Heterodimers Are an Integral Component of Chemokine Signaling Repertoire
Abstract
1. Introduction
2. Biophysical Basis for Chemokine Heterodimerization
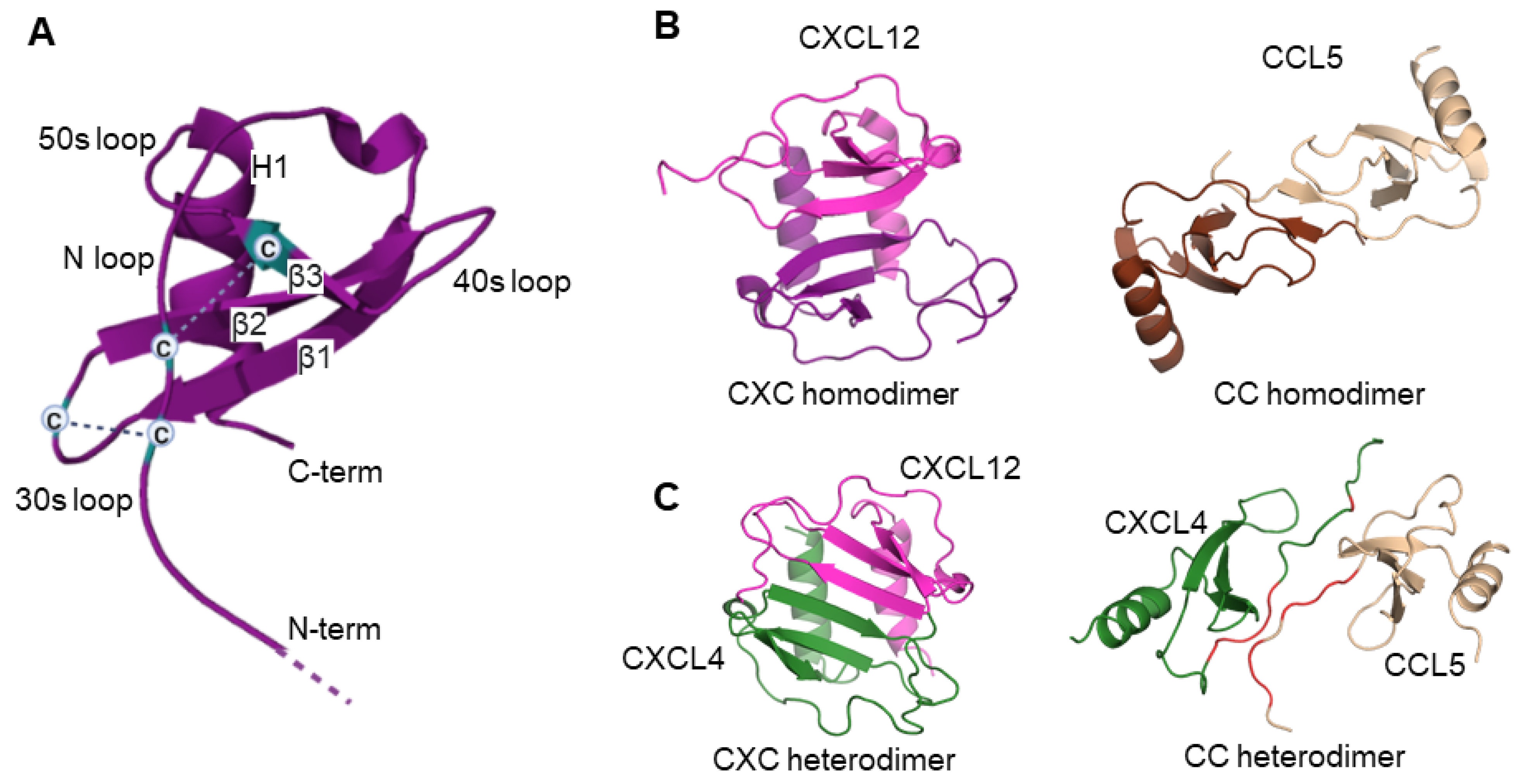
2.1. Chemokine Monomers and Homooligomers
2.2. Chemokine Heterodimers
3. Chemokine Mixtures Trigger Functional Responses Different from Individual Chemokines
4. Obligate Chemokine Heterodimers as Tools to Study Heterodimer Function
4.1. Experimental Approaches to Form Obligate Chemokine Heterodimers
4.2. Functional Activity of Obligate Chemokine Heterodimers
4.3. Limitations of Using Obligate Heterodimers
5. Molecular Mechanisms of Chemokine Heterodimers
5.1. Chemokine Heterodimers Can Bind and Activate Chemokine Receptors
Possible Receptor Binding Modes of a Chemokine Heterodimer
5.2. Interactions with Glycosaminoglycans (GAGs)
6. Summary and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef] [PubMed]
- Baggiolini, M. Chemokines and leukocyte traffic. Nature 1998, 392, 565–568. [Google Scholar] [CrossRef]
- Baggiolini, M. Chemokines in pathology and medicine. J. Intern Med. 2001, 250, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Luster, A.D. Chemokines—Chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 1998, 338, 436–445. [Google Scholar] [CrossRef]
- Mackay, C.R. Chemokines: Immunology’s high impact factors. Nat. Immunol. 2001, 2, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Raman, D.; Sobolik-Delmaire, T.; Richmond, A. Chemokines in health and disease. Exp. Cell Res. 2011, 317, 575–589. [Google Scholar] [CrossRef]
- Zlotnik, A. Chemokines and cancer. Int. J. Cancer 2006, 119, 2026–2029. [Google Scholar] [CrossRef]
- Belperio, J.A.; Keane, M.P.; Arenberg, D.A.; Addison, C.L.; Ehlert, J.E.; Burdick, M.D.; Strieter, R.M. CXC chemokines in angiogenesis. J. Leukoc. Biol. 2000, 68, 1–8. [Google Scholar] [CrossRef]
- Chow, M.T.; Luster, A.D. Chemokines in cancer. Cancer Immunol. Res. 2014, 2, 1125–1131. [Google Scholar] [CrossRef]
- Vilgelm, A.E.; Richmond, A. Chemokines Modulate Immune Surveillance in Tumorigenesis, Metastasis, and Response to Immunotherapy. Front. Immunol. 2019, 10, 333. [Google Scholar] [CrossRef]
- Kohli, K.; Pillarisetty, V.G.; Kim, T.S. Key chemokines direct migration of immune cells in solid tumors. Cancer Gene Ther. 2022, 29, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Proudfoot, A.E.; Uguccioni, M. Modulation of Chemokine Responses: Synergy and Cooperativity. Front. Immunol. 2016, 7, 183. [Google Scholar] [PubMed]
- Graham, G.J.; Handel, T.M.; Proudfoot, A.E.I. Leukocyte Adhesion: Reconceptualizing Chemokine Presentation by Glycosaminoglycans. Trends Immunol. 2019, 40, 472–481. [Google Scholar] [PubMed]
- Thelen, M. Dancing to the tune of chemokines. Nat. Immunol. 2001, 2, 129–134. [Google Scholar] [CrossRef]
- Bachelerie, F.; Ben-Baruch, A.; Burkhardt, A.M.; Combadiere, C.; Farber, J.M.; Graham, G.J.; Horuk, R.; Sparre-Ulrich, A.H.; Locati, M.; Luster, A.D.; et al. International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol. Rev. 2014, 66, 1–79. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. Chemokines: A new classification system and their role in immunity. Immunity 2000, 12, 121–127. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. The chemokine superfamily revisited. Immunity 2012, 36, 705–716. [Google Scholar] [CrossRef]
- Rossi, D.; Zlotnik, A. The biology of chemokines and their receptors. Annu. Rev. Immunol. 2000, 18, 217–242. [Google Scholar] [CrossRef]
- Koenen, R.R.; von Hundelshausen, P.; Nesmelova, I.V.; Zernecke, A.; Liehn, E.A.; Sarabi, A.; Kramp, B.K.; Piccinini, A.M.; Paludan, S.R.; Kowalska, M.A.; et al. Disrupting functional interactions between platelet chemokines inhibits atherosclerosis in hyperlipidemic mice. Nat. Med. 2009, 15, 97–103. [Google Scholar] [CrossRef]
- Nesmelova, I.V.; Sham, Y.; Dudek, A.Z.; van Eijk, L.I.; Wu, G.; Slungaard, A.; Mortari, F.; Griffioen, A.W.; Mayo, K.H. Platelet factor 4 and interleukin-8 CXC chemokine heterodimer formation modulates function at the quaternary structural level. J. Biol. Chem. 2005, 280, 4948–4958. [Google Scholar] [CrossRef]
- Crown, S.E.; Yu, Y.; Sweeney, M.D.; Leary, J.A.; Handel, T.M. Heterodimerization of CCR2 chemokines and regulation by glycosaminoglycan binding. J. Biol. Chem. 2006, 281, 25438–25446. [Google Scholar] [CrossRef] [PubMed]
- Sepuru, K.M.; Rajarathnam, K. Structural basis of a chemokine heterodimer binding to glycosaminoglycans. Biochem. J. 2021, 478, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Joseph, P.R.; Sawant, K.V.; Rajarathnam, K. Chemokine CXCL7 Heterodimers: Structural Insights, CXCR2 Receptor Function, and Glycosaminoglycan Interactions. Int. J. Mol. Sci. 2017, 18, 748. [Google Scholar] [CrossRef] [PubMed]
- von Hundelshausen, P.; Agten, S.M.; Eckardt, V.; Blanchet, X.; Schmitt, M.M.; Ippel, H.; Neideck, C.; Bidzhekov, K.; Leberzammer, J.; Wichapong, K.; et al. Chemokine interactome mapping enables tailored intervention in acute and chronic inflammation. Sci. Transl. Med. 2017, 9, eaah6650. [Google Scholar] [CrossRef]
- Nguyen, K.T.P.; Druhan, L.J.; Avalos, B.R.; Zhai, L.; Rauova, L.; Nesmelova, I.V.; Dreau, D. CXCL12-CXCL4 heterodimerization prevents CXCL12-driven breast cancer cell migration. Cell. Signal. 2020, 66, 109488. [Google Scholar] [CrossRef]
- Rosenbaum, D.M.; Rasmussen, S.G.; Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 2009, 459, 356–363. [Google Scholar] [CrossRef]
- Fredriksson, R.; Lagerstrom, M.C.; Lundin, L.G.; Schioth, H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef]
- Murphy, P.M.; Baggiolini, M.; Charo, I.F.; Hebert, C.A.; Horuk, R.; Matsushima, K.; Miller, L.H.; Oppenheim, J.J.; Power, C.A. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol. Rev. 2000, 52, 145–176. [Google Scholar]
- Eiger, D.S.; Boldizsar, N.; Honeycutt, C.C.; Gardner, J.; Rajagopal, S. Biased agonism at chemokine receptors. Cell. Signal. 2021, 78, 109862. [Google Scholar] [CrossRef]
- Amarandi, R.M.; Hjorto, G.M.; Rosenkilde, M.M.; Karlshoj, S. Probing Biased Signaling in Chemokine Receptors. Methods Enzymol. 2016, 570, 155–186. [Google Scholar]
- Marchese, A. Endocytic trafficking of chemokine receptors. Curr. Opin. Cell Biol. 2014, 27, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Reiter, E.; Lefkowitz, R.J. GRKs and beta-arrestins: Roles in receptor silencing, trafficking and signaling. Trends Endocrinol. Metab. 2006, 17, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.S.; Alagesan, P.; Desai, N.K.; Pack, T.F.; Wu, J.H.; Inoue, A.; Freedman, N.J.; Rajagopal, S. C-X-C Motif Chemokine Receptor 3 Splice Variants Differentially Activate Beta-Arrestins to Regulate Downstream Signaling Pathways. Mol. Pharmacol. 2017, 92, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Freitas, C.; Desnoyer, A.; Meuris, F.; Bachelerie, F.; Balabanian, K.; Machelon, V. The relevance of the chemokine receptor ACKR3/CXCR7 on CXCL12-mediated effects in cancers with a focus on virus-related cancers. Cytokine Growth Factor Rev. 2014, 25, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Bachelerie, F.; Graham, G.J.; Locati, M.; Mantovani, A.; Murphy, P.M.; Nibbs, R.; Rot, A.; Sozzani, S.; Thelen, M. New nomenclature for atypical chemokine receptors. Nat. Immunol. 2014, 15, 207–208. [Google Scholar] [CrossRef]
