optical Spectra for 2-Iron Ferredoxin Models by LCAO-X.Alpha. Valence Bond Theory. J. Am. Chem. Soc. 1984, 106, 2316–2327. [Google Scholar] [CrossRef]
- Banci, L.; Camponeschi, F.; Ciofi-Baffoni, S.; Piccioli, M. The NMR Contribution to Protein-Protein Networking in Fe-S Protein Maturation. J. Biol. Inorg. Chem. JBIC A Publ. Soc. Biol. Inorg. Chem. 2018, 23, 665–685. [Google Scholar] [CrossRef] [PubMed]
- Cameron, J.M.; Janer, A.; Levandovskiy, V.; Mackay, N.; Rouault, T.A.; Tong, W.-H.; Ogilvie, I.; Shoubridge, E.A.; Robinson, B.H. Mutations in Iron-Sulfur Cluster Scaffold Genes NFU1 and BOLA3 Cause a Fatal Deficiency of Multiple Respiratory Chain and 2-Oxoacid Dehydrogenase Enzymes. Am. J. Hum. Genet. 2011, 89, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.R.; Friederich, M.W.; Swanson, M.A.; Shaikh, T.; Bhattacharya, K.; Scharer, G.H.; Aicher, J.; Creadon-Swindell, G.; Geiger, E.; MacLean, K.N.; et al. Variant Non Ketotic Hyperglycinemia Is Caused by Mutations in LIAS, BOLA3 and the Novel Gene GLRX5. Brain 2014, 137, 366–379. [Google Scholar] [CrossRef]
- Stutterd, C.A.; Lake, N.J.; Peters, H.; Lockhart, P.J.; Taft, R.J.; van der Knaap, M.S.; Vanderver, A.; Thorburn, D.R.; Simons, C.; Leventer, R.J. Severe Leukoencephalopathy with Clinical Recovery Caused by Recessive BOLA3 Mutations. JIMD Rep. 2018, 43, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Braymer, J.J.; Freibert, S.A.; Rakwalska-Bange, M.; Lill, R. Mechanistic Concepts of Iron-Sulfur Protein Biogenesis in Biology. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118863. [Google Scholar] [CrossRef]
- Russell, O.M.; Gorman, G.S.; Lightowlers, R.N.; Turnbull, D.M. Mitochondrial Diseases: Hope for the Future. Cell 2020, 181, 168–188. [Google Scholar] [CrossRef]
- Sali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Patt, S.L.; Sykes, B.D. Water Eliminated Fourier Transform NMR Spectroscopy. J. Chem. Phys. 1972, 56, 3182–3184. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using Circular Dichroism Collected as a Function of Temperature to Determine the Thermodynamics of Protein Unfolding and Binding Interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bargagna, B.; Banci, L.; Camponeschi, F. Understanding the Molecular Basis of the Multiple Mitochondrial Dysfunctions Syndrome 2: The Disease-Causing His96Arg Mutation of BOLA3. Int. J. Mol. Sci. 2023, 24, 11734. https://doi.org/10.3390/ijms241411734
Bargagna B, Banci L, Camponeschi F. Understanding the Molecular Basis of the Multiple Mitochondrial Dysfunctions Syndrome 2: The Disease-Causing His96Arg Mutation of BOLA3. International Journal of Molecular Sciences. 2023; 24(14):11734. https://doi.org/10.3390/ijms241411734
Chicago/Turabian StyleBargagna, Beatrice, Lucia Banci, and Francesca Camponeschi. 2023. "Understanding the Molecular Basis of the Multiple Mitochondrial Dysfunctions Syndrome 2: The Disease-Causing His96Arg Mutation of BOLA3" International Journal of Molecular Sciences 24, no. 14: 11734. https://doi.org/10.3390/ijms241411734
APA StyleBargagna, B., Banci, L., & Camponeschi, F. (2023). Understanding the Molecular Basis of the Multiple Mitochondrial Dysfunctions Syndrome 2: The Disease-Causing His96Arg Mutation of BOLA3. International Journal of Molecular Sciences, 24(14), 11734. https://doi.org/10.3390/ijms241411734