- Bonecchi, R.; Graham, G.J. Atypical Chemokine Receptors and Their Roles in the Resolution of the inflammatory Response. Front. Immunol. 2016, 7, 224. [Google Scholar] [CrossRef] [PubMed]
- Stone, M.J.; Hayward, J.A.; Huang, C.; Huma, Z.E.; Sanchez, J. Mechanisms of Regulation of the Chemokine-Receptor Network. Int. J. Mol. Sci. 2017, 18, 342. [Google Scholar] [CrossRef]
- Johnson, Z.; Proudfoot, A.E.; Handel, T.M. Interaction of chemokines and glycosaminoglycans: A new twist in the regulation of chemokine function with opportunities for therapeutic intervention. Cytokine Growth Factor Rev. 2005, 16, 625–636. [Google Scholar] [CrossRef]
- Proudfoot, A.E.; Handel, T.M.; Johnson, Z.; Lau, E.K.; LiWang, P.; Clark-Lewis, I.; Borlat, F.; Wells, T.N.; Kosco-Vilbois, M.H. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc. Natl. Acad. Sci. USA 2003, 100, 1885–1890. [Google Scholar] [CrossRef]
- Handel, T.M.; Johnson, Z.; Crown, S.E.; Lau, E.K.; Proudfoot, A.E. Regulation of protein function by glycosaminoglycans—As exemplified by chemokines. Annu. Rev. Biochem. 2005, 74, 385–410. [Google Scholar] [CrossRef]
- Crijns, H.; Vanheule, V.; Proost, P. Targeting Chemokine-Glycosaminoglycan Interactions to Inhibit Inflammation. Front. Immunol. 2020, 11, 483. [Google Scholar] [CrossRef] [PubMed]
- Rajarathnam, K.; Desai, U.R. Structural Insights Into How Proteoglycans Determine Chemokine-CXCR1/CXCR2 Interactions: Progress and Challenges. Front. Immunol. 2020, 11, 660. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fuster, M.; Sriramarao, P.; Esko, J.D. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat. Immunol. 2005, 6, 902–910. [Google Scholar] [CrossRef]
- Middleton, J.; Neil, S.; Wintle, J.; Clark-Lewis, I.; Moore, H.; Lam, C.; Auer, M.; Hub, E.; Rot, A. Transcytosis and surface presentation of IL-8 by venular endothelial cells. Cell 1997, 91, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Palmer, A.C.; Banerjee, B.; Fritchley, S.J.; Kirby, J.A. Examination of the function of RANTES, MIP-1alpha, and MIP-1beta following interaction with heparin-like glycosaminoglycans. J. Biol. Chem. 2000, 275, 11721–11727. [Google Scholar] [CrossRef]
- O’Boyle, G.; Mellor, P.; Kirby, J.A.; Ali, S. Anti-inflammatory therapy by intravenous delivery of non-heparan sulfate-binding CXCL12. FASEB J. 2009, 23, 3906–3916. [Google Scholar] [CrossRef]
- Ali, S.; Robertson, H.; Wain, J.H.; Isaacs, J.D.; Malik, G.; Kirby, J.A. A non-glycosaminoglycan-binding variant of CC chemokine ligand 7 (monocyte chemoattractant protein-3) antagonizes chemokine-mediated inflammation. J. Immunol. 2005, 175, 1257–1266. [Google Scholar] [CrossRef]
- Gangavarapu, P.; Rajagopalan, L.; Kolli, D.; Guerrero-Plata, A.; Garofalo, R.P.; Rajarathnam, K. The monomer-dimer equilibrium and glycosaminoglycan interactions of chemokine CXCL8 regulate tissue-specific neutrophil recruitment. J. Leukoc. Biol. 2012, 91, 259–265. [Google Scholar] [CrossRef]
- Rek, A.; Brandner, B.; Geretti, E.; Kungl, A.J. A biophysical insight into the RANTES-glycosaminoglycan interaction. Biochim. Biophys. Acta 2009, 1794, 577–582. [Google Scholar] [CrossRef]
- Ellyard, J.I.; Simson, L.; Bezos, A.; Johnston, K.; Freeman, C.; Parish, C.R. Eotaxin selectively binds heparin. An interaction that protects eotaxin from proteolysis and potentiates chemotactic activity in vivo. J. Biol. Chem. 2007, 282, 15238–15247. [Google Scholar] [CrossRef]
- Proudfoot, A.E.; Fritchley, S.; Borlat, F.; Shaw, J.P.; Vilbois, F.; Zwahlen, C.; Trkola, A.; Marchant, D.; Clapham, P.R.; Wells, T.N. The BBXB motif of RANTES is the principal site for heparin binding and controls receptor selectivity. J. Biol. Chem. 2001, 276, 10620–10626. [Google Scholar] [CrossRef] [PubMed]
- Sheng, G.J.; Oh, Y.I.; Chang, S.K.; Hsieh-Wilson, L.C. Tunable heparan sulfate mimetics for modulating chemokine activity. J. Am. Chem. Soc. 2013, 135, 10898–10901. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.C.; Tyler, R.C.; Peterson, F.C.; Dyer, D.P.; Zhang, F.; Linhardt, R.J.; Handel, T.M.; Volkman, B.F. Examination of Glycosaminoglycan Binding Sites on the XCL1 Dimer. Biochemistry 2016, 55, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.P.; Johnson, Z.; Borlat, F.; Zwahlen, C.; Kungl, A.; Roulin, K.; Harrenga, A.; Wells, T.N.; Proudfoot, A.E. The X-ray structure of RANTES: Heparin-derived disaccharides allows the rational design of chemokine inhibitors. Structure 2004, 12, 2081–2093. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.; Andaya, A.; Bleiholder, C.; Leary, J.A. Differentiation of CC vs CXC chemokine dimers with GAG octasaccharide binding partners: An ion mobility mass spectrometry approach. J. Am. Chem. Soc. 2013, 135, 4325–4332. [Google Scholar] [CrossRef]
- Mayo, K.H.; Ilyina, E.; Roongta, V.; Dundas, M.; Joseph, J.; Lai, C.K.; Maione, T.; Daly, T.J. Heparin binding to platelet factor-4. An NMR and site-directed mutagenesis study: Arginine residues are crucial for binding. Biochem. J. 1995, 312 Pt 2, 357–365. [Google Scholar] [CrossRef]
- Mikhailov, D.; Young, H.C.; Linhardt, R.J.; Mayo, K.H. Heparin dodecasaccharide binding to platelet factor-4 and growth-related protein-alpha. Induction of a partially folded state and implications for heparin-induced thrombocytopenia. J. Biol. Chem. 1999, 274, 25317–25329. [Google Scholar] [CrossRef]
- Hoogewerf, A.J.; Kuschert, G.S.; Proudfoot, A.E.; Borlat, F.; Clark-Lewis, I.; Power, C.A.; Wells, T.N. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry 1997, 36, 13570–13578. [Google Scholar] [CrossRef]
- Dyer, D.P.; Salanga, C.L.; Volkman, B.F.; Kawamura, T.; Handel, T.M. The dependence of chemokine-glycosaminoglycan interactions on chemokine oligomerization. Glycobiology 2016, 26, 312–326. [Google Scholar] [CrossRef]
- Wang, X.; Watson, C.; Sharp, J.S.; Handel, T.M.; Prestegard, J.H. Oligomeric structure of the chemokine CCL5/RANTES from NMR, MS, and SAXS data. Structure 2011, 19, 1138–1148. [Google Scholar] [CrossRef]
- Poluri, K.M.; Joseph, P.R.B.; Sawant, K.V.; Rajarathnam, K. Molecular basis of glycosaminoglycan heparin binding to the chemokine CXCL1 dimer. J. Biol. Chem. 2013, 288, 25143–25153. [Google Scholar] [CrossRef] [PubMed]
- Sawant, K.V.; Poluri, K.M.; Dutta, A.K.; Sepuru, K.M.; Troshkina, A.; Garofalo, R.P.; Rajarathnam, K. Chemokine CXCL1 mediated neutrophil recruitment: Role of glycosaminoglycan interactions. Sci. Rep. 2016, 6, 33123. [Google Scholar] [CrossRef] [PubMed]
- Sadir, R.; Baleux, F.; Grosdidier, A.; Imberty, A.; Lortat-Jacob, H. Characterization of the stromal cell-derived factor-1alpha-heparin complex. J. Biol. Chem. 2001, 276, 8288–8296. [Google Scholar] [CrossRef] [PubMed]
- Roscic-Mrkic, B.; Fischer, M.; Leemann, C.; Manrique, A.; Gordon, C.J.; Moore, J.P.; Proudfoot, A.E.I.; Trkola, A. RANTES (CCL5) uses the proteoglycan CD44 as an auxiliary receptor to mediate cellular activation signals and HIV-1 enhancement. Blood 2003, 102, 1169–1177. [Google Scholar] [CrossRef]
- MacGregor, H.J.; Kato, Y.; Marshall, L.J.; Nevell, T.G.; Shute, J.K. A copper-hydrogen peroxide redox system induces dityrosine cross-links and chemokine oligomerisation. Cytokine 2011, 56, 669–675. [Google Scholar] [CrossRef]
- Boittier, E.D.; Gandhi, N.S.; Ferro, V.; Coombe, D.R. Cross-Species Analysis of Glycosaminoglycan Binding Proteins Reveals Some Animal Models Are “More Equal” than Others. Molecules 2019, 24, 924. [Google Scholar] [CrossRef]
- Jansma, A.L.; Kirkpatrick, J.P.; Hsu, A.R.; Handel, T.M.; Nietlispach, D. NMR analysis of the structure, dynamics, and unique oligomerization properties of the chemokine CCL27. J. Biol. Chem. 2010, 285, 14424–14437. [Google Scholar] [CrossRef]
- Liang, W.G.; Triandafillou, C.G.; Huang, T.Y.; Zulueta, M.M.; Banerjee, S.; Dinner, A.R.; Hung, S.C.; Tang, W.J. Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3. Proc. Natl. Acad. Sci. USA 2016, 113, 5000–5005. [Google Scholar] [CrossRef]
- Johnson, Z.; Kosco-Vilbois, M.H.; Herren, S.; Cirillo, R.; Muzio, V.; Zaratin, P.; Carbonatto, M.; Mack, M.; Smailbegovic, A.; Rose, M.; et al. Interference with heparin binding and oligomerization creates a novel anti-inflammatory strategy targeting the chemokine system. J. Immunol. 2004, 173, 5776–5785. [Google Scholar] [CrossRef]
- Stringer, S.E.; Gallagher, J.T. Specific binding of the chemokine platelet factor 4 to heparan sulfate. J. Biol. Chem. 1997, 272, 20508–20514. [Google Scholar] [CrossRef]
- Mayo, K.H.; Chen, M.J. Human platelet factor 4 monomer-dimer-tetramer equilibria investigated by 1H NMR spectroscopy. Biochemistry 1989, 28, 9469–9478. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.J.; Mayo, K.H. Human platelet factor 4 subunit association/dissociation thermodynamics and kinetics. Biochemistry 1991, 30, 6402–6411. [Google Scholar] [CrossRef]
- Swaminathan, G.J.; Holloway, D.E.; Colvin, R.A.; Campanella, G.K.; Papageorgiou, A.C.; Luster, A.D.; Acharya, K.R. Crystal structures of oligomeric forms of the IP-10/CXCL10 chemokine. Structure 2003, 11, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Handel, T.M.; Johnson, Z.; Rodrigues, D.H.; Dos Santos, A.C.; Cirillo, R.; Muzio, V.; Riva, S.; Mack, M.; Deruaz, M.; Borlat, F.; et al. An engineered monomer of CCL2 has anti-inflammatory properties emphasizing the importance of oligomerization for chemokine activity in vivo. J. Leukoc. Biol. 2008, 84, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Lubkowski, J.; Bujacz, G.; Boque, L.; Domaille, P.J.; Handel, T.M.; Wlodawer, A. The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions. Nat. Struct. Biol. 1997, 4, 64–69. [Google Scholar] [CrossRef]
- Yang, Y.; Mayo, K.H.; Daly, T.J.; Barry, J.K.; La Rosa, G.J. Subunit association and structural analysis of platelet basic protein and related proteins investigated by 1H NMR spectroscopy and circular dichroism. J. Biol. Chem. 1994, 269, 20110–20118. [Google Scholar] [CrossRef]
- Mayo, K.H.; Roongta, V.; Ilyina, E.; Milius, R.; Barker, S.; Quinlan, C.; La Rosa, G.; Daly, T.J. NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: A symmetric tetramer. Biochemistry 1995, 34, 11399–11409. [Google Scholar] [CrossRef]
- Burrows, S.D.; Doyle, M.L.; Murphy, K.P.; Franklin, S.G.; White, J.R.; Brooks, I.; McNulty, D.E.; Scott, M.O.; Knutson, J.R.; Porter, D.; et al. Determination of the monomer-dimer equilibrium of interleukin-8 reveals it is a monomer at physiological concentrations. Biochemistry 1994, 33, 12741–12745. [Google Scholar] [CrossRef]
- Skelton, N.J.; Aspiras, F.; Ogez, J.; Schall, T.J. Proton NMR assignments and solution conformation of RANTES, a chemokine of the C-C type. Biochemistry 1995, 34, 5329–5342. [Google Scholar] [CrossRef]
- Joseph, P.R.; Rajarathnam, K. Solution NMR characterization of WT CXCL8 monomer and dimer binding to CXCR1 N-terminal domain. Protein Sci. 2015, 24, 81–92. [Google Scholar] [CrossRef]
- Brown, A.J.; Sepuru, K.M.; Rajarathnam, K. Structural Basis of Native CXCL7 Monomer Binding to CXCR2 Receptor N-Domain and Glycosaminoglycan Heparin. Int. J. Mol. Sci. 2017, 18, 508. [Google Scholar] [CrossRef]
- Sepuru, K.M.; Poluri, K.M.; Rajarathnam, K. Solution structure of CXCL5—A novel chemokine and adipokine implicated in inflammation and obesity. PLoS ONE 2014, 9, e93228. [Google Scholar] [CrossRef]
- Chan, D.I.; Hunter, H.N.; Tack, B.F.; Vogel, H.J. Human macrophage inflammatory protein 3alpha: Protein and peptide nuclear magnetic resonance solution structures, dimerization, dynamics, and anti-infective properties. Antimicrob. Agents. Chemother. 2008, 52, 883–894. [Google Scholar] [CrossRef]
- McCornack, M.A.; Boren, D.M.; LiWang, P.J. Glycosaminoglycan disaccharide alters the dimer dissociation constant of the chemokine MIP-1 beta. Biochemistry 2004, 43, 10090–10101. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, C.T.; Peterson, F.C.; Pelzek, A.J.; Volkman, B.F. The monomer-dimer equilibrium of stromal cell-derived factor-1 (CXCL 12) is altered by pH, phosphate, sulfate, and heparin. Protein Sci. 2005, 14, 1071–1081. [Google Scholar] [CrossRef]
- Nesmelova, I.V.; Idiyatullin, D.; Mayo, K.H. Measuring protein self-diffusion in protein-protein mixtures using a pulsed gradient spin-echo technique with WATERGATE and isotope filtering. J. Magn. Reson. 2004, 166, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Laurence, J.S.; Blanpain, C.; Burgner, J.W.; Parmentier, M.; LiWang, P.J. CC chemokine MIP-1 beta can function as a monomer and depends on Phe13 for receptor binding. Biochemistry 2000, 39, 3401–3409. [Google Scholar] [CrossRef]
- Hanzawa, H.; Haruyama, H.; Konishi, K.; Watanabe, K.; Tsurufuji, S. Subunit association and monomer structure of CINC/Gro revealed by 1H-NMR. J. Biochem. 1997, 121, 835–841. [Google Scholar] [CrossRef]
- Rajarathnam, K.; Kay, C.M.; Dewald, B.; Wolf, M.; Baggiolini, M.; Clark-Lewis, I.; Sykes, B.D. Neutrophil-activating peptide-2 and melanoma growth-stimulatory activity are functional as monomers for neutrophil activation. J. Biol. Chem. 1997, 272, 1725–1729. [Google Scholar] [CrossRef]
- Clark-Lewis, I.; Kim, K.S.; Rajarathnam, K.; Gong, J.H.; Dewald, B.; Moser, B.; Baggiolini, M.; Sykes, B.D. Structure-activity relationships of chemokines. J. Leukoc. Biol. 1995, 57, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Lowman, H.B.; Fairbrother, W.J.; Slagle, P.H.; Kabakoff, R.; Liu, J.; Shire, S.; Hebert, C.A. Monomeric variants of IL-8: Effects of side chain substitutions and solution conditions upon dimer formation. Protein Sci. 1997, 6, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Paolini, J.F.; Willard, D.; Consler, T.; Luther, M.; Krangel, M.S. The chemokines IL-8, monocyte chemoattractant protein-1, and I-309 are monomers at physiologically relevant concentrations. J. Immunol. 1994, 153, 2704–2717. [Google Scholar] [CrossRef] [PubMed]
- Desikan, R.; Jayachandran, P. CYTOCON DB: A versatile database of human cell and molecule concentrations for accelerating model development. CPT Pharmacomet. Syst. Pharmacol. 2023, 12, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Leonov, V.; Mogilevskaya, E.; Gerasimuk, E.; Gizzatkulov, N.; Demin, O. CYTOCON: The manually curated database of human in vivo cell and molecule concentrations. CPT Pharmacomet. Syst. Pharmacol. 2023, 12, 41–49. [Google Scholar] [CrossRef]
- Tanaka, Y.; Adams, D.H.; Hubscher, S.; Hirano, H.; Siebenlist, U.; Shaw, S. T-cell adhesion induced by proteoglycan-immobilized cytokine MIP-1 beta. Nature 1993, 361, 79–82. [Google Scholar] [CrossRef]
- Rot, A. Binding of neutrophil attractant/activation protein-1 (interleukin 8) to resident dermal cells. Cytokine 1992, 4, 347–352. [Google Scholar] [CrossRef]
- Rot, A. Endothelial cell binding of NAP-1/IL-8: Role in neutrophil emigration. Immunol. Today 1992, 13, 291–294. [Google Scholar] [CrossRef]
- Brandt, E.; Petersen, F.; Ludwig, A.; Ehlert, J.E.; Bock, L.; Flad, H.D. The beta-thromboglobulins and platelet factor 4: Blood platelet-derived CXC chemokines with divergent roles in early neutrophil regulation. J. Leukoc. Biol. 2000, 67, 471–478. [Google Scholar] [CrossRef]
- Schenk, B.I.; Petersen, F.; Flad, H.D.; Brandt, E. Platelet-derived chemokines CXC chemokine ligand (CXCL)7, connective tissue-activating peptide III, and CXCL4 differentially affect and cross-regulate neutrophil adhesion and transendothelial migration. J. Immunol. 2002, 169, 2602–2610. [Google Scholar] [CrossRef]
- Wiesner, T.; Bugl, S.; Mayer, F.; Hartmann, J.T.; Kopp, H.G. Differential changes in platelet VEGF, Tsp, CXCL12, and CXCL4 in patients with metastatic cancer. Clin. Exp. Metastasis 2010, 27, 141–149. [Google Scholar] [CrossRef]
- Glenister, K.M.; Payne, K.A.; Sparrow, R.L. Proteomic analysis of supernatant from pooled buffy-coat platelet concentrates throughout 7-day storage. Transfusion 2008, 48, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Files, J.C.; Malpass, T.W.; Yee, E.K.; Ritchie, J.L.; Harker, L.A. Studies of human plate alpha-granule release in vivo. Blood 1981, 58, 607–618. [Google Scholar] [CrossRef]
- Dawson, J.; Miltz, W.; Mir, A.K.; Wiessner, C. Targeting monocyte chemoattractant protein-1 signalling in disease. Expert Opin. Ther. Targets 2003, 7, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Spaks, A. Role of CXC group chemokines in lung cancer development and progression. J. Thorac. Dis. 2017, 9, S164–S171. [Google Scholar] [CrossRef] [PubMed]
- Orimo, A.; Gupta, P.B.; Sgroi, D.C.; Arenzana-Seisdedos, F.; Delaunay, T.; Naeem, R.; Carey, V.J.; Richardson, A.L.; Weinberg, R.A. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005, 121, 335–348. [Google Scholar] [CrossRef]
- Dabrowska, E.; Przylipiak, A.; Zajkowska, M.; Piskor, B.M.; Sidorkiewicz, I.; Szmitkowski, M.; Lawicki, S. Possible Diagnostic Application of CXCL12 and CXCR4 as Tumor Markers in Breast Cancer Patients. Anticancer Res. 2020, 40, 3221–3229. [Google Scholar] [CrossRef]
- Zucker, M.B.; Katz, I.R. Platelet factor 4: Production, structure, and physiologic and immunologic action. Proc. Soc. Exp. Biol. Med. 1991, 198, 693–702. [Google Scholar] [CrossRef]
- Rajarathnam, K.; Schnoor, M.; Richardson, R.M.; Rajagopal, S. How do chemokines navigate neutrophils to the target site: Dissecting the structural mechanisms and signaling pathways. Cell. Signal. 2019, 54, 69–80. [Google Scholar] [CrossRef]
- Cambier, S.; Gouwy, M.; Proost, P. The chemokines CXCL8 and CXCL12: Molecular and functional properties, role in disease and efforts towards pharmacological intervention. Cell Mol. Immunol. 2023, 20, 217–251. [Google Scholar] [CrossRef]
- Wang, X.; Sharp, J.S.; Handel, T.M.; Prestegard, J.H. Chemokine oligomerization in cell signaling and migration. Prog. Mol. Biol. Transl. Sci. 2013, 117, 531–578. [Google Scholar]
- Chen, Y.P.; Wu, H.L.; Boye, K.; Pan, C.Y.; Chen, Y.C.; Pujol, N.; Lin, C.W.; Chiu, L.Y.; Billottet, C.; Alves, I.D.; et al. Oligomerization State of CXCL4 Chemokines Regulates G Protein-Coupled Receptor Activation. ACS Chem. Biol. 2017, 12, 2767–2778. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P.R.; Sarmiento, J.M.; Mishra, A.K.; Das, S.T.; Garofalo, R.P.; Navarro, J.; Rajarathnam, K. Probing the role of CXC motif in chemokine CXCL8 for high affinity binding and activation of CXCR1 and CXCR2 receptors. J. Biol. Chem. 2010, 285, 29262–29269. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, A.; Sawant, K.V.; Sarmiento, J.; Navarro, J.; Rajarathnam, K. Chemokine CXCL1 dimer is a potent agonist for the CXCR2 receptor. J. Biol. Chem. 2013, 288, 12244–12252. [Google Scholar] [CrossRef]
- Das, S.T.; Rajagopalan, L.; Guerrero-Plata, A.; Sai, J.; Richmond, A.; Garofalo, R.P.; Rajarathnam, K. Monomeric and dimeric CXCL8 are both essential for in vivo neutrophil recruitment. PLoS ONE 2010, 5, e11754. [Google Scholar] [CrossRef]
- Leong, S.R.; Lowman, H.B.; Liu, J.; Shire, S.; Deforge, L.E.; Gillece-Castro, B.L.; McDowell, R.; Hebert, C.A. IL-8 single-chain homodimers and heterodimers: Interactions with chemokine receptors CXCR1, CXCR2, and DARC. Protein Sci. 1997, 6, 609–617. [Google Scholar] [CrossRef]
- Nasser, M.W.; Raghuwanshi, S.K.; Grant, D.J.; Jala, V.R.; Rajarathnam, K.; Richardson, R.M. Differential activation and regulation of CXCR1 and CXCR2 by CXCL8 monomer and dimer. J. Immunol. 2009, 183, 3425–3432. [Google Scholar] [CrossRef] [PubMed]
- Drury, L.J.; Ziarek, J.J.; Gravel, S.; Veldkamp, C.T.; Takekoshi, T.; Hwang, S.T.; Heveker, N.; Volkman, B.F.; Dwinell, M.B. Monomeric and dimeric CXCL12 inhibit metastasis through distinct CXCR4 interactions and signaling pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 17655–17660. [Google Scholar] [CrossRef]
- Williams, M.A.; Cave, C.M.; Quaid, G.; Robinson, C.; Daly, T.J.; Witt, D.; Lentsch, A.B.; Solomkin, J.S. Interleukin 8 dimerization as a mechanism for regulation of neutrophil adherence-dependent oxidant production. Shock 2005, 23, 371–376. [Google Scholar] [CrossRef]
- Gutjahr, J.C.; Crawford, K.S.; Jensen, D.R.; Naik, P.; Peterson, F.C.; Samson, G.P.B.; Legler, D.F.; Duchene, J.; Veldkamp, C.T.; Rot, A.; et al. The dimeric form of CXCL12 binds to atypical chemokine receptor 1. Sci. Signal. 2021, 14, eabc9012. [Google Scholar] [CrossRef]
- Dudek, A.Z.; Nesmelova, I.; Mayo, K.; Verfaillie, C.M.; Pitchford, S.; Slungaard, A. Platelet factor 4 promotes adhesion of hematopoietic progenitor cells and binds IL-8: Novel mechanisms for modulation of hematopoiesis. Blood 2003, 101, 4687–4694. [Google Scholar] [CrossRef]
- von Hundelshausen, P.; Koenen, R.R.; Sack, M.; Mause, S.F.; Adriaens, W.; Proudfoot, A.E.; Hackeng, T.M.; Weber, C. Heterophilic interactions of platelet factor 4 and RANTES promote monocyte arrest on endothelium. Blood 2005, 105, 924–930. [Google Scholar] [CrossRef]
- Guan, E.; Wang, J.; Norcross, M.A. Identification of human macrophage inflammatory proteins 1alpha and 1beta as a native secreted heterodimer. J. Biol. Chem. 2001, 276, 12404–12409. [Google Scholar] [CrossRef] [PubMed]
- Kufareva, I.; Salanga, C.L.; Handel, T.M. Chemokine and chemokine receptor structure and interactions: Implications for therapeutic strategies. Immunol. Cell Biol. 2015, 93, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.C.; Mayo, K.H. Chemokines from a Structural Perspective. Int. J. Mol. Sci. 2017, 18, 2088. [Google Scholar] [CrossRef] [PubMed]
- Rajarathnam, K.; Sykes, B.D.; Dewald, B.; Baggiolini, M.; Clark-Lewis, I. Disulfide bridges in interleukin-8 probed using non-natural disulfide analogues: Dissociation of roles in structure from function. Biochemistry 1999, 38, 7653–7658. [Google Scholar] [CrossRef]
- Fernandez, E.J.; Lolis, E. Structure, function, and inhibition of chemokines. Annu. Rev. Pharmacol. Toxicol. 2002, 42, 469–499. [Google Scholar] [CrossRef]
- Herring, C.A.; Singer, C.M.; Ermakova, E.A.; Khairutdinov, B.I.; Zuev, Y.F.; Jacobs, D.J.; Nesmelova, I.V. Dynamics and thermodynamic properties of CXCL7 chemokine. Proteins 2015, 83, 1987–2007. [Google Scholar] [CrossRef]
- Ye, J.; Mayer, K.L.; Stone, M.J. Backbone dynamics of the human CC-chemokine eotaxin. J. Biomol. NMR 1999, 15, 115–124. [Google Scholar] [CrossRef]
- Ye, J.; Mayer, K.L.; Mayer, M.R.; Stone, M.J. NMR solution structure and backbone dynamics of the CC chemokine eotaxin-3. Biochemistry 2001, 40, 7820–7831. [Google Scholar] [CrossRef]
- Mayer, K.L.; Stone, M.J. Backbone dynamics of the CC-chemokine eotaxin-2 and comparison among the eotaxin group chemokines. Proteins 2003, 50, 184–191. [Google Scholar] [CrossRef]
- Szpakowska, M.; D’Uonnolo, G.; Luis, R.; Alonso Bartolome, A.; Thelen, M.; Legler, D.F.; Chevigne, A. New pairings and deorphanization among the atypical chemokine receptor family—Physiological and clinical relevance. Front. Immunol. 2023, 14, 1133394. [Google Scholar] [CrossRef]
- Urvas, L.; Kellenberger, E. Structural Insights into Molecular Recognition and Receptor Activation in Chemokine-Chemokine Receptor Complexes. J. Med. Chem. 2023, 66, 7070–7085. [Google Scholar] [CrossRef]
- Gustavsson, M. New insights into the structure and function of chemokine receptor:chemokine complexes from an experimental perspective. J. Leukoc. Biol. 2020, 107, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Kleist, A.B.; Getschman, A.E.; Ziarek, J.J.; Nevins, A.M.; Gauthier, P.A.; Chevigne, A.; Szpakowska, M.; Volkman, B.F. New paradigms in chemokine receptor signal transduction: Moving beyond the two-site model. Biochem. Pharmacol. 2016, 114, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P.R.; Sawant, K.V.; Isley, A.; Pedroza, M.; Garofalo, R.P.; Richardson, R.M.; Rajarathnam, K. Dynamic conformational switching in the chemokine ligand is essential for G-protein-coupled receptor activation. Biochem. J. 2013, 456, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.; Huma, Z.E.; Lane, J.R.; Liu, X.; Bridgford, J.L.; Payne, R.J.; Canals, M.; Stone, M.J. Evaluation and extension of the two-site, two-step model for binding and activation of the chemokine receptor CCR1. J. Biol. Chem. 2019, 294, 3464–3475. [Google Scholar] [CrossRef] [PubMed]
- Clore, G.M.; Gronenborn, A.M. Three-dimensional structures of alpha and beta chemokines. FASEB J 1995, 9, 57–62. [Google Scholar] [CrossRef]
- Malik, Z.A.; Tack, B.F. Structure of human MIP-3alpha chemokine. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006, 62, 631–634. [Google Scholar] [CrossRef]
- Tuinstra, R.L.; Peterson, F.C.; Kutlesa, S.; Elgin, E.S.; Kron, M.A.; Volkman, B.F. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc. Natl. Acad. Sci. USA 2008, 105, 5057–5062. [Google Scholar] [CrossRef]
- Tyler, R.C.; Murray, N.J.; Peterson, F.C.; Volkman, B.F. Native-state interconversion of a metamorphic protein requires global unfolding. Biochemistry 2011, 50, 7077–7079. [Google Scholar] [CrossRef]
- Malkowski, M.G.; Wu, J.Y.; Lazar, J.B.; Johnson, P.H.; Edwards, B.F. The crystal structure of recombinant human neutrophil-activating peptide-2 (M6L) at 1.9-A resolution. J. Biol. Chem. 1995, 270, 7077–7087. [Google Scholar] [CrossRef] [PubMed]
- St Charles, R.; Walz, D.A.; Edwards, B.F. The three-dimensional structure of bovine platelet factor 4 at 3.0-A resolution. J. Biol. Chem. 1989, 264, 2092–2099. [Google Scholar] [CrossRef] [PubMed]
- Young, H.; Roongta, V.; Daly, T.J.; Mayo, K.H. NMR structure and dynamics of monomeric neutrophil-activating peptide 2. Biochem. J. 1999, 338 Pt 3, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.G.; Ren, M.; Zhao, F.; Tang, W.J. Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme. J. Mol. Biol. 2015, 427, 1345–1358. [Google Scholar] [CrossRef]
- LiWang, A.C.; Cao, J.J.; Zheng, H.; Lu, Z.; Peiper, S.C.; LiWang, P.J. Dynamics study on the anti-human immunodeficiency virus chemokine viral macrophage-inflammatory protein-II (VMIP-II) reveals a fully monomeric protein. Biochemistry 1999, 38, 442–453. [Google Scholar] [CrossRef]
- Rajarathnam, K.; Li, Y.; Rohrer, T.; Gentz, R. Solution structure and dynamics of myeloid progenitor inhibitory factor-1 (MPIF-1), a novel monomeric CC chemokine. J. Biol. Chem. 2001, 276, 4909–4916. [Google Scholar] [CrossRef]
- Brown, A.J.; Sepuru, K.M.; Sawant, K.V.; Rajarathnam, K. Platelet-Derived Chemokine CXCL7 Dimer Preferentially Exists in the Glycosaminoglycan-Bound Form: Implications for Neutrophil-Platelet Crosstalk. Front. Immunol. 2017, 8, 1248. [Google Scholar] [CrossRef]
- Lau, E.K.; Paavola, C.D.; Johnson, Z.; Gaudry, J.P.; Geretti, E.; Borlat, F.; Kungl, A.J.; Proudfoot, A.E.; Handel, T.M. Identification of the glycosaminoglycan binding site of the CC chemokine, MCP-1: Implications for structure and function in vivo. J. Biol. Chem. 2004, 279, 22294–22305. [Google Scholar] [CrossRef]
- Salanga, C.L.; Handel, T.M. Chemokine oligomerization and interactions with receptors and glycosaminoglycans: The role of structural dynamics in function. Exp. Cell Res. 2011, 317, 590–601. [Google Scholar] [CrossRef]
- Weber, C.; Koenen, R.R. Fine-tuning leukocyte responses: Towards a chemokine ‘interactome’. Trends Immunol. 2006, 27, 268–273. [Google Scholar] [CrossRef]
- Venetz, D.; Ponzoni, M.; Schiraldi, M.; Ferreri, A.J.; Bertoni, F.; Doglioni, C.; Uguccioni, M. Perivascular expression of CXCL9 and CXCL12 in primary central nervous system lymphoma: T-cell infiltration and positioning of malignant B cells. Int. J. Cancer 2010, 127, 2300–2312. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, S.; Petkovic, V.; Sebastiani, S.; Danelon, M.G.; Uguccioni, M.; Gerber, B.O. A rich chemokine environment strongly enhances leukocyte migration and activities. Blood 2005, 105, 3405–3412. [Google Scholar] [CrossRef] [PubMed]
- Carlson, J.; Baxter, S.A.; Dreau, D.; Nesmelova, I.V. The heterodimerization of platelet-derived chemokines. Biochim. Biophys. Acta 2013, 1834, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Giri, J.; Das, R.; Nylen, E.; Chinnadurai, R.; Galipeau, J. CCL2 and CXCL12 Derived from Mesenchymal Stromal Cells Cooperatively Polarize IL-10+ Tissue Macrophages to Mitigate Gut Injury. Cell Rep. 2020, 30, 1923–1934.e1924. [Google Scholar] [CrossRef]
- Koenen, R.R.; Weber, C. Co-immunoprecipitation of Platelet Factor 4 and RANTES from human platelets. Protoc. Exch. 2009. [Google Scholar] [CrossRef]
- Mayo, K.H.; Dings, R.P.; Flader, C.; Nesmelova, I.; Hargittai, B.; van der Schaft, D.W.; van Eijk, L.I.; Walek, D.; Haseman, J.; Hoye, T.R.; et al. Design of a partial peptide mimetic of anginex with antiangiogenic and anticancer activity. J. Biol. Chem. 2003, 278, 45746–45752. [Google Scholar] [CrossRef]
- Nesmelova, I.V.; Sham, Y.; Gao, J.; Mayo, K.H. CXC and CC chemokines form mixed heterodimers: Association free energies from molecular dynamics simulations and experimental correlations. J. Biol. Chem. 2008, 283, 24155–24166. [Google Scholar] [CrossRef]
- Rajagopal, P.; Waygood, E.B.; Reizer, J.; Saier, M.H., Jr.; Klevit, R.E. Demonstration of protein-protein interaction specificity by NMR chemical shift mapping. Protein Sci. 1997, 6, 2624–2627. [Google Scholar] [CrossRef]
- Nguyen, K.T.P.; Volkman, B.; Dréau, D.; Nesmelova, I.V. A new obligate CXCL4–CXCL12 heterodimer for studying chemokine heterodimer activities and mechanisms. Sci. Rep. 2022, 12, 17204. [Google Scholar] [CrossRef]
- Agten, S.M.; Koenen, R.R.; Ippel, H.; Eckardt, V.; von Hundelshausen, P.; Mayo, K.H.; Weber, C.; Hackeng, T.M. Probing Functional Heteromeric Chemokine Protein-Protein Interactions through Conformation-Assisted Oxime Ligation. Angew. Chem. Int. Ed. Engl. 2016, 55, 14963–14966. [Google Scholar] [CrossRef]
- Gouwy, M.; Schiraldi, M.; Struyf, S.; Van Damme, J.; Uguccioni, M. Possible mechanisms involved in chemokine synergy fine tuning the inflammatory response. Immunol. Lett. 2012, 145, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Struyf, S.; Stoops, G.; Van Coillie, E.; Gouwy, M.; Schutyser, E.; Lenaerts, J.P.; Fiten, P.; Van Aelst, I.; Proost, P.; Opdenakker, G.; et al. Gene cloning of a new plasma CC chemokine, activating and attracting myeloid cells in synergy with other chemoattractants. Biochemistry 2001, 40, 11715–11722. [Google Scholar] [CrossRef] [PubMed]
- Gouwy, M.; Struyf, S.; Catusse, J.; Proost, P.; Van Damme, J. Synergy between proinflammatory ligands of G protein-coupled receptors in neutrophil activation and migration. J. Leukoc. Biol. 2004, 76, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, S.; Danelon, G.; Gerber, B.; Uguccioni, M. CCL22-induced responses are powerfully enhanced by synergy inducing chemokines via CCR4: Evidence for the involvement of first beta-strand of chemokine. Eur. J. Immunol. 2005, 35, 746–756. [Google Scholar] [CrossRef]
- Gouwy, M.; Struyf, S.; Noppen, S.; Schutyser, E.; Springael, J.Y.; Parmentier, M.; Proost, P.; Van Damme, J. Synergy between coproduced CC and CXC chemokines in monocyte chemotaxis through receptor-mediated events. Mol. Pharmacol. 2008, 74, 485–495. [Google Scholar] [CrossRef]
- Kuscher, K.; Danelon, G.; Paoletti, S.; Stefano, L.; Schiraldi, M.; Petkovic, V.; Locati, M.; Gerber, B.O.; Uguccioni, M. Synergy-inducing chemokines enhance CCR2 ligand activities on monocytes. Eur. J. Immunol. 2009, 39, 1118–1128. [Google Scholar] [CrossRef]
- Broxmeyer, H.E.; Sherry, B.; Cooper, S.; Lu, L.; Maze, R.; Beckmann, M.P.; Cerami, A.; Ralph, P. Comparative analysis of the human macrophage inflammatory protein family of cytokines (chemokines) on proliferation of human myeloid progenitor cells. Interacting effects involving suppression, synergistic suppression, and blocking of suppression. J. Immunol. 1993, 150, 3448–3458. [Google Scholar] [CrossRef]
- Broxmeyer, H.E.; Cooper, S.; Hangoc, G.; Kim, C.H. Stromal cell-derived factor-1/CXCL12 selectively counteracts inhibitory effects of myelosuppressive chemokines on hematopoietic progenitor cell proliferation in vitro. Stem. Cells Dev. 2005, 14, 199–203. [Google Scholar] [CrossRef]
- Gijsbers, K.; Gouwy, M.; Struyf, S.; Wuyts, A.; Proost, P.; Opdenakker, G.; Penninckx, F.; Ectors, N.; Geboes, K.; Van Damme, J. GCP-2/CXCL6 synergizes with other endothelial cell-derived chemokines in neutrophil mobilization and is associated with angiogenesis in gastrointestinal tumors. Exp. Cell Res. 2005, 303, 331–342. [Google Scholar] [CrossRef]
- Williams, A.E.; Jose, R.J.; Mercer, P.F.; Brealey, D.; Parekh, D.; Thickett, D.R.; O’Kane, C.; McAuley, D.F.; Chambers, R.C. Evidence for chemokine synergy during neutrophil migration in ARDS. Thorax 2017, 72, 66–73. [Google Scholar] [CrossRef]
- Poltavets, V.; Faulkner, J.W.; Dhatrak, D.; Whitfield, R.J.; McColl, S.R.; Kochetkova, M. CXCR4-CCR7 Heterodimerization Is a Driver of Breast Cancer Progression. Life 2021, 11, 1049. [Google Scholar] [CrossRef] [PubMed]
- Krug, A.; Uppaluri, R.; Facchetti, F.; Dorner, B.G.; Sheehan, K.C.F.; Schreiber, R.D.; Cella, M.; Colonna, M. Cutting edge: IFN-producing cells respond to CXCR3 ligands in the presence of CXCL12 and secrete inflammatory chemokines upon activation. J. Immunol. 2002, 169, 6079–6083. [Google Scholar] [CrossRef] [PubMed]
- Vanbervliet, B.; Bendriss-Vermare, N.; Massacrier, C.; Homey, B.; de Bouteiller, O.; Briere, F.; Trinchieri, G.; Caux, C. The inducible CXCR3 ligands control plasmacytoid dendritic cell responsiveness to the constitutive chemokine stromal cell-derived factor 1 (SDF-1)/CXCL12. J. Exp. Med. 2003, 198, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Meller, S.; Lauerma, A.I.; Kopp, F.M.; Winterberg, F.; Anthoni, M.; Muller, A.; Gombert, M.; Haahtela, A.; Alenius, H.; Rieker, J.; et al. Chemokine responses distinguish chemical-induced allergic from irritant skin inflammation: Memory T cells make the difference. J. Allergy Clin. Immun. 2007, 119, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Mellado, M.; Rodriguez-Frade, J.M.; Vila-Coro, A.J.; Fernandez, S.; de Ana, A.M.; Jones, D.R.; Toran, J.L.; Martinez-A, C. Chemokine receptor homo- or heterodimerization activates distinct signaling pathways. EMBO J. 2001, 20, 2497–2507. [Google Scholar] [CrossRef] [PubMed]
- Iellem, A.; Mariani, M.; Lang, R.; Recalde, H.; Panina-Bordignon, P.; Sinigaglia, F.; D’Ambrosio, D. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J. Exp. Med. 2001, 194, 847–853. [Google Scholar] [CrossRef]
- Basu, S.; Broxmeyer, H.E. CCR5 ligands modulate CXCL12-induced chemotaxis, adhesion, and Akt phosphorylation of human cord blood CD34+ cells. J. Immunol. 2009, 183, 7478–7488. [Google Scholar] [CrossRef]
- Bai, Z.; Hayasaka, H.; Kobayashi, M.; Li, W.; Guo, Z.; Jang, M.H.; Kondo, A.; Choi, B.I.; Iwakura, Y.; Miyasaka, M. CXC chemokine ligand 12 promotes CCR7-dependent naive T cell trafficking to lymph nodes and Peyer’s patches. J. Immunol. 2009, 182, 1287–1295. [Google Scholar] [CrossRef]
- Zwijnenburg, P.J.; Polfliet, M.M.; Florquin, S.; van den Berg, T.K.; Dijkstra, C.D.; van Deventer, S.J.; Roord, J.J.; van der Poll, T.; van Furth, A.M. CXC-chemokines KC and macrophage inflammatory protein-2 (MIP-2) synergistically induce leukocyte recruitment to the central nervous system in rats. Immunol. Lett. 2003, 85, 1–4. [Google Scholar] [CrossRef]
- Stanford, M.M.; Issekutz, T.B. The relative activity of CXCR3 and CCR5 ligands in T lymphocyte migration: Concordant and disparate activities in vitro and in vivo. J. Leukoc. Biol. 2003, 74, 791–799. [Google Scholar] [CrossRef]
- Vajen, T.; Koenen, R.R.; Werner, I.; Staudt, M.; Projahn, D.; Curaj, A.; Sonmez, T.T.; Simsekyilmaz, S.; Schumacher, D.; Mollmann, J.; et al. Blocking CCL5-CXCL4 heteromerization preserves heart function after myocardial infarction by attenuating leukocyte recruitment and NETosis. Sci. Rep. 2018, 8, 10647. [Google Scholar] [CrossRef] [PubMed]
- Iida, Y.; Xu, B.; Xuan, H.; Glover, K.J.; Tanaka, H.; Hu, X.; Fujimura, N.; Wang, W.; Schultz, J.R.; Turner, C.R.; et al. Peptide inhibitor of CXCL4-CCL5 heterodimer formation, MKEY, inhibits experimental aortic aneurysm initiation and progression. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 718–726. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Xiong, X.; Zhang, Y.; Yan, D.; Jian, Z.; Xu, B.; Zhao, H. MKEY, a Peptide Inhibitor of CXCL4-CCL5 Heterodimer Formation, Protects Against Stroke in Mice. J. Am. Heart Assoc. 2016, 5, e003615. [Google Scholar] [CrossRef] [PubMed]
- Struyf, S.; Gouwy, M.; Dillen, C.; Proost, P.; Opdenakker, G.; Van Damme, J. Chemokines synergize in the recruitment of circulating neutrophils into inflamed tissue. Eur. J. Immunol. 2005, 35, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Van Raemdonck, K.; Gouwy, M.; Lepers, S.A.; Van Damme, J.; Struyf, S. CXCL4L1 and CXCL4 signaling in human lymphatic and microvascular endothelial cells and activated lymphocytes: Involvement of mitogen-activated protein (MAP) kinases, Src and p70S6 kinase. Angiogenesis 2014, 17, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Sarabi, A.; Kramp, B.K.; Drechsler, M.; Hackeng, T.M.; Soehnlein, O.; Weber, C.; Koenen, R.R.; Von Hundelshausen, P. CXCL4L1 inhibits angiogenesis and induces undirected endothelial cell migration without affecting endothelial cell proliferation and monocyte recruitment. J. Thromb. Haemost. 2011, 9, 209–219. [Google Scholar] [CrossRef]
- Kramp, B.K.; Sarabi, A.; Koenen, R.R.; Weber, C. Heterophilic chemokine receptor interactions in chemokine signaling and biology. Exp. Cell Res. 2011, 317, 655–663. [Google Scholar] [CrossRef]
- Yan, Y.; Thakur, M.; van der Vorst, E.P.C.; Weber, C.; Doring, Y. Targeting the chemokine network in atherosclerosis. Atherosclerosis 2021, 330, 95–106. [Google Scholar]
- Rajarathnam, K.; Prado, G.N.; Fernando, H.; Clark-Lewis, I.; Navarro, J. Probing receptor binding activity of interleukin-8 dimer using a disulfide trap. Biochemistry 2006, 45, 7882–7888. [Google Scholar] [CrossRef]
- Tan, J.H.; Canals, M.; Ludeman, J.P.; Wedderburn, J.; Boston, C.; Butler, S.J.; Carrick, A.M.; Parody, T.R.; Taleski, D.; Christopoulos, A.; et al. Design and receptor interactions of obligate dimeric mutant of chemokine monocyte chemoattractant protein-1 (MCP-1). J. Biol. Chem. 2012, 287, 14692–14702. [Google Scholar] [CrossRef]
- Takekoshi, T.; Ziarek, J.J.; Volkman, B.F.; Hwang, S.T. A locked, dimeric CXCL12 variant effectively inhibits pulmonary metastasis of CXCR4-expressing melanoma cells due to enhanced serum stability. Mol. Cancer Ther. 2012, 11, 2516–2525. [Google Scholar]
- Ravindran, A.; Joseph, P.R.; Rajarathnam, K. Structural basis for differential binding of the interleukin-8 monomer and dimer to the CXCR1 N-domain: Role of coupled interactions and dynamics. Biochemistry 2009, 48, 8795–8805. [Google Scholar] [CrossRef]
- Jin, H.; Shen, X.; Baggett, B.R.; Kong, X.; LiWang, P.J. The human CC chemokine MIP-1beta dimer is not competent to bind to the CCR5 receptor. J. Biol. Chem. 2007, 282, 27976–27983. [Google Scholar] [CrossRef]
- Kufareva, I.; Gustavsson, M.; Holden, L.G.; Qin, L.; Zheng, Y.; Handel, T.M. Disulfide Trapping for Modeling and Structure Determination of Receptor: Chemokine Complexes. Methods Enzymol. 2016, 570, 389–420. [Google Scholar]
- Tang, W.; Becker, M.L. “Click” reactions: A versatile toolbox for the synthesis of peptide-conjugates. Chem. Soc. Rev. 2014, 43, 7013–7039. [Google Scholar]
- Ulrich, S.; Boturyn, D.; Marra, A.; Renaudet, O.; Dumy, P. Oxime ligation: A chemoselective click-type reaction for accessing multifunctional biomolecular constructs. Chemistry 2014, 20, 34–41. [Google Scholar]
- Zhang, Y.; Rollins, B.J. A dominant negative inhibitor indicates that monocyte chemoattractant protein 1 functions as a dimer. Mol. Cell Biol. 1995, 15, 4851–4855. [Google Scholar] [CrossRef]
- Paavola, C.D.; Hemmerich, S.; Grunberger, D.; Polsky, I.; Bloom, A.; Freedman, R.; Mulkins, M.; Bhakta, S.; McCarley, D.; Wiesent, L.; et al. Monomeric monocyte chemoattractant protein-1 (MCP-1) binds and activates the MCP-1 receptor CCR2B. J. Biol. Chem. 1998, 273, 33157–33165. [Google Scholar] [CrossRef]
- Ziarek, J.J.; Kleist, A.B.; London, N.; Raveh, B.; Montpas, N.; Bonneterre, J.; St-Onge, G.; DiCosmo-Ponticello, C.J.; Koplinski, C.A.; Roy, I.; et al. Structural basis for chemokine recognition by a G protein-coupled receptor and implications for receptor activation. Sci. Signal. 2017, 10, eaah5756. [Google Scholar] [CrossRef]
- Ahuja, S.K.; Lee, J.C.; Murphy, P.M. CXC chemokines bind to unique sets of selectivity determinants that can function independently and are broadly distributed on multiple domains of human interleukin-8 receptor B—Determinants of high affinity binding and receptor activitation are distinct. J. Biol. Chem. 1996, 271, 225–232. [Google Scholar] [CrossRef]
- Loetscher, P.; Seitz, M.; Clarklewis, I.; Baggiolini, M.; Moser, B. Both Interleukin-8 Receptors Independently Mediate Chemotaxis—Jurkat Cells Transfected with Il-8r1 or Il-8r2 Migrate in Response to Il-8, Gro-Alpha and Nap-2. FEBS Lett. 1994, 341, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, C.T.; Seibert, C.; Peterson, F.C.; De la Cruz, N.B.; Haugner, J.C., 3rd; Basnet, H.; Sakmar, T.P.; Volkman, B.F. Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci. Signal. 2008, 1, ra4. [Google Scholar] [CrossRef] [PubMed]
- Kufareva, I.; Stephens, B.S.; Holden, L.G.; Qin, L.; Zhao, C.; Kawamura, T.; Abagyan, R.; Handel, T.M. Stoichiometry and geometry of the CXC chemokine receptor 4 complex with CXC ligand 12: Molecular modeling and experimental validation. Proc. Natl. Acad. Sci. USA 2014, 111, E5363–E5372. [Google Scholar] [CrossRef] [PubMed]
- Lasagni, L.; Francalanci, M.; Annunziato, F.; Lazzeri, E.; Giannini, S.; Cosmi, L.; Sagrinati, C.; Mazzinghi, B.; Orlando, C.; Maggi, E.; et al. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J. Exp. Med. 2003, 197, 1537–1549. [Google Scholar]
- Rajagopalan, L.; Rajarathnam, K. Structural basis of chemokine receptor function--a model for binding affinity and ligand selectivity. Biosci. Rep. 2006, 26, 325–339. [Google Scholar]
- Bhusal, R.P.; Foster, S.R.; Stone, M.J. Structural basis of chemokine and receptor interactions: Key regulators of leukocyte recruitment in inflammatory responses. Protein Sci. 2020, 29, 420–432. [Google Scholar] [CrossRef]
- Kufareva, I.; Gustavsson, M.; Zheng, Y.; Stephens, B.S.; Handel, T.M. What Do Structures Tell Us About Chemokine Receptor Function and Antagonism? Annu. Rev. Biophys. 2017, 46, 175–198. [Google Scholar]
- Arimont, M.; Sun, S.L.; Leurs, R.; Smit, M.; de Esch, I.J.P.; de Graaf, C. Structural Analysis of Chemokine Receptor-Ligand Interactions. J. Med. Chem. 2017, 60, 4735–4779. [Google Scholar] [CrossRef]
- Skelton, N.J.; Quan, C.; Reilly, D.; Lowman, H. Structure of a CXC chemokine-receptor fragment in complex with interleukin-8. Structure 1999, 7, 157–168. [Google Scholar] [CrossRef]
- Clubb, R.T.; Omichinski, J.G.; Clore, G.M.; Gronenborn, A.M. Mapping the binding surface of interleukin-8 complexed with an N-terminal fragment of the type 1 human interleukin-8 receptor. FEBS Lett. 1994, 338, 93–97. [Google Scholar] [CrossRef]
- Williams, G.; Borkakoti, N.; Bottomley, G.A.; Cowan, I.; Fallowfield, A.G.; Jones, P.S.; Kirtland, S.J.; Price, G.J.; Price, L. Mutagenesis studies of interleukin-8. Identification of a second epitope involved in receptor binding. J. Biol. Chem. 1996, 271, 9579–9586. [Google Scholar] [CrossRef] [PubMed]
- Booth, V.; Keizer, D.W.; Kamphuis, M.B.; Clark-Lewis, I.; Sykes, B.D. The CXCR3 binding chemokine IP-10/CXCL10: Structure and receptor interactions. Biochemistry 2002, 41, 10418–10425. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wu, L.; Yuan, S.; Wu, M.; Xu, Y.; Sun, Q.; Li, S.; Zhao, S.; Hua, T.; Liu, Z.J. Structural basis of CXC chemokine receptor 2 activation and signalling. Nature 2020, 585, 135–140. [Google Scholar] [CrossRef]
- Stephens, B.S.; Ngo, T.; Kufareva, I.; Handel, T.M. Functional anatomy of the full-length CXCR4-CXCL12 complex systematically dissected by quantitative model-guided mutagenesis. Sci. Signal. 2020, 13, eaay5024. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Munoz, L.; Villares, R.; Rodriguez-Fernandez, J.L.; Rodriguez-Frade, J.M.; Mellado, M. Remodeling our concept of chemokine receptor function: From monomers to oligomers. J. Leukoc. Biol. 2018, 104, 323–331. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, G.; Garcia-Cuesta, E.M.; Gomariz, R.P.; Rodriguez-Frade, J.M.; Mellado, M. The multilayered complexity of the chemokine receptor system. Biochem. Biophys. Res. Commun. 2020, 528, 347–358. [Google Scholar] [CrossRef]
- Vila-Coro, A.J.; Mellado, M.; de Ana, A.M.; Lucas, P.; del Real, G.; Martinez, C.; Rodriguez-Frade, J.M. HIV-1 infection through the CCR5 receptor is blocked by receptor dimerization. Proc. Natl. Acad. Sci. USA 2000, 97, 3388–3393. [Google Scholar] [CrossRef]
- Rodriguez-Frade, J.M.; Vila-Coro, A.J.; de Ana, A.M.; Albar, J.P.; Martinez, A.C.; Mellado, M. The chemokine monocyte chemoattractant protein-1 induces functional responses through dimerization of its receptor CCR2. Proc. Natl. Acad. Sci. USA 1999, 96, 3628–3633. [Google Scholar] [CrossRef]
- Springael, J.Y.; Le Minh, P.N.; Urizar, E.; Costagliola, S.; Vassart, G.; Parmentier, M. Allosteric modulation of binding properties between units of chemokine receptor homo- and hetero-oligomers. Mol. Pharmacol. 2006, 69, 1652–1661. [Google Scholar] [CrossRef]
- Hassan, S.; Buchanan, M.; Jahan, K.; Aguilar-Mahecha, A.; Gaboury, L.; Muller, W.J.; Alsawafi, Y.; Mourskaia, A.A.; Siegel, P.M.; Salvucci, O.; et al. CXCR4 peptide antagonist inhibits primary breast tumor growth, metastasis and enhances the efficacy of anti-VEGF treatment or docetaxel in a transgenic mouse model. Int. J. Cancer 2011, 129, 225–232. [Google Scholar] [CrossRef]
- Portella, L.; Vitale, R.; De Luca, S.; D’Alterio, C.; Ierano, C.; Napolitano, M.; Riccio, A.; Polimeno, M.N.; Monfregola, L.; Barbieri, A.; et al. Preclinical development of a novel class of CXCR4 antagonist impairing solid tumors growth and metastases. PLoS ONE 2013, 8, e74548. [Google Scholar] [CrossRef] [PubMed]
- von Hundelshausen, P.; Wichapong, K.; Gabius, H.J.; Mayo, K.H. The marriage of chemokines and galectins as functional heterodimers. Cell Mol. Life Sci. 2021, 78, 8073–8095. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, V.; Miller, M.C.; Blanchet, X.; Duan, R.; Leberzammer, J.; Duchene, J.; Soehnlein, O.; Megens, R.T.; Ludwig, A.K.; Dregni, A.; et al. Chemokines and galectins form heterodimers to modulate inflammation. EMBO Rep. 2020, 21, e47852. [Google Scholar] [CrossRef] [PubMed]


| Chemokines | Biological Activity Tested | Effect |
|---|---|---|
| CXC mixtures | ||
| CXCL1-CXCL2 * | Leukocyte recruitment in rats | enhancement [179] |
| CXCL4-CXCL8 | CXCL8-stimulated HUVEC proliferation | inhibition [20] |
| CXCL4-CXCL8 | CXCR2-transfected Ba/F3 cells chemotaxis | enhancement [20] |
| CXCL4-CXCL8 | CXCR1-transfected Ba/F3 cells or neutrophil chemotaxis | no effect [20,163] |
| CXCL4-CXCL12 | MDA-MB-231 breast cancer cells chemotaxis | inhibition [25] |
| CXCL7-CXCL8 | Neutrophil chemotaxis | no effect [163] |
| CXCL8-CXCL12 | Neutrophil chemotaxis | enhancement [163] |
| CXCL9/10/11-CXCL12 | DLBCL lymphoma cells, pDC chemotaxis | enhancement [151,172,173] |
| CC mixtures | ||
| CCL2-CCL19 | Monocyte chemotaxis | no effect [166] |
| CCL2-CCL21 | Monocyte chemotaxis | no effect [166] |
| CCL3-CCL5 | T lymphoblast chemotaxis in rats | no effect [180] |
| CCL7-CCL19 | Monocyte or CCR7+ dendritic cell chemotaxis | enhancement [166] |
| CCL7-CCL21 | Monocyte or CCR7+ dendritic cell chemotaxis | enhancement [166] |
| CXC-CC mixtures | ||
| CXCL4-CCL5 | Monocyte adhesion on HUVEC | enhancement [121] |
| CXCL4-CCL5 * | Monocyte and neutrophil recruitment in mice Inhibition of atherosclerosis and aortic aneurysm, preservation of heart function after myocardial infarction, protection against stroke by disrupting CXCL4-CCL5 interaction | enhancement [19,181,182,183] |
| CXCL6-CCL7 * | Neutrophil recruitment to inflamed tissue in mice | enhancement [184] |
| CXCL8-CCL2 | Neutrophil and monocyte chemotaxis | enhancement [163,165,170] |
| CXCL8-CCL7 | Neutrophil and monocyte chemotaxis | enhancement [163,165,170] |
| CXCL8-CCL8 | Neutrophil and monocyte chemotaxis | enhancement [163,165,170] |
| CXCL10-CCL3 * | T lymphoblast chemotaxis in rats | no effect [180] |
| CXCL10-CCL5 * | T lymphoblast chemotaxis in rats | enhancement [180] |
| CXCL10-CCL22 | T lymphocyte chemotaxis | enhancement [164] ** |
| CXCL12-CCL2 | Monocyte chemotaxis | enhancement [165] |
| CXCL12-CCL2 * | IL-10 expression by CCR2+ macrophages, tissue macrophage IL-10 polarization in mice | enhancement [154] |
| CXCL13-CCL19 | CCR7+ or CXCR5+ leukocytes chemotaxis | enhancement [152] ** |
| CXCL13-CCL21 | CCR7+ or CXCR5+ leukocytes chemotaxis | enhancement [152] ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaffashi, K.; Dréau, D.; Nesmelova, I.V. Heterodimers Are an Integral Component of Chemokine Signaling Repertoire. Int. J. Mol. Sci. 2023, 24, 11639. https://doi.org/10.3390/ijms241411639
Kaffashi K, Dréau D, Nesmelova IV. Heterodimers Are an Integral Component of Chemokine Signaling Repertoire. International Journal of Molecular Sciences. 2023; 24(14):11639. https://doi.org/10.3390/ijms241411639
Chicago/Turabian StyleKaffashi, Kimia, Didier Dréau, and Irina V. Nesmelova. 2023. "Heterodimers Are an Integral Component of Chemokine Signaling Repertoire" International Journal of Molecular Sciences 24, no. 14: 11639. https://doi.org/10.3390/ijms241411639
APA StyleKaffashi, K., Dréau, D., & Nesmelova, I. V. (2023). Heterodimers Are an Integral Component of Chemokine Signaling Repertoire. International Journal of Molecular Sciences, 24(14), 11639. https://doi.org/10.3390/ijms241411639







